ABSTRACT
Gene rearrangement of the mixed lineage leukemia (MLL) gene causes leukemia by inducing the constitutive expression of a gene subset normally expressed only in the immature haematopoietic progenitor cells. MLL gene rearrangements often generate fusion products of MLL and a component of the AF4 family/ENL family/P-TEFb (AEP) complex. MLL-AEP fusion proteins have the potential of constitutively recruiting the P-TEFb elongation complex. Thus, it is hypothesized that relieving the promoter proximal pausing of RNA polymerase II is the rate-limiting step of MLL fusion-dependent transcription. AEP also has the potential to recruit the mediator complex via MED26. We recently showed that AEP activates transcription initiation by facilitating TBP loading to the TATA element through the SL1 complex. In the present study, we show that the key activity responsible for the oncogenic property of MLL-AEP fusion proteins is the TBP loading activity, and not the mediator recruitment or transcriptional elongation activities. Thus, we propose that TBP loading by AF4 through SL1 is the major rate-limiting step in MLL fusion-dependent transcription.
KEYWORDS: AF4, leukemia, mediator, MLL, SL1, TBP, transcription
Introduction
Transcription is an intricate process composed of multiple steps including TBP loading, pre-initiation complex (PIC) formation, and transcription elongation. The TATA-binding protein (TBP) is loaded onto the TATA box whose consensus sequence is TATAWAWR (W indicates A/T; R indicates A/G) to initiate the first step of transcription.1 Although many promoters do not contain the TATA box, it appears that the TBP still needs to be loaded onto a similar sequence with the aid of various co-activator complexes such as the TFIID complex2 to initiate transcription.
The AF4 protein family—comprising AF4 (also known as AFF1), AF5Q31 (also known as AFF4), LAF4 (also known as AFF3), and FMR2 (also known as AFF2)3— associates with the positive transcription elongation factor b (P-TEFb) complex and ELL family proteins,4-8 both of which facilitate transcription elongation.9,10 Therefore, the AF4 protein complex is thought to play a significant role in releasing paused RNA polymerase II (RNAP2), which is necessary for entry into the transcription elongation phase. Notably, the AF4 family protein is involved in various biological processes such as heat shock response7 and transcription of the human immunodeficiency virus (HIV),11,12 in which the Tat protein of HIV binds to the P-TEFb-bound AF4 complex to facilitate target RNA recognition and transcription elongation.13,14
The AF4 family protein has an evolutionarily conserved motif in the ALF domain, which contains a binding motif for the seven in absentia homolog (SIAH) proteins.15,16 SIAH proteins are ubiquitin ligases that promote proteasome-dependent degradation. Multiple germline mutations in this motif of AF5Q31 were found in patients with CHOPS syndrome,17 who exhibit developmental defects similar to those of Cornelia de Lange syndrome. The mutations appear to be gain-of-function mutations as the mutant proteins are resistant to the SIAH-dependent degradation.
The AF4 family protein also associates with the ENL family protein, which is composed of ENL (also known as MLLT1) and AF9 (also known as MLLT3). The ENL family proteins have a YEATS domain at its N-terminus, which associates with the acetylated histone H3 lysine 9/27,18 and the ANC1 homology domain (AHD) at its C-terminus,19 which binds to AF4 family proteins. Biochemically stable complexes containing AF4 family proteins, ENL family proteins, and P-TEFb have been purified independently by multiple groups6,7,11,12 and are known by several aliases including AEP and SEC (super elongation complex). It has been reported that AEP associates with components of the PAF1 complex20 and the mediator complex,21 which are presumed to associate with RNAP2 at the promoter-proximal pausing phase. These notions led to models in which AEP is recruited to the higher-order RNAP2 complex paused at the promoter-proximal regions to activate transcription elongation. Different AF4 family proteins constitute slightly different complexes in compositions and appear to have non-redundant functions.22,23 For instance, the AF4 complex plays a dominant role in the viral genome transcription of HIV, while AF5Q31 is more heavily involved in the activation of the HSP70 gene.24
Genes in the AF4 and the ENL gene families are frequently rearranged to generate fusion genes with the MLL gene (also kwon as KMT2A) in leukemia.25 MLL is an ortholog of Drosophila trithorax, which is a core member of the TRX group of proteins that maintain homeobox (HOX) gene expression.26-28 MLL is required for the maintenance of Hox gene expression during embryogenesis in mice,29,30 and loss of Mll attenuates Hox gene expression to result in embryonic lethality at E10–14.29,31,32 Moreover, MLL is required for the expression of posterior Hoxa genes such as Hoxa7, Hoxa9, and Hoxa10 in the haematopoietic lineage.31,33 Expression of posterior Hoxa genes is normally maintained at high levels in the immature progenitor compartments but is progressively downregulated as the cells differentiate.34,35 The posterior HOXA proteins facilitate the expansion of immature haematopoietic progenitors.36-41 Therefore, gene knockout of Mll causes a severe reduction in the immature haematopoietic progenitor compartments.32,33,42-44
MLL fusion proteins induce the constitutive expression of a gene subset that includes HOXA9 and MEIS1, which are normally expressed specifically in immature haematopoietic progenitors—such as the haematopoietic stem cells (HSCs).38 MLL fusion proteins also maintain the RNA expression levels of CDK6,45,46 a key regulator for the exit from quiescence of HSCs.47 Because these gene products confer a HSC-like self-renewal potential to the haematopoietic progenitors, MLL fusion-expressing cells remain undifferentiated and aberrantly self-renew to cause leukemia.36,37,48 Based on the known transcriptional elongation function of the AEP complex, it was thought that the MLL-AEP fusion proteins (MLL-ENL, MLL-AF9, MLL-AF4, and MLL-AF5Q31) aberrantly activate transcription by facilitating transcription elongation through interactions with the AEP components.49
MLL fusion proteins form a complex with menin through the MLL portion.50,51 Interaction between the MLL fusion protein and menin is required for the subsequent association with the lens epithelium-derived growth factor (LEDGF also known as PSIP1).52 Thus, menin is an adaptor protein that tethers the MLL fusion protein to LEDGF, which binds chromatin containing di-/tri-methylated histone H3 lysine 36 (H3K36me2/3) through its PWWP domain.53,54 Both H3K36me2 and H3K36me3 are associated with active transcription.55,56 Moreover, the MLL fusion protein complex associates with non-methylated CpGs through its CXXC domain.57-60 Our structure/function analysis showed that the PWWP domain and the CXXC domain are necessary and sufficient for the MLL fusion proteins to target the HOXA9 promoter.53 Thus, the PWWP domain and the CXXC domain are the minimum targeting module (MTM) of MLL fusion proteins. Because H3K36me2/3 is enriched in actively transcribed genes and non-methylated CpGs are clustered in the non-suppressed promoters, MLL fusion proteins broadly target previously active CpG-rich promoters, where MLL-AEP fusion proteins constitutively recruit AEP components to activate transcription.6,61
However, MLL-AEP-dependent gene activation does not explain all aspects of leukemogenesis. For instance, DOT1-like histone H3K79 methyltransferase (DOT1L) binds specifically to ENL family members6,61,62 and plays an important role in this process by maintaining MLL-AEP-dependent gene activation.63-67 In addition, wildtype MLL is reportedly required for MLL-AF9-dependent leukemogenesis,68 suggesting that MLL-AF9 and wildtype MLL collaborate to induce oncogenic transcription. The mechanism of MLL-AF4-dependent leukemogenesis appears to be more complex since Bursen et al. reported that its reciprocal fusion protein, AF4-MLL, induced acute lymphoblastic leukemia (ALL) in mice in the absence of MLL-AF4, raising the possibility that AF4-MLL activates an its own oncogenic pathway.69 Conversely, knockdown experiments using human cell leukemia cell lines harboring t(4;11) translocations showed that the AF4-MLL protein is dispensable for cell proliferation or survival, whereas MLL-AF4 is required.70 This result was corroborated with an MLL-AF4 knock-in mouse that developed ALL in vivo in the absence of AF4-MLL.71 Further, it has also been suggested that MLL-AF4 and AF4-MLL cooperatively activate the RUNX1 axis to facilitate leukemogenesis.72 These reports provide significant insights into MLL pathogenesis, but have left many unsolved questions regarding the necessity of MLL-AEP fusion proteins and other factors. Nevertheless, it is clear that MLL-AEP fusion proteins constitutively activate their target genes—such as Hoxa9— to confer unrestrained proliferative capacity to haematopoietic progenitors, which we believe is one of the most important aspects in leukemogenesis. In this article, we focus on the mechanisms of gene activation by MLL-AEP fusion proteins and propose a novel model based on our newly acquired data.
AEP associates with SL1 to activate transcription by loading TBP to the TATA element
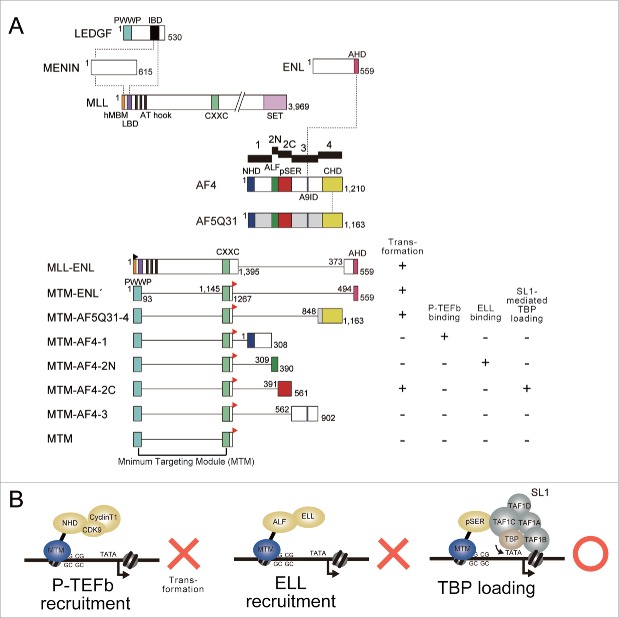
The transforming potential of MLL-AEP fusion proteins can be evaluated by the myeloid progenitor transformation assays.73 In this assay, various MLL fusion constructs were transduced by retrovirus to myeloid progenitors derived from murine bone marrow and the cells were cultured in a semi-solid media. Normally, myeloid progenitors proliferate ex vivo in the early passages of culture but differentiate quickly. Hence, normal cells do not form colonies in the third and fourth passages of culture. However, cells transduced with MLL fusion genes maintain high-level expression of MLL target genes—such as Hoxa9—and continue to form colonies in the later rounds. An artificial protein composed of MTM and AHD of ENL or the C-terminal homology domain (CHD) of AF5Q31 can transform myeloid progenitors74 (Fig. 1A). Because both AHD and CHD are binding platforms for AF4, it was thought that AF4 confers transformation abilities to MLL fusion proteins. To identify the potential functions responsible for transformation, we generated various constructs in which MTM was fused to a subdivided domain of AF4, and tested their ability to transform myeloid progenitors. Among all AF4 subdomains tested, only the pSER domain conferred transforming ability74 (Fig. 1A). In contrast, MTM fusion proteins with domains that recruit elongation effectors—such as the N-terminal homology domain (NHD) that recruits P-TEFb, and ALF, which recruits ELL—did not result in myeloid progenitor transformation. Hence, the function of the pSER domain appears to be critical in the transcriptional activation of the MLL target genes, and not the transcriptional elongation activity.
Figure 1.
The pSER domain, but not the modules that recruit elongation factors, confer transforming ability to the MLL fusion proteins. (A) The structure of MLL fusion constructs. The properties of each construct are summarized on the right. MTM: the minimum structure of the PWWP domain of LEDGF and the minimum structure of the CXXC domain of MLL and its C-terminal basic region; black flag: FLAG epitope; red flag: the HA epitope. (B) Schematic representation of the modules required for MLL fusion-dependent transformation.
By analyzing the factors that associate with the pSER domain on chromatin, we identified selectivity factor 1 (SL1) as a specific pSER domain binder.74 SL1 is a protein complex composed of TBP and 4 TBP-associated factors RNA polymerase I subunits (TAFIs) (TAF1A/TAFI48, TAF1B/TAFI63, TAF1C/TAFI110, and TAF1D/TAFI41). It has been shown that SL1 is a core component of the PIC of RNA polymerase I (RNAP1).75-78 Upstream binding factor recruits SL1 onto the promoters of rRNA genes to initiate RNAP1-dependent transcription.79 However, the role of SL1 in RNAP2-dependent transcription was previously unclear. Our results indicate that AF4 recruits SL1 to load TBP onto the TATA element to activate transcription initiation.74 Supporting this notion, deletion of the TATA box sequence in the promoter resulted in a substantial decrease of transactivation activity mediated by the pSER domain while SL1 recruitment was not impaired.
These results indicate that the MLL-AEP fusion proteins aberrantly activate transcription by utilizing the TBP-loading function, whereas recruitment of P-TEFb or ELL elongation factors does not confer transforming ability (Fig. 1B). Thus, TBP loading, but not transcription elongation, is the rate-limiting step of gene activation of MLL target genes.
MED26 is recruited through the DLXLS motif to potentiate transcriptional activation
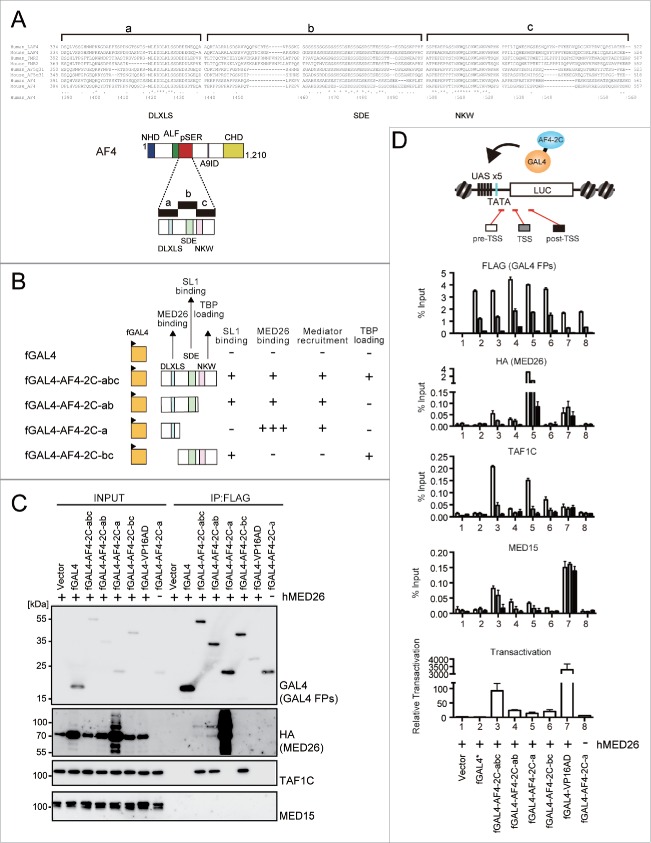
Mediator plays a central role in gene activation mainly by facilitating the PIC formation through direct association with RNAP2.80 Mediator is a large protein complex composed of some 30 distinct subunits and exists in a variety of subunit compositions. Although mediator complexes are evolutionarily conserved from yeast to metazoans, certain components—such as MED26—are specific only to the metazoans.
MED26 has been shown to associate with AEP, other mediator complex components, and TFIID components.21 In order to evaluate the significance of the MED26 interaction in AF4-dependent gene activation, we first mapped the interaction domain between AF4 and MED26. Notably, the pSER domain of AF4 can be further subdivided into 3 evolutionarily conserved regions (designated a, b, and c) (Fig. 2A). Each subdomain contains a conserved motif, including (a) the DLXLS motif, (b) the SDE motif, and (c) the NKW motif. Thus, we generated a series of constructs in which various subdivided portions of the pSER domain were fused to the FLAG-tagged GAL4 DNA binding domain (fGAL4) (Fig. 2B) and tested their ability to bind to MED26 (Fig. 2C). Immunoprecipitation (IP) analysis showed that the constructs containing the “a” domain co-precipitated with MED26 (Fig. 2C), suggesting that AF4 associates with MED26 through the DLXLS motif. Moreover, fGAL4 fused with the “a” domain (fGAL4-AF4-2C-a) exhibited stronger association with MED26 compared to fGAL4 fused with the “a” and “b” domains (fGAL4-AF4-2C-ab) that also associates with SL1 through the SDE motif, suggesting that its association with SL1 hinders the MED26 interaction with the DLXLS motif. WB analysis on the input samples also showed that exogenous MED26 proteins are highly stabilized in the presence of fGAL4-AF4-2C-a. Notably, traces of heavily modified MED26 proteins and degraded MED26 proteins were observed in the upper and lower sections of the MED26 protein in denaturing gel, suggestive of degradation-related modification—possibly ubiquitination. Strong association between MED26 and fGAL4-AF4-2C-a was confirmed by ChIP-qPCR analysis on a 293T cell line (293T-LUC) harboring the reporter containing 5 GAL4-responsive elements and the minimal promoter upstream of the luciferase gene in its genome74 (Fig. 2D). The “a” domain strongly recruited MED26 while inclusion of the “b” domain attenuated MED26 recruitment, supporting the hypothesis that SL1 association hinders the association between MED26 and AF4. Interestingly, TAF1C was recruited to the chromatin by fGAL4-AF4-2C-a specifically in the presence of excess MED26 (Fig. 2D, lanes 5 and 8), despite of its inability to directly bind to SL1 in IP-WB analysis (Fig. 2B). This suggests that AF4 recruits the mediator complex via MED26, where AF4 subsequently dissociates the MED26-bound mediator complex to allow it to recruit SL1. Supporting this hypothesis, MED15—a component of the mediator complex—was recruited to chromatin by the “a” domain-containing fGAL4 fusion proteins (Fig. 2D), but this interaction was not observed in co-IP analysis (Fig. 2C). fGAL4 fused with the VP16 activation domain (fGAL4-VP16AD) served as a positive control of mediator-recruiting factors in ChIP-qPCR analysis (Fig. 2D). Significantly, fGAL4-VP16AD recruited TAF1C in the presence of excess MED26, supporting the hypothesis that the MED26-bound mediator complex recruits SL1.
Figure 2.
The roles of 3 conserved motifs in the pSER domain. (A) Sequence alignments of the pSER domain. The pSER domain can be divided into 3 parts, each of which contains one conserved motif such as the DLXLS, SDE, and NKW motifs. (B) The structure of the FLAG-tagged GAL4-fusion constructs harboring various subdomains of the pSER domain. The properties of each construct are summarized on the right. (C) The domain responsible for association with MED26. Various FLAG-tagged GAL4 fusion constructs and HA-tagged MED26 construct were transfected into 293T-LUC cells and analyzed by IP-western blotting (WB). Each protein was visualized using antibodies for the indicated proteins/tags. (D) The domain responsible for recruitment of various factors and transactivation. Various FLAG-tagged GAL4 constructs and HA-tagged MED26 construct were transfected into 293T-LUC cells and analyzed by ChIP-qPCR for indicated proteins. The same set of FLAG-tagged GAL4 constructs and pRL-tk construct were transfected into 293T-LUC cells and analyzed for their transcriptional activation activity.
Next, we analyzed the transcriptional activation activity of these GAL4 fusion proteins in the 293T-LUC cells (Fig. 2D, bottom panel). As shown previously,74 fGAL4-AF4-2C-bc exhibited transcriptional activation activity. fGAL4 fused with the entire pSER domain (fGAL4-AF4-2C-abc) showed stronger activity than fGAL4-AF4-2C-bc, indicating that MED26 association facilitates transcriptional activation. Deletion of the “c” domain resulted in a substantial decrease in the transactivation as it is responsible for TBP loading to the TATA element.74 These results suggest that SL1 recruitment via the “b” domain and TBP loading via the “c” domain are critical in transcriptional activation while MED26 potentiates it by facilitating SL1 recruitment and PIC formation.
Direct MED26 recruitment by MLL fusion is dispensable for transformation
To examine whether the mediator complex is recruited to MLL target genes in MLL-rearranged leukemia cells, we performed ChIP-qPCR analysis using HB1119 cells that endogenously express the MLL-ENL fusion protein. Notably, the ChIP signal of MED15 was observed at the MLL target loci, co-localizing with those of MLL, AF4, and TAF1C (Fig. 3), confirming that the mediator complex is recruited to MLL-ENL-occupied loci.
Figure 3.
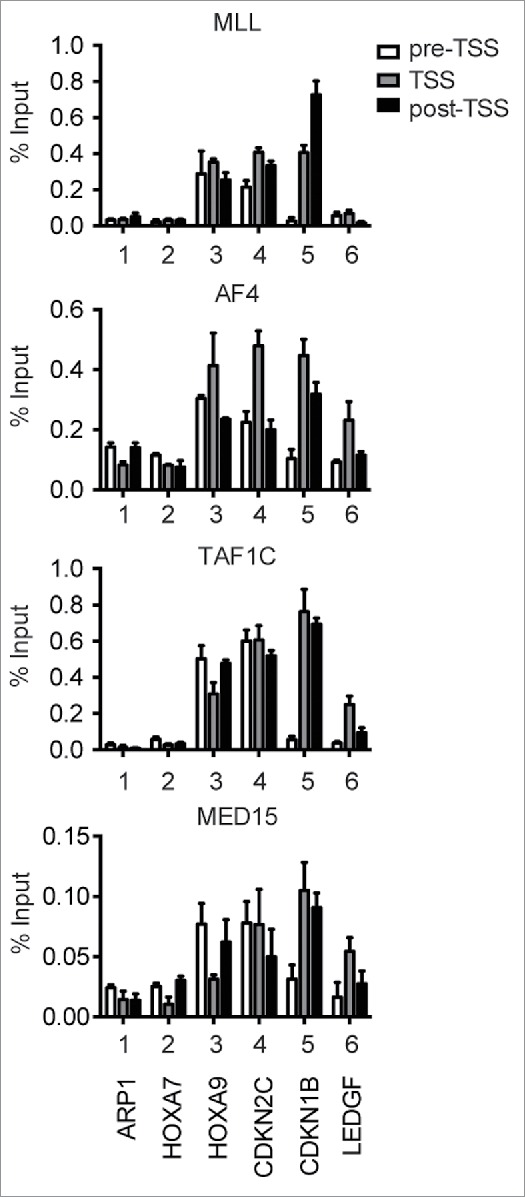
The mediator complex co-localizes with MLL-AEP/SL1 complex in MLL leukemia cells. Localization of MLL, AF4, TAF1C, and MED15 at various loci in HB1119 cells. The genomic localization of each protein was determined by ChIP-qPCR. Precipitated DNA was analyzed using specific probes for the pre-TSS (−1.0 to −0.5 kb from the TSS), TSS (0 to +0.5 kb from the TSS), and post-TSS (+1.0 to +1.5 kb from the TSS) regions of the indicated genes. The ChIP signals are expressed as the percent input with error bars (SD of PCR triplicates). The data are partially redundant with those published previously.74
To further elucidate the role of MED26 in MLL fusion-dependent transformation, we performed myeloid progenitor transformation assays with various constructs in which MTM was fused to various fragments of the pSER domain (Fig. 4A and B). MTM fused with the pSER domain (MTM-AF4-2C-abc) transformed myeloid progenitors as previously reported.74 MTM fused with the “b” and “c” domains (MTM-AF4-2C-bc) transformed in a manner similar to MTM-AF4-2C-abc. On the other hand, MTM fused with the “a” and “b” domains (MTM-AF4-2C-ab) or just the “a” domain (MTM-AF4-2C-a) capable of recruiting MED26, failed to induce transformation. These results indicate that SL1 binding and TBP loading mediated by the SDE and NKW motifs are critically required for transformation by MLL fusion proteins, while MED26 recruitment is not. Although all of the steps including TBP loading, PIC formation, and elongation must occur in gene activation of MLL target genes, TBP loading is the rate-limiting step while the other 2 steps may be adequately supported by other endogenous factors.
Figure 4.
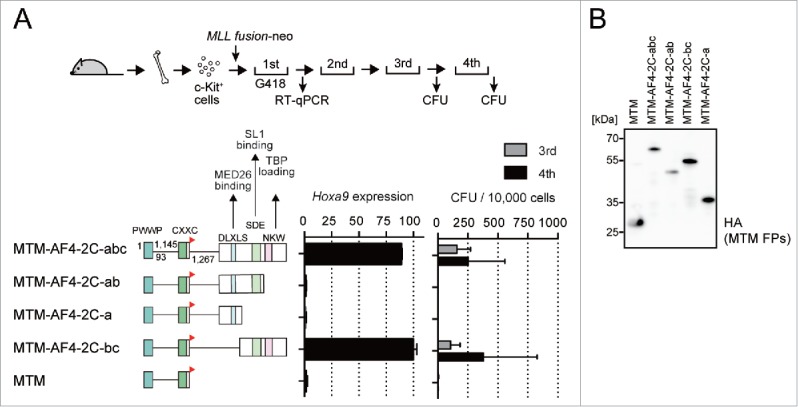
MED26 recruitment by MLL fusion proteins is dispensable for myeloid transformation. (A) Transforming ability of the MTM fusion constructs with various subdomains of the pSER domain. A schematic of the myeloid progenitor transformation assay is shown on top. Hoxa9 expression in the first round of colonies normalized to Gapdh expression is shown as the relative value of MTM-AF4-2C-bc (arbitrarily set at 100%) with error bars (SD of PCR triplicates). The number of colony-forming units (CFUs) of third and fourth rounds is shown with error bars (SD was calculated from data arising from more than 3 independent experiments). (B) Protein expression of various MTM fusion proteins in virus packaging cells. The whole cell extracts of the virus-packaging cells were analyzed by WB using anti-HA antibody.
Conclusion
Our recently published paper74 and this study show that the AEP complex potentially activates multiple steps of transcription, including TBP loading, PIC formation, and transcription elongation. It is widely believed that the AF4 family protein complex plays a role specialized to transcription elongation and therefore MLL-AEP fusion proteins activate transcription by relieving the promoter-proximal pausing of RNAP2.49
Although it was known that AF4 proteins have transactivation property,6,81-83 its significance had been overlooked and unaccounted for. We recently showed that this transactivation activity was mediated by SL1, which presumably provides TBP to the TATA element.74 The current working model postulates that a sequence of events occurs in the following order: (1) AF4 family proteins tethered to the active chromatin through ENL recruit the mediator complex via MED26; (2) and the MED26-bound mediator complex then dissociates from AF4 proteins and recruits SL1; and (3) the mediator complex situates SL1 to the SDE motif of AF4 proteins, which subsequently loads TBP to the TATA element with the help of the NKW motif; and (4) the mediator complex aids PIC formation of RNAP2; and (5) the P-TEFb complex (and possibly ELL family proteins) relieves the promoter proximal-pausing of RNAP2 (Fig. 5). This view is quite different from the prevailing views in that AF4 protein complexes are recruited only to relieve the pausing RNAP2 complex.20,21 Of note, the N-terminal portion of AF4 (1-360 aa) has the potential to recruit TFIIH complex—a key component of RNAP2-PIC84—further supporting the notion that AF4 facilitates multiple phases of transcription initiation.
Figure 5.
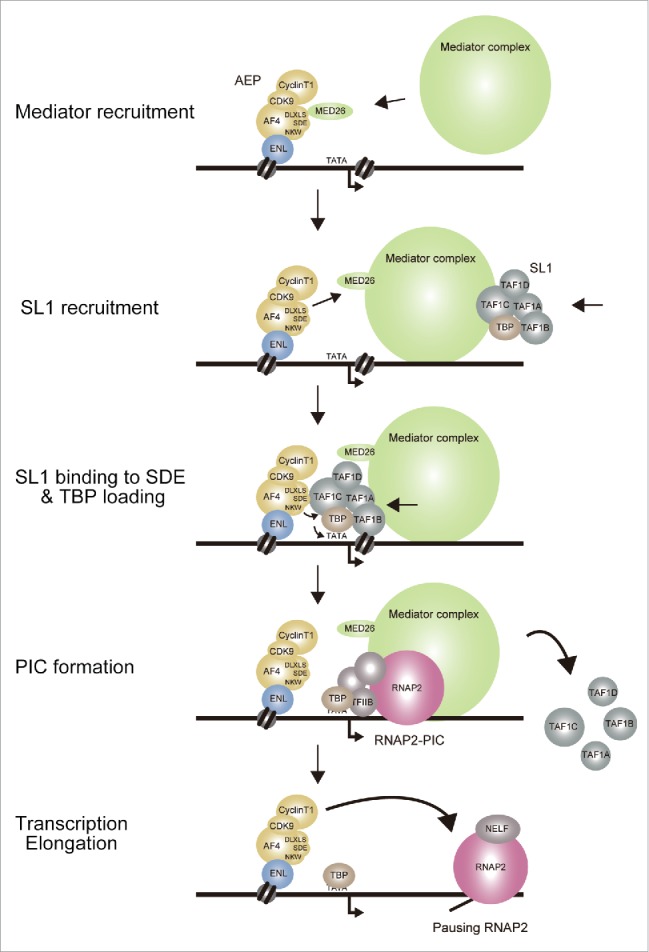
AEP activates gene expression by facilitating TBP loading, PIC formation, and transcriptional elongation. Working model of AEP-dependent transcriptional activation.
The significance of AEP-driven TBP loading is supported by the results of structure/function analysis of the MLL fusion proteins (Fig. 1). If relieving the pausing RNAP2 were the major function of AEP in gene activation, MLL fusion proteins harboring transcription elongation activity would activate MLL target genes to transform haematopoietic progenitors. However, the artificial constructs in which MTM is tethered to NHD or ALF, both of which potentially recruit elongation activity, failed to transform myeloid progenitors while MTM fused with the pSER domain successfully activated HOXA9 and immortalized myeloid progenitors. These results indicate that the main activity that needs to be aberrantly recruited by MLL fusion proteins is the TBP loading activity but not the elongation activity. In this study, we also showed that the activity to recruit the mediator complex is also dispensable. Interestingly, while AF4 facilitates SL1 recruitment through the MED26-bound mediator complex, this activity is not absolutely required for MLL-AEP fusion protein-dependent gene expression—perhaps because MLL-AEP fusion proteins can sufficiently recruit SL1 through a direct association. It is thought that the major function of the mediator complex is to facilitate PIC formation; however, an MLL-AF4 mutant lacking the MED26 binding motif (MTM-AF4-2C-bc) activated Hoxa9 expression and immortalized haematopoietic progenitors, suggesting that mediator-dependent PIC formation is either dispensable or compensated by non-MLL-AEP fusion factors. These results suggest that the PIC formation and transcription elongation can proceed without the aid of MLL fusion proteins once TBP is loaded onto the TATA element. Hence, TBP loading through AF4 is the major rate-limiting step in MLL fusion-dependent gene activation (Fig. 6). Thus, we propose that the major function of MLL-AEP fusion proteins is to load TBP to the TATA element, and not activate transcription elongation or recruit the mediator complex.
Figure 6.
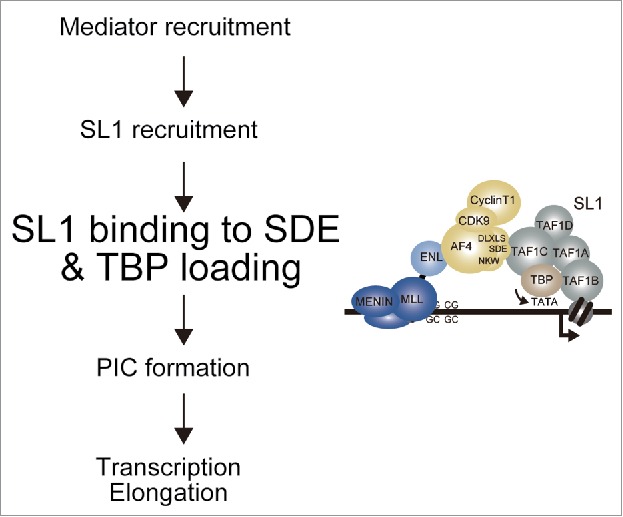
TBP loading is the critical rate-limiting step activated by MLL-AEP fusion proteins. Working model of MLL-AEP-dependent transcriptional activation.
Materials and methods
Vector construction
The pMSCV-neo-MTM-AF4 and pCMV5- GAL4-AF4 constructs were generated as per previous reports74 or newly generated through restriction enzyme digestion/PCR-based mutagenesis.
Cells and cell culture
293T-LUC cells were generated by lentiviral transduction with the pLKO-puro-FR-LUC reporter as previously reported.74 PLAT-E cells and 293T-LUC cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (PS). The HB1119 human leukemia cell line51 was cultured in RPMI 1640 medium, supplemented with 10% FBS and PS.
IP-western blotting and ChIP-qPCR
IP and ChIP-qPCR were performed using fractionation-assisted native chromatin immunoprecipitation method, which was described in detail previously.74 Specific antibodies for MLLN (rpN1),85 AF4 (sc-49350, Santacruz Biotech), TAF1C (A303-698A, Bethyl Laboratories), MED15 (A302-422A, Bethyl Laboratories), FLAG (M2, F-3165, Sigma), and HA (3F10, Roche) were used for ChIP analysis.
Transactivation assay
Transactivation assays were performed with 293T-LUC cells harboring the GAL4-responsive luciferase reporter. 293T-LUC cells were transfected with the expression vectors for the various GAL4 fusion proteins and the pRL-TK plasmid and the luciferase activity was measured using the dual luciferase reporter kit (Promega) 24 h after transfection. Luciferase activity values were normalized to the Renilla luciferase activity and expressed as the mean and standard deviation of triplicate samples.
Myeloid progenitor transformation assay
The myeloid progenitor transformation assay was performed as previously reported.74 In brief, cells were harvested from the femurs and tibiae of 5-week-old female C57BL/6 mice.53 C-Kit-positive cells were enriched with an anti-c-Kit antibody (Miltenyi Biotech, 130-091-224). Ecotropic retrovirus particles were produced using PLAT-E packaging cells86 and were used for viral transduction. The cells were transduced with a recombinant retrovirus by spinoculation and then plated onto a methylcellulose medium (Iscove's modified Dulbecco's medium, 20% FBS, 1.6% methylcellulose, 100 µM β-mercaptoethanol) containing murine stem cell factors, interleukin-3, and granulocyte-macrophage colony-stimulating factors (10 ng mL−1 of each). G418 (1 mg mL−1) was used to select the transduced cells. Hoxa9 expression was quantified using RT-qPCR after the first round of culture. Colony-forming units (CFUs) at the third and fourth rounds were quantified per 104 plated cells, after 4–6 d in culture. Experiments were approved by the Kyoto University Institutional Animal Care and Use Committee.
RT-qPCR
RT-qPCR was performed as previously reported.74 Gene expression was confirmed with qPCR, using the TaqMan probes for Hoxa9 (Mm00439364_m1) and Gapdh (Mm99999915_g1) (Life Technologies). The expression level of Hoxa9 normalized to that of Gapdh and was determined using a standard curve and the relative quantification method, as described in ABI User Bulletin #2 (Applied Biosystems).
Disclosure of potential conflicts of interest
This work was supported in part by research funding from Dainippon Sumitomo Pharma Co., Ltd.
Acknowledgments
We thank Drs. Toshio Kitamura for providing the PLAT-E cells and Hidehisa Takahashi for providing the MED26 expression vector and critical reading this manuscript.
Notes on contributors
A.Y. and H.O. conceived the project. H.O., S.T., and A.Y. performed all of the experiments. A.T. provided essential reagents. A.Y. wrote the manuscript.
Funding
This work was supported by JSPS KAKENNHI grants to H.O. (number 25870373) and A.Y. (number 16H05337).
References
- [1].Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004; 116:699-709; PMID:15006352; http://dx.doi.org/ 10.1016/S0092-8674(04)00205-3 [DOI] [PubMed] [Google Scholar]
- [2].Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012; 483:295-301; PMID:22258509; http://dx.doi.org/ 10.1038/nature10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nilson I, Reichel M, Ennas MG, Greim R, Knorr C, Siegler G, Greil J, Fey GH, Marschalek R. Exon/intron structure of the human AF-4 gene, a member of the AF-4/LAF-4/FMR-2 gene family coding for a nuclear protein with structural alterations in acute leukaemia. Br J Haematol 1997; 98:157-69; PMID:9233580; http://dx.doi.org/ 10.1046/j.1365-2141.1997.1522966.x [DOI] [PubMed] [Google Scholar]
- [4].Estable MC, Naghavi MH, Kato H, Xiao H, Qin J, Vahlne A, Roeder RG. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J Biomed Sci 2002; 9:234-45; PMID:12065898; http://dx.doi.org/ 10.1007/BF02256070 [DOI] [PubMed] [Google Scholar]
- [5].Benedikt A, Baltruschat S, Scholz B, Bursen A, Arrey TN, Meyer B, Varagnolo L, Muller AM, Karas M, Dingermann T, et al.. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia 2011; 25:135-44; PMID:21030982; http://dx.doi.org/ 10.1038/leu.2010.249 [DOI] [PubMed] [Google Scholar]
- [6].Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010; 17:198-212; PMID:20153263; http://dx.doi.org/ 10.1016/j.ccr.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular cell 2010; 37:429-37; PMID:20159561; http://dx.doi.org/ 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 2007; 16:92-106; PMID:17135274; http://dx.doi.org/ 10.1093/hmg/ddl444 [DOI] [PubMed] [Google Scholar]
- [9].Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 1996; 271:1873-6; PMID:8596958; http://dx.doi.org/ 10.1126/science.271.5257.1873 [DOI] [PubMed] [Google Scholar]
- [10].Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell 2006; 23:297-305; PMID:16885020; http://dx.doi.org/ 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- [11].Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Molecular cell 2010; 38:439-51; PMID:20471949; http://dx.doi.org/ 10.1016/j.molcel.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Molecular cell 2010; 38:428-38; PMID:20471948; http://dx.doi.org/ 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schulze-Gahmen U, Lu H, Zhou Q, Alber T. AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. Elife 2014; 3:e02375; PMID:24843025; http://dx.doi.org/ 10.7554/eLife.02375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gu J, Babayeva ND, Suwa Y, Baranovskiy AG, Price DH, Tahirov TH. Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4. Cell cycle 2014; 13:1788-97; PMID:24727379; http://dx.doi.org/ 10.4161/cc.28756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Oliver PL, Bitoun E, Clark J, Jones EL, Davies KE. Mediation of Af4 protein function in the cerebellum by Siah proteins. Proc Natl Acad Sci U S A 2004; 101:14901-6; PMID:15459319; http://dx.doi.org/ 10.1073/pnas.0406196101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bursen A, Moritz S, Gaussmann A, Moritz S, Dingermann T, Marschalek R. Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene 2004; 23:6237-49; PMID:15221006; http://dx.doi.org/ 10.1038/sj.onc.1207837 [DOI] [PubMed] [Google Scholar]
- [17].Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, et al.. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet 2015; 47:338-44; PMID:25730767; http://dx.doi.org/ 10.1038/ng.3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, et al.. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014; 159:558-71; PMID:25417107; http://dx.doi.org/ 10.1016/j.cell.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al.. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007; 110:4445-54; PMID:17855633; http://dx.doi.org/ 10.1182/blood-2007-05-090514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A 2011; 108:E636-45; PMID:21873227; http://dx.doi.org/ 10.1073/pnas.1107107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al.. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011; 146:92-104; PMID:21729782; http://dx.doi.org/ 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol 2012; 32:2608-17; PMID:22547686; http://dx.doi.org/ 10.1128/MCB.00182-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Luo Z, Lin C, Woodfin AR, Bartom ET, Gao X, Smith ER, Shilatifard A. Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev 2016; 30:92-101; PMID:26728555; http://dx.doi.org/ 10.1101/gad.270413.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lu H, Li Z, Zhang W, Schulze-Gahmen U, Xue Y, Zhou Q. Gene target specificity of the Super Elongation Complex (SEC) family: how HIV-1 Tat employs selected SEC members to activate viral transcription. Nucleic Acids Res 2015; 43:5868-79; PMID:26007649; http://dx.doi.org/ 10.1093/nar/gkv541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007; 7:823-33; PMID:17957188; http://dx.doi.org/ 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- [26].Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 1992; 71:691-700; PMID:1423624; http://dx.doi.org/ 10.1016/0092-8674(92)90602-9 [DOI] [PubMed] [Google Scholar]
- [27].Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 1992; 71:701-8; PMID:1423625; http://dx.doi.org/ 10.1016/0092-8674(92)90603-A [DOI] [PubMed] [Google Scholar]
- [28].Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet 1992; 2:113-8; PMID:1303259; http://dx.doi.org/ 10.1038/ng1092-113 [DOI] [PubMed] [Google Scholar]
- [29].Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995; 378:505-8; PMID:7477409; http://dx.doi.org/ 10.1038/378505a0 [DOI] [PubMed] [Google Scholar]
- [30].Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A 1998; 95:10632-6; PMID:9724755; http://dx.doi.org/ 10.1073/pnas.95.18.10632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 1998; 92:108-17; PMID:9639506 [PubMed] [Google Scholar]
- [32].Yokoyama A, Ficara F, Murphy MJ, Meisel C, Naresh A, Kitabayashi I, Cleary ML. Proteolytically cleaved MLL subunits are susceptible to distinct degradation pathways. J Cell Sci 2011; 124:2208-19; PMID:21670200; http://dx.doi.org/ 10.1242/jcs.080523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 2007; 1:324-37; PMID:18371366; http://dx.doi.org/ 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 2006; 10:257-68; PMID:17045204; http://dx.doi.org/ 10.1016/j.ccr.2006.08.020 [DOI] [PubMed] [Google Scholar]
- [35].Yokoyama A, Ficara F, Murphy MJ, Meisel C, Hatanaka C, Kitabayashi I, Cleary ML. MLL becomes functional through intra-molecular interaction not by proteolytic processing. PloS one 2013; 8:e73649; PMID:24040009; http://dx.doi.org/ 10.1371/journal.pone.0073649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J 1998; 17:3714-25; PMID:9649441; http://dx.doi.org/ 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 2000; 19:608-16; PMID:10698505; http://dx.doi.org/ 10.1038/sj.onc.1203371 [DOI] [PubMed] [Google Scholar]
- [38].Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al.. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006; 442:818-22; PMID:16862118; http://dx.doi.org/ 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- [39].Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 2005; 106:3988-94; PMID:16091451; http://dx.doi.org/ 10.1182/blood-2005-05-2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 1997; 89:1922-30; PMID:9058712 [PubMed] [Google Scholar]
- [41].Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 2002; 99:121-9; PMID:11756161; http://dx.doi.org/ 10.1182/blood.V99.1.121 [DOI] [PubMed] [Google Scholar]
- [42].Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 1997; 90:1799-806; PMID:9292512 [PubMed] [Google Scholar]
- [43].Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell 2004; 6:437-43; PMID:15030765; http://dx.doi.org/ 10.1016/S1534-5807(04)00061-9 [DOI] [PubMed] [Google Scholar]
- [44].McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 2007; 1:338-45; PMID:18371367; http://dx.doi.org/ 10.1016/j.stem.2007.07.002 [DOI] [PubMed] [Google Scholar]
- [45].Placke T, Faber K, Nonami A, Putwain SL, Salih HR, Heidel FH, Kramer A, Root DE, Barbie DA, Krivtsov AV, et al.. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 2014; 124:13-23; PMID:24764564; http://dx.doi.org/ 10.1182/blood-2014-02-558114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].van der Linden MH, Willekes M, van Roon E, Seslija L, Schneider P, Pieters R, Stam RW. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL-rearranged infant ALL. Cell cycle 2014; 13:834-44; PMID:24736461; http://dx.doi.org/ 10.4161/cc.27757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S, Neumann A, Plumb I, Doulatov S, Chen J, et al.. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 2015; 16:302-13; PMID:25704240; http://dx.doi.org/ 10.1016/j.stem.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev 2003; 17:2298-307; PMID:12952893; http://dx.doi.org/ 10.1101/gad.1111603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 2010; 10:721-8; PMID:20844554; http://dx.doi.org/ 10.1038/nrc2915 [DOI] [PubMed] [Google Scholar]
- [50].Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 2005; 123:207-18; PMID:16239140; http://dx.doi.org/ 10.1016/j.cell.2005.09.025 [DOI] [PubMed] [Google Scholar]
- [51].Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 2004; 24:5639-49; PMID:15199122; http://dx.doi.org/ 10.1128/MCB.24.13.5639-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 2008; 14:36-46; PMID:18598942; http://dx.doi.org/ 10.1016/j.ccr.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Okuda H, Kawaguchi M, Kanai A, Matsui H, Kawamura T, Inaba T, Kitabayashi I, Yokoyama A. MLL fusion proteins link transcriptional coactivators to previously active CpG-rich promoters. Nucleic Acids Res 2014; 42:4241-56; PMID:24465000; http://dx.doi.org/ 10.1093/nar/gkt1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, et al.. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res 2013; 41:3924-36; PMID:23396443; http://dx.doi.org/ 10.1093/nar/gkt074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823-37; PMID:17512414; http://dx.doi.org/ 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- [56].Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al.. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Molecular cell 2011; 44:609-20; PMID:22099308; http://dx.doi.org/ 10.1016/j.molcel.2011.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 2002; 30:958-65; PMID:11842107; http://dx.doi.org/ 10.1093/nar/30.4.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol 2004; 24:10470-8; PMID:15542854; http://dx.doi.org/ 10.1128/MCB.24.23.10470-10478.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 2010; 17:62-8; PMID:20010842; http://dx.doi.org/ 10.1038/nsmb.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J 2006; 25:4503-12; PMID:16990798; http://dx.doi.org/ 10.1038/sj.emboj.7601340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A 2011; 108:15751-6; PMID:21896721; http://dx.doi.org/ 10.1073/pnas.1111498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure 2013; 21:176-83; PMID:23260655; http://dx.doi.org/ 10.1016/j.str.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al.. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 2011; 20:66-78; PMID:21741597; http://dx.doi.org/ 10.1016/j.ccr.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chen L, Deshpande AJ, Banka D, Bernt KM, Dias S, Buske C, Olhava EJ, Daigle SR, Richon VM, Pollock RM, et al.. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia 2013; 27:813-22; PMID:23138183; http://dx.doi.org/ 10.1038/leu.2012.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 2011; 117:4759-68; PMID:21398221; http://dx.doi.org/ 10.1182/blood-2010-12-327668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 2011; 117:6912-22; PMID:21521783; http://dx.doi.org/ 10.1182/blood-2011-02-334359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen CW, Koche RP, Sinha AU, Deshpande AJ, Zhu N, Eng R, Doench JG, Xu H, Chu SH, Qi J, et al.. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med 2015; 21:335-43; PMID:25822366; http://dx.doi.org/ 10.1038/nm.3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al.. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 2010; 17:148-59; PMID:20159607; http://dx.doi.org/ 10.1016/j.ccr.2009.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, Dingermann T, Marschalek R. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood 2010; 115:3570-9; PMID:20194896; http://dx.doi.org/ 10.1182/blood-2009-06-229542 [DOI] [PubMed] [Google Scholar]
- [70].Kumar AR, Yao Q, Li Q, Sam TA, Kersey JH. t(4;11) leukemias display addiction to MLL-AF4 but not to AF4-MLL. Leuk Res 2011; 35:305-9; PMID:20869771; http://dx.doi.org/ 10.1016/j.leukres.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al.. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008; 14:355-68; PMID:18977325; http://dx.doi.org/ 10.1016/j.ccr.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wilkinson AC, Ballabio E, Geng H, North P, Tapia M, Kerry J, Biswas D, Roeder RG, Allis CD, Melnick A, et al.. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell reports 2013; 3:116-27; PMID:23352661; http://dx.doi.org/ 10.1016/j.celrep.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J 1997; 16:4226-37; PMID:9250666; http://dx.doi.org/ 10.1093/emboj/16.14.4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Okuda H, Kanai A, Ito S, Matsui H, Yokoyama A. AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription. Nat Commun 2015; 6:8869; PMID:26593443; http://dx.doi.org/ 10.1038/ncomms9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Learned RM, Cordes S, Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol 1985; 5:1358-69; PMID:3929071; http://dx.doi.org/ 10.1128/MCB.5.6.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 1994; 266:1966-72; PMID:7801123; http://dx.doi.org/ 10.1126/science.7801123 [DOI] [PubMed] [Google Scholar]
- [77].Eberhard D, Tora L, Egly JM, Grummt I. A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res 1993; 21:4180-6; PMID:8414971; http://dx.doi.org/ 10.1093/nar/21.18.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 2007; 26:1560-8; PMID:17318177; http://dx.doi.org/ 10.1038/sj.emboj.7601601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Sub-cellular biochemistry 2013; 61:211-36; PMID:23150253; http://dx.doi.org/ 10.1007/978-94-007-4525-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010; 11:761-72; PMID:20940737; http://dx.doi.org/ 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce CM, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci U S A 1995; 92:12160-4; PMID:8618864; http://dx.doi.org/ 10.1073/pnas.92.26.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Morrissey JJ, Raney S, Cleary ML. The FEL (AF-4) protein donates transcriptional activation sequences to Hrx-Fel fusion proteins in leukemias containing T(4;11)(Q21;Q23) chromosomal translocations. Leuk Res 1997; 21:911-7; PMID:9403001; http://dx.doi.org/ 10.1016/S0145-2126(97)00012-X [DOI] [PubMed] [Google Scholar]
- [83].Ma C, Staudt LM. LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood 1996; 87:734-45; PMID:8555498 [PubMed] [Google Scholar]
- [84].Scholz B, Kowarz E, Rossler T, Ahmad K, Steinhilber D, Marschalek R. AF4 and AF4N protein complexes: recruitment of P-TEFb kinase, their interactome and potential functions. Am J Blood Res 2015; 5:10-24; PMID:26171280 [PMC free article] [PubMed] [Google Scholar]
- [85].Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood 2002; 100:3710-8; PMID:12393701; http://dx.doi.org/ 10.1182/blood-2002-04-1015 [DOI] [PubMed] [Google Scholar]
- [86].Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000; 7:1063-6; PMID:10871756; http://dx.doi.org/ 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]