Abstract
In skeletal regeneration approaches using human bone marrow derived mesenchymal stromal cells (hBM-MSC), functional evaluation before implantation has traditionally used biomarkers identified using fetal bovine serum-based osteogenic induction media and time courses of at least two weeks. However, emerging pre-clinical evidence indicates donor-dependent discrepancies between these ex vivo measurements and the ability to form bone, calling for improved tests. Therefore, we adopted a multiparametric approach aiming to generate an osteogenic potency assay with improved correlation. hBM-MSC populations from six donors, each expanded under clinical-grade (cGMP) conditions, showed heterogeneity for ex vivo growth response, mineralization and bone-forming ability in a murine xenograft assay. A subset of literature-based biomarker genes was reproducibly upregulated to a significant extent across all populations as cells responded to two different osteogenic induction media. These 12 biomarkers were also measurable in a one-week assay, befitting clinical cell expansion time frames and cGMP growth conditions. They were selected for further challenge using a combinatorial approach aimed at determining ex vivo and in vivo consistency. We identified five globally relevant osteogenic signature genes, notably TGF-ß1 pathway interactors; ALPL, COL1A2, DCN, ELN and RUNX2. Used in agglomerative cluster analysis, they correctly grouped the bone-forming cell populations as distinct. Although donor #6 cells were correlation slope outliers, they contrastingly formed bone without showing ex vivo mineralization. Mathematical expression level normalization of the most discrepantly upregulated signature gene COL1A2, sufficed to cluster donor #6 with the bone-forming classification. Moreover, attenuating factors causing genuine COL1A2 gene down-regulation, restored ex vivo mineralization. This suggested that the signature gene had an osteogenically influential role; nonetheless no single biomarker was fully deterministic whereas all five signature genes together led to accurate cluster analysis. We show proof of principle for an osteogenic potency assay providing early characterization of primary cGMP-hBM-MSC cultures according to their donor-specific bone-forming potential.
Introduction
Severe bone fractures often heal slowly with clinically challenging morbidity. Multipotent human Bone Marrow Mesenchymal Stromal Cells (hBM-MSC), frequently referred to as Mesenchymal Stem Cells, can be combined with biomaterial to help improve bone regeneration [1, 2]. A growing number of options are available for this approach, involving mesenchymal stem cells from different tissue sources [3], but concerns that alternative sources are not necessarily equivalent support choice of bone marrow derived hBM-MSC for bone therapy [4].
A discrepancy between the limited number of sourced autogenic hMSC to be found in the bone marrow and the number required for therapy, is nowadays resolved by expanding the cell population in culture according to current Good Manufacturing Practice (cGMP) [5]. To minimize risk of xenogenic immune incompatibility and prion infection, replacement of fetal bovine serum (FBS) with non-animal growth factors, e.g. human serum [6] or human platelet lysate (PL) [7, 8] is recommended.
Deteriorated cell function from the onset of senescence and concern for phenotypic drift mean that minimal timelines are recommended for cGMP production of hBM-MSC [9]. Though ex vivo expansion of primary hMSC populations obtained from the bone marrow is inherently finite [10–12], advances in culture methods allow cGMP facilities to grow 200 million stromal cells from a bone marrow sample within three weeks; a quantity considered sufficient for autologous therapy [13]. Nevertheless, beyond cell expansion limits, clinical outcomes can be thwarted by donor-specific heterogeneity in hBM-MSC functional potency [14].
A key prerequisite for hBM-MSC bone healing is retention of the specific potential to differentiate to osteoblasts rather than simply form stromal scar tissue [15]. Differentiating hBM-MSC mature to osteoblasts via a temporal cascade of selectively expressed regulatory transcription factors and osteogenic genes governing matrix deposition and mineralization [16]; such molecules and transition phenotypes may serve as readily detectable time-dependent osteogenic biomarkers [17]. Ideally, their measurement would provide indication of the status of a broad set of cellular parameters and bone forming competence. However, correlations between ex vivo expression of osteogenic biomarkers and bone formation in vivo have not been straightforward. Beyond early examples where only hBM-MSC strains with high levels of osteogenic markers ex vivo subsequently formed bone [18, 19], most studies over the past decade reveal surprisingly little direct correlation between bone forming potential and canonical biomarkers of ex vivo osteogenic differentiation, including mRNA expression levels of pro-collagen type I, alpha 1 (COL1A1), osteopontin (SPP1), alkaline phosphatase (ALPL) or runt related transcription factor 2 (RUNX2) [20–24].
Despite the above caveats, recent studies have aimed to correlate ex vivo measurements with bone formation, seeking more specifically informative indicators than proliferation [25]. Cell models that permitted genome-wide comparison of telomerized hMSC-TERT clones with different bone-forming ability, revealed that clone-specific bone-forming potential corresponded particularly well with the ex vivo gene expression of specific extracellular matrix proteins [26]. Notably, decorin (DCN), tetranectin (CLEC3B), collagen type-I, alpha 2 (COL1A2) and elastin (ELN) were bone-predictive genes induced by treatment of hMSC-TERT cells with osteogenic medium (OM) [27]. This agreed with prior views that the onset of differentiation was coupled to proliferation [28] and that the early rate of primarily type I collagen matrix production governed fracture healing [29].
However, the applicability of these correlative osteogenic biomarkers identified in telomerase-immortalized cells in FBS-based culture media to primary hBM-MSC grown in cGMP PL-based culture media has not been determined and prior gene expression assays have not necessarily conformed to the restricted time frames of preclinical cell expansion. Here, we developed a multiparametric phenotype-driven strategy to accommodate inter-donor heterogeneity whilst determining whether ex vivo osteogenic biomarker expression could indicate the subsequent bone-forming potential of cGMP-hBM-MSC from individual donors. Among donor-specific hBM-MSC populations that positively responded to OM with metabolic activation and matrix mineralization, we first verified expression of osteogenic biomarker genes in cGMP-hBM-MSC treated with OM containing FBS and then tested whether similar results were obtainable in OM containing PL (OM-PL). To be consistent with previous osteogenic biomarker studies, gene expression was first measured at comparable two-week time points. Then, to better match cGMP protocol timelines, we measured osteogenic biomarker expression after only one week of OM treatment.
We reasoned that cluster analysis seeking to correlate ex vivo gene expression with in vivo bone formation would need to be based on genes whose upregulation was statistically significant in all contexts. The bone-forming cGMP-hBM-MSC populations treated with OM-PL for one week shared seven upregulated osteogenic biomarker genes. Five of these genes were also consistently up regulated in cells positive for all our ex vivo tests of osteogenic differentiation. These five “signature genes” represented the varied cellular functions of matrix mineralization (ALPL), extracellular matrix synthesis (COL1A2, DCN and ELN) and transcriptional regulation (RUNX2). We here describe how using comparative cluster analysis the signature genes could promptly help discriminate heterogeneous donor-specific cGMP-hBM-MSC strains according to their bone-forming potential.
Materials and Methods
Cell Culture
cGMP facilities; Etablissement Français du Sang, Toulouse (France), Institute of Clinical Transfusion Medicine and Immunogenetics Ulm (Germany) provided donor-specific strains (n = 6, labelled #1-#6) of human bone marrow derived mesenchymal stem cells (hBM-MSCs) each population expandable to single clinical doses of at least 100 x106 cGMP-hBM-MSC. The two-step protocol for unprocessed bone marrow cells involved seeding at an initial density of 50,000 white blood cells/cm2 in 300 mL complete medium in CellStack™ (Corning, Belgium) tissue culture vessels using PL based, animal-serum free alphaMEM medium (Lonza, Gaithersburg USA) [30]. Informed written consent from all six donors conformed to the Declaration of Helsinki and project approval by local ethical committees included testing of BM donors according to blood product guidelines.
Single passage cGMP-hBM-MSCs were shipped in frozen vials and thawed cells were seeded at 6 x103 cells/cm2 in T75 flasks (Greiner Bio-one, Germany) incubated at 37°C with 5% humidified CO2 using maintenance medium (MM) consisting of Minimum Essential Medium (MEM) Alpha without nucleosides (Gibco® Invitrogen, UK), supplemented with 8% (v/v) human Platelet lysate (PL), [31] 1% (v/v) L-Glutamine (Gibco® Invitrogen, Belgium), 1 UI/mL heparin (Sigma-Aldrich, USA) and 10 μg/mL ciprofloxacin (HIKMA, Portugal). The cGMP-hBM-MSCs were replenished with fresh MM twice weekly and at 80–85% confluence were detached using trypsin 0.05%/EDTA 0.02% (PAA Laboratories, Austria) or TrypLE (Gibco® Invitrogen, Belgium). The cGMP-hBM-MSCs were immunophenotypically and functionally characterized in the cGMP facilities ensuring high viability before shipping (Table 1).
Table 1. Characterisation of donor-specific cGMP-hBM-MSC population.
| Donor | Gender | Age | CFU-F | CD34+ | CD45+ | CD73+ | CD90+ | CD105+ | HLA-DR, DP, DQ+ |
|---|---|---|---|---|---|---|---|---|---|
| #1 | M | 54 | 196 | 0.02 | 1.94 | 99.90 | 96.9 | 99.72 | 1.28 |
| #2 | F | 51 | 30 | ND | 0.97 | 100 | 100 | ND | 2.9 |
| #3 | F | 24 | 550 | 0.10 | 0.14 | 97.43 | 99.81 | 98.5 | 2.58 |
| #4 | M | 42 | 68 | 0.00 | 4.4 | 100 | 100 | 99.76 | 0.4 |
| #5 | F | 24 | 92 | 0.17 | 0.07 | 98.82 | 99.69 | 99.56 | 2.85 |
| #6 | M | 21 | 98 | 0.03 | 0.05 | 99.9 | 100 | 93.14 | 0.28 |
*The number of Colony forming Unit-Fibroblast per 106 mononuclear cells scored after the initial plating of the bone marrow sample. The % immunophenotype positivity was determined at passage 1 before shipment. CD34, Hematopoietic Progenitor Cell Antigen CD34; CD45, leukocyte common antigen; CD73, Ecto-5'-nucleotidase; CD90, Thymocyte antigen 1; CD105, Endoglin; HLA-DR, DP, DQ, Human leukocyte antigens. ND: not determined.
Induction of ex-vivo osteogenic differentiation
Cells were seeded concurrently at a density of 104/cm2 in 24-well multiwell plates (Greiner Bio-one) for Alizarin Red and Von Kossa staining and in T25 flasks (Greiner Bio-one) for RNA extraction. All culture vessels were incubated at 37°C with 5% CO2 in a humidified incubator (Thermo Scientific, Italy). At 85–90% cell confluence (≈3 days post seeding), we induced osteogenic differentiation. Two alternative differentiation protocol time courses were compared. Firstly, a previously established two-week (2W) protocol whereby cells were treated for the first week using osteogenic medium (OM) containing the inducing agents 10mM β-Glycerophosphate (ß-GP) (Sigma-Aldrich), 0.1 mM ascorbic acid-2-phosphate (AA) (Sigma-Aldrich), 10 nM Dexamethasone (Dex) (Sigma-Aldrich) supplemented with either either 10% (v/v) defined FBS (OM-FBS) or 8% (v/v) PL (OM-PL). For the second week, the OM-FBS or OM-PL was additionally supplemented with 100 ng/ml rhBMP-2 (Peprotech, UK). A two-week protocol facilitated biomarker data comparison with previous literature [26, 27, 32] and provided a more comprehensive comparison of FBS versus PL based OM. Secondly, to create an assay protocol more consistent with preclinical cell expansion timelines, we explored a one-week (1W) cell treatment protocol, whereby OM-FBS or OM-PL was supplemented with all the above inducing agents, including rhBMP-2 from the outset.
Matrix mineralization
After OM treatment for one or two weeks, the extent of hBM-MSC matrix mineralization was characterised by Alizarin red S (ALZ) and Von Kossa (VK) staining. For the former, cells were washed at room temperature (RT) in PBS (1X), fixed in ice-cold methanol (100% v/v), washed with distilled water and stained with Alizarin Red S (Sigma) (1.5% v/v, pH 4.2) for 5 minutes to detect calcium precipitation. Stained monolayers were visualized by brightfield 10X magnification with an inverted microscope (Zeiss). For stain quantification, the plates were incubated at room temperature (RT) in the dark for 15 minutes with 500 μl of Cetylpyridinium Chloride (CPC) added to each well. The eluted dye solution was transferred to a fresh microcentrifuge tube, diluted ten times with PBS (1X) and dispensed as triplicate aliquots into a transparent 96-well plate (200 μL/well). Each measurement was performed at 562 nm on an ELISA reader (GDV, Roma).
Von Kossa staining to visualize phosphate and carbonate anions [33] was performed two weeks after OM treatment. Cell monolayers were washed in PBS (1X) at RT for 5 minutes and fixed in ice-cold methanol (100% v/v) for 4 minutes. The cells were rinsed twice in distilled water and incubated with 0.8 μm-filtered 1% silver nitrate (Sigma) for 30 minutes under a UV lamp. Stained samples were washed twice in distilled water and visualized with 10X magnification using an inverted microscope (Zeiss). Dark positive VK stained areas were quantified as a percentage of the total area using Image J software (http://rsb.info.nih.gov/ij/).
Cell cycle activation biomarker MKI67+ expression
A primer set (Table 2) amplifying a 129 bp sequence spanning exons 13 and 14 of the MKI67 gene recognised the short and long isoform splice variants encoding the Ki-67 nuclear protein associated with cell cycle activation. RNA extraction and quantitative real time reverse-transcriptase polymerase chain reaction (qRT-PCR) analysis was performed as described below.
Table 2. Primer sequences for polymerase chain reactions.
| Gene | Primer Sequence | Amplified Length |
|---|---|---|
| ACTB | 5'-ACCTTCTACAATGAGCTGCG-3' (sense) | 148 bp |
| 5'-CCTGGATAGCAACGTACATGG-3' (antisense) | ||
| ALPL | 5'-GATGTGGAGTATGAGAGTGACG-3' (sense) | 142 bp |
| 5'-GGTCAAGGGTCA GGAGTTC-3' (antisense) | ||
| BGLAP | 5'-CAGCGAGGTAGTGAAGAGAC-3' (sense) | 144 bp |
| 5'-TGAAAGCCGATGTGGTCAG-3' (antisense) | ||
| BGN | 5'-TGGAGAACAGTGGCTTTGAAC-3' (sense) | 134 bp |
| 5'-GTTGTGGTCTAGGTGGAGTTC-3' (antisense) | ||
| CADM-1 | 5'-CCAGCGGTATCTAGAAGTACAG-3' (sense) | 146 bp |
| 5'-TCACCCAAGTTACCATCACAG-3' (antisense) | ||
| CLEC3B | 5'-TCCTCCTCTGCCTCTTCTC-3' (sense) | 136 bp |
| 5'-GTGTCCAGACGGCTCTTG-3' (antisense) | ||
| COL1A1 | 5'-CCCCTGGAAAGAATGGAGATG-3' (sense) | 148 bp |
| 5'-TCCAAACCACTGAAACCTCTG-3' (antisense) | ||
| COL1A2 | 5'-AGGACAAGAAACACGTCTGG-3' (sense) | 146 bp |
| 5'-GGTGATGTTCTGAGAGGCATAG-3' (antisense) | ||
| DCN | 5'-AAAATGCCCAAAACTCTTCAGG-3' (sense) | 146 bp |
| 5'-GCCCCATTTTCAATTCCTGAG-3' (antisense) | ||
| DLX5 | 5'-CTTATGCCGACTATAGCTACGC-3' (sense) | 124 bp |
| 5'-CCATTCACCATTCTCACCTCG-3' (antisense) | ||
| ELN | 5'-CCTGGCTTCGGATTGTCTC-3' (sense) | 148 bp |
| 5'-CAAAGGGTTTACATTCTCCACC-3' (antisense) | ||
| MKI67 | 5'-AAAAGAATTGAACCTGCGGAAG-3' (sense) | 129 bp |
| 5'-AGTCTTATTTTGGCGTCTGGAG-3' (antisense) | ||
| MSX2 | 5'-CGGTCAAGTCGGAAAATTCAG-3' (sense) | 149 bp |
| 5'-GGATGTGGTAAAGGGCGTG-3' (antisense) | ||
| PTH1R | 5'-ATGCTCTTCAACTCCTTCCAG-3' (sense) | 126 bp |
| 5'-CTTTCGCTTGAAGTCCAGTG-3' (antisense) | ||
| RUNX2 | 5'-TTCACCTTGACCATAACCGTC-3' (sense) | 148 bp |
| 5'-GGCGGTCAGAGAACAAACTAG-3' (antisense) | ||
| SFRP1 | 5'-AAGTGTGACAAGTTCCCCG-3' (sense) | 127 bp |
| 5'-TGGCCTCAGATTTCAACTCG-3' (antisense) | ||
| SPP1 | 5'-CAGTGATTTGCTTTTGCCTCC-3' (sense) | 149 bp |
| 5'-ATTCTGCTTCTGAGATGGGTC-3' (antisense) | ||
| SP7 | 5'-GCCAGAAGCTGTGAAACCTC-3' (sense) | 141 bp |
| 5'-GCTGCAAGCTCTCCATAACC-3' (antisense) | ||
| TAZ | 5'-CGAATTCCTGCGTTTCAAGTG-3' (sense) | 147 bp |
| 5'-GTGATTTTCTGTCCAAAGCGG-3' (antisense) |
ACTB, Beta-actin; ALPL, alkaline phosphatase; BGLAP, osteocalcin; BGN, biglycan; CADM-1, Cell adhesion molecule 1; CLEC3B, tetranectin; COL1A1, collagen type I alpha1; COL1A2, collagen type I alpha2; DCN, decorin; DLX5, distal-less homeo box 5; ELN, Elastin; MKI67, antigen identified by monoclonal antibody Ki-67; MSX2, Msh homeobox 2; PTH1R, Parathyroid hormone 1 receptor; RUNX2, Runt-related transcription factor 2; SFRP1, Secreted frizzled-related protein 1; SPP1, Osteopontin; SP7, Osterix; TAZ, Tafazzin.
RNA extraction and quantitative Real-Time PCR (qRT-PCR)
Total cellular RNA was isolated using a single-step method with TRIzol (Invitrogen) according to the manufacturer's instructions. First-strand complementary cDNA was synthesized from 1 μg of total RNA using a revertAid H minus first-strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. The reaction was terminated by heating at 70°C for 5 minutes. The single strand cDNA was quantified by spectrophotometer (Beckman Coulter DU® 730) so as to use 10 ng of cDNA in each Real-Time PCR well.
Quantitative real-time PCR was performed using the Applied Biosystems StepOne™ Real-Time PCR System and the Fast SYBR® Green Master Mix reagent. The quantification of gene expression for each target gene and reference gene was performed in separate tubes. Forward and reverse primers were designed using IDT PrimerQuest® (http://eu.idtdna.com/PrimerQuest/Home/Index) ensuring they spanned an intron sequence to be specific for mRNA rather than genomic DNA. The relative expression level of the target gene was normalized to that of the endogenous reference ß-actin (ACTB) gene and the 2-ΔΔCt cycle threshold method was used to calculate the relative expression levels of the target genes defined by the primers (Table 2).
In-vivo Xenograft Bone Formation (BF) in immunodeficient mice
With University of Modena and Reggio Emilia ethical committee approval, the bone forming potential of the cGMP-hBM-MSC from each donor was tested using four implantation sites shared between two 8-week old NOD.CB17-Prkdcscid/J mice. Previously described methods were modified to maintain the same cell/scaffold ratio as proposed for a clinical trial. Briefly, animals were anaesthetized with 3.6% isofluorane and the dorsal skin was shaved and cleaned. Incisions of ≈1 cm in length were performed on upper and lower dorsal regions on the back of each mouse. Blunt dissection was used to form a 3 cm long pocket and the graft was implanted therein. A total of 1.6 x106 cells were mixed with 40 mg (≈40 granules) of hydroxyapatite β tricalcium phosphate (HA-βTCP 20:10, 1-2mm, Biomatlante, Vigneux de Bretagne, France) and implanted subcutaneously into the upper and lower dorsal flank of each mouse. The control samples were implanted with 40 mg HA-βTCP scaffold granules alone and the incisions were closed with sutures of ethicon vicryl rapide 5–0 (Johnson & Johnson). Cell/scaffold xenografts and control samples recovered from sacrificed animals after six weeks were transferred to 4% neutral buffered formalin for two days. Two PBS washes were followed by decalcification for 2–3 weeks in buffered 15% EDTA (pH 7.4). The decalcified HA-ßTCP implants were embedded in paraffin and 3 μm sections were stained with Hematoxylin and Eosin (H&E) for assessment of bone tissue formation. The percentage of bone tissue formed per total area [34] was quantified by two independent observers estimating the percentage of pink stained osteoid tissue relative to the total amount of scaffold and diversely stained surrounding stroma. At least 45 fields of view were examined for each donor, quantifying bone within near-equivalent total areas of scaffold.
Phenotype driven selection of osteogenic biomarker “signature genes”
Eighteen candidate biomarker genes selected from a literature search were first assayed to determine time-dependent expression in response to one or two week osteogenic induction protocols. Genes showing consistent and significant upregulation in response to OM treatment were then examined in subsets of phenotypically functional cGMP-hBM-MSC populations to determine the average extent of gene upregulation according to each phenotype; ALZ+, VK+, MKI67+ and BF+. Genes significantly upregulated in all phenotypic contexts were classified as globally relevant osteogenic “signature genes”.
Functional interaction networks
The Search Tool for the Retrieval of Interacting Genes (STRING) database (v9.05) explored possible relationships between the osteogenic signature gene products and whether they might be collectively associated with a particular regulatory network [35]. To maintain strict relevance, the three most confidently predicted functional partners were chosen to identify associated signaling pathways.
Agglomerative Cluster Analysis
To determine inter-donor similarity according to signature gene expression patterns in OM treated cGMP-hBM-MSC, open-source software Cluster 3.0 (C clustering Library 1.50) visualized using Java Treeview v1.1.6r2, was used for agglomerative cluster analysis of the mRNA expression data. The best-fit cluster algorithm for our continuous variables was determined by comparing different linkage methods and distance measures. The averaged metric of Pearson centroid linkage with uncentered correlation distance was compared to more specific Single linkage with Euclidian distance metrics. Sample means were set to zero since our gene expression values represented a fold-increase in mRNA expression above a zero reference state determined by the control samples.
Pearson centroid linkage calculated a vector point from an average of all the items contained within the cluster, minimizing the effect of outlier values. Uncentered correlation distance provided a metric for the strength of linear association between two variables calculated from the sample values and their standard deviations. Euclidian distance, a geometric distance in multidimensional space, was appropriate for continuous variables sharing the same scale and assimilated the gene expression data better by taking the magnitude of changes more into account. The single linkage nearest-neighbor method may have drawbacks if the data has many points that bridge between clusters, but for our purposes it advantageously emphasized dissimilarity.
To understand the significance with which cluster analysis could distinguish donor-specific cGMP-hBM-MSC populations according to their extent of bone formation, the correlation coefficient indicating the degree of similarity between donors was plotted against the average amount of bone (mean%) formed by the respective members of the correlation.
Biomarker signature gene verification; a role in outlier phenotypes
Cluster analysis of cells from outlier donor #6 that formed bone despite no apparent ex vivo mineralization, highlighted unusually high type I collagen gene upregulation. We adopted methods from evidence that inhibition of TGF-ß1 signaling corrected abnormal COL1A1-to-COL1A2 gene expression ratios and increased alizarin red staining [36] to explore whether similar intervention could influence expression of the signature gene COL1A2 and restore the ex vivo mineralization function to donor#6 cells. Cells were plated at a seeding density of 104/cm2 in 24-well multiwell plates (Greiner Bio-one) for Alizarin Red staining and in T25 flasks (Greiner Bio-one) for RNA extraction. All culture vessels were incubated at 37°C with 5% CO2 in a humidified incubator. During the three days needed for cells to reach 85–90% confluence before OM treatment, MM was supplemented with either 40 mU/mL interferon-gamma (IFN-γ), a cytokine antagonistic to TGF-ß1 mediated collagen synthesis [37, 38] or 2 μM SB-431542, an inhibitor of the TGF-ß1 receptor activin receptor-like kinase (ALK5), [39]. The cells were then treated with OM-PL to induce matrix mineralization, harvesting mRNA for COL1A2 gene expression analysis after one week and staining for Alizarin red after two weeks in OM-PL as described above.
Statistical analysis
For the Real Time PCR analysis a two-tailed paired t test was applied to analyse differentially expressed genes between the control and induced groups in the OM-FBS or OM-PL inductions. Genes with >3-fold upregulation and p values less than 0.05 were considered significant. Individual gene expression graphs show standard error mean (S.E.M.) bars as a measure of the uncertainty in estimate of the mean. Graphs showing gene expression averaged from several donors show the standard deviation (S.D.) as a measure of the variability between donors.
Results
OM-PL promoted prompt induction of cGMP-hBM-MSC matrix mineralization
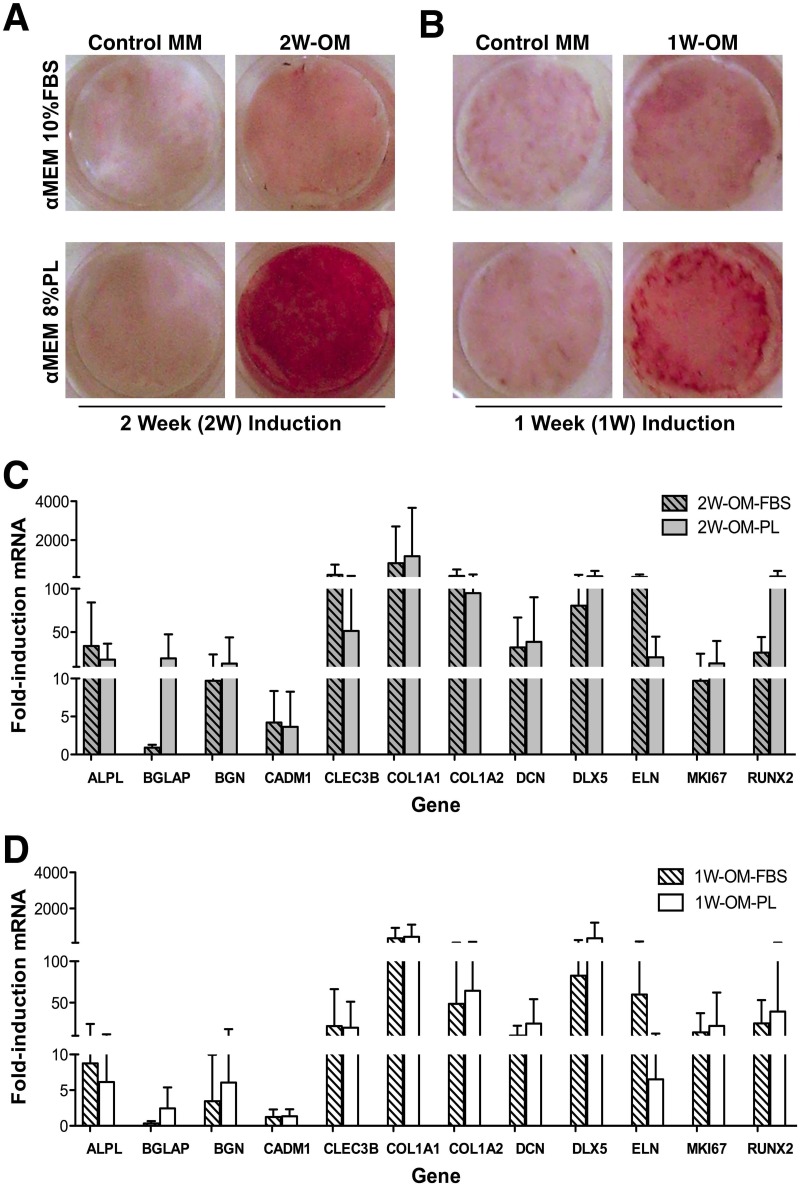
Pilot studies tested induction of differentiation in the six donor-specific cGMP-hBM-MSC populations, detecting matrix mineralization by ALZ staining after OM-PL or OM-FBS treatment. The low level ALZ staining of the control cells in MM (Fig 1A and 1B left panels) was increased when using OM-FBS and consistently more so using OM-PL for both 1W-OM and 2W-OM time points (Fig 1A and 1B lower right panels).
Fig 1. Osteogenic induction: Alizarin Red S staining and Osteogenic Biomarker expression.
Representative photomicrographs of 24-well plate wells stained with Alizarin Red S after (A) two weeks or (B) one week of treatment with osteogenic medium (OM) based on Fetal Bovine Serum (FBS) or Platelet Lysate (PL). Parallel samples cultured in maintenance medium (MM) lacking osteogenic factors served as controls for spontaneous differentiation. Gene upregulation of 12 osteogenic biomarker genes in cGMP-hBM-MSC populations derived from six donors was measured after treatment with OM-FBS (hatched columns) or OM-PL (plain columns) for (C) two weeks or (D) one week. Measurements from triplicate determinations were all statistically significant (p < 0.05).
OM-PL reproduced osteogenic biomarker gene induction seen with OM-FBS
Given positive differentiation outcomes from our protocols, we explored whether literature-mined osteogenic biomarker genes (often obtained with OM-FBS) were applicable to cells cultured with PL. We also needed to learn whether such biomarkers remained relevant for 1W-OM protocols. Of the 18 osteogenic biomarker genes initially selected, 6 genes; msh homeobox homologue 2 (MSX2), (parathyroid hormone receptor 1 (PTH1R), secreted frizzle-related protein 1 (SFRP1), osteopontin/secreted phosphoprotein 1 (OPN/SPP1), osterix (SP7) and tafazzin (TAZ) failed to show an appreciable fold-increase with respect to control samples in most of the six donor-specific cGMP-hBM-MSC populations (data not shown). In contrast, the remaining twelve osteogenic biomarker genes were robustly upregulated in cells treated with 2W-OM-FBS or 2W-OM-PL, in many cases over ten-fold (Fig 1C). Comparing OM-FBS with OM-PL the average level of gene upregulation was broadly equivalent, though exceptions included more upregulation of BGLAP (≈20X) and RUNX2 (≈5X) using OM-PL and more upregulation of ELN (≈5X) with OM-FBS. Nonetheless, more significant underlying donor-specific variation meant that most gene expression differences between OM-FBS and OM-PL were not statistically significant. After OM treatment for just one week, the osteogenic biomarker genes remained significantly measureable, despite an overall trend for lower levels of upregulation (Fig 1D), data in S1 Table. There remained a tendency for more upregulation of BGLAP (≈7X) using OM-PL and more upregulation of ELN (≈9X) using OM-FBS, but underlying donor heterogeneity again meant that these differences were not statistically significant. These osteogenic biomarker expression patterns confirmed cGMP-hBM-MSC differentiation in response to the 1W-OM-PL protocol.
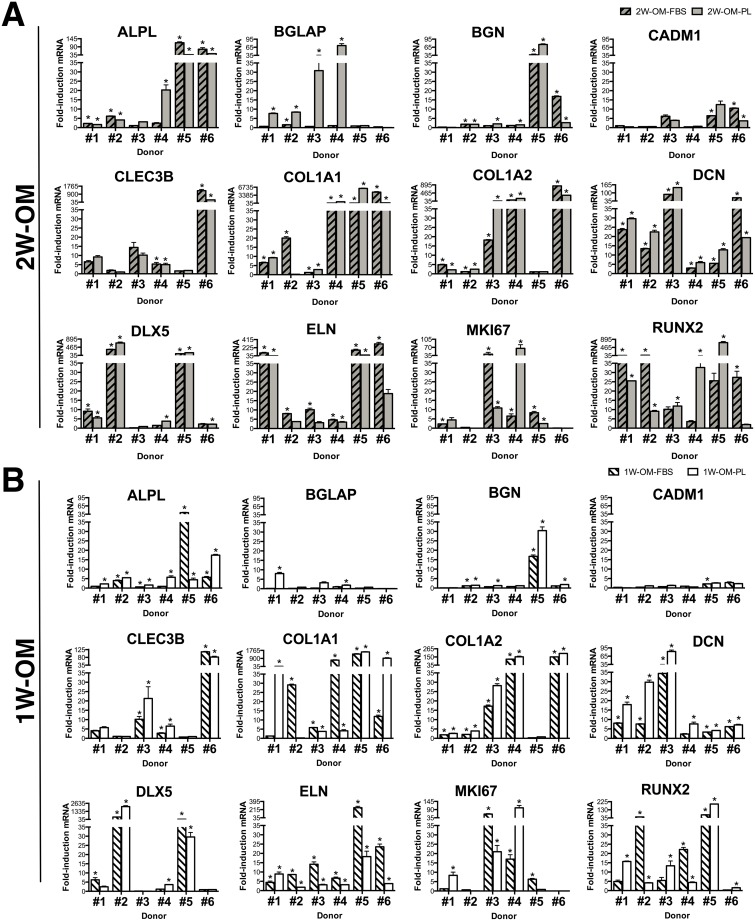
Osteogenic biomarker gene expression revealed inter-donor heterogeneity
Individual cGMP-hBM-MSC populations (donors #1-#6) showed striking donor-specific gene expression patterns after OM treatment. Nonetheless, the two-week data (Fig 2A) showed close parity with data from the one-week differentiation protocol (Fig 2B), data in S1 Table. A comparison of geometric means indicated that most genes were upregulated to a greater extent in OM-PL than OM-FBS e.g. BGLAP (4.5x), DCN (2.15x), BGN (1.74x), COL1A2 (1.63x), ALPL (1.62x), CLEC3B (1.37x), RUNX2 (1.1x) and DLX5 (1.05x). The most consistently upregulated genes (p <0.05) in OM-FBS or OM-PL were ALPL, BGN, CLEC3B, COL1A2, DCN and DLX5 after two weeks and BGN, CADM1, CLEC3B, COL1A2, DCN, DLX5 and ELN after one week of OM treatment (Table 3). One notable difference between use of FBS or PL was that in OM-PL RUNX2 expression was more significantly covariant with other biomarker genes, namely BGN, CADM1, COL1A1 and ELN at two weeks and BGN, COL1A1 and ELN at one week (Table 4).
Fig 2. Inter-donor heterogeneity for osteogenic biomarker expression.
The level of named gene upregulation in response to osteogenic medium treatment for (A) two weeks (2W-OM) or (B) one week (1W-OM) determined from triplicate measurements was tested to ensure statistical significance (*p < 0.05), error bars indicating standard error of the mean.
Table 3. Correlations for osteogenic biomarker gene upregulation in cGMP-hBM-MSC treated with OM-FBS or OM-PL for two weeks (2W) or one week (1W).
| Gene | 2W-OM-FBS versus 2W-OM-PL | 1W-OM-FBS versus 1W-OM-PL | ||
|---|---|---|---|---|
| (r2) | p-value | (r2) | p-value | |
| ALPL | 0.851 | 0.032 | -0.033 | ns |
| BGLAP | 0.186 | ns | -0.084 | ns |
| BGN | 0.911 | 0.0116 | 1.00 | 6.81 E-5 |
| CADM1 | 0.554 | ns | 0.846 | 0.034 |
| CLEC3B | 1.00 | 1.518 E-7 | 0.987 | 2.67 E-4 |
| COL1A1 | -0.185 | ns | 0.664 | ns |
| COL1A2 | 0.924 | 0.008 | 1.00 | 3.55 E-7 |
| DCN | 0.818 | 0.047 | 0.975 | 0.001 |
| DLX5 | 0.995 | 4.28 E-5 | 0.991 | 1.32 E-4 |
| ELN | 0.560 | ns | 0.910 | 0.012 |
| MKI67 | 0.045 | ns | 0.244 | ns |
| RUNX2 | -0.026 | ns | 0.776 | ns |
(r2): coefficient of determination. ns: not significant
Table 4. Covariantly upregulated genes in cGMP-hBM-MSC treated with OM-FBS or OM-PL for two weeks (2W) or one week (1W).
| Differentiation | Covariant Genes | Correlation | p-value |
|---|---|---|---|
| Protocol | (r2) | ||
| 2W-OM-FBS | ALPL-BGN | 0.987 | 2.50E-04 |
| BGLAP-DLX5 | 0.850 | 0.032 | |
| CLEC3B-COL1A1 | 1.000 | 1.60E-07 | |
| CLEC3B-COLIA2 | 0.994 | 4.90E-05 | |
| COL1A1-COL1A2 | 0.995 | 3.80E-05 | |
| 2W-OM-PL | BGLAP-MKI67 | 0.960 | 0.002 |
| BGN-CADM1 | 0.946 | 0.004 | |
| BGN-COL1A1 | 0.995 | 3.40E-05 | |
| BGN-ELN | 0.832 | 0.040 | |
| BGN-RUNX2 | 0.998 | 3.90E-06 | |
| CADM1-COL1A1 | 0.927 | 0.008 | |
| CADM1-RUNX2 | 0.932 | 0.007 | |
| CLEC3B-COL1A2 | 0.878 | 0.021 | |
| COL1A1-ELN | 0.822 | 0.045 | |
| COL1A1-RUNX2 | 0.996 | 2.60E-05 | |
| ELN-RUNX2 | 0.846 | 0.034 | |
| 1W-OM-FBS | ALPL-BGN | 0.995 | 3.00E-05 |
| ALPL-COL1A1 | 0.840 | 0.037 | |
| ALPL-ELN | 0.995 | 4.00E-05 | |
| BGN-COL1A1 | 0.868 | 0.025 | |
| BGN-ELN | 0.999 | 3.20E-06 | |
| COL1A1-ELN | 0.859 | 0.029 | |
| DCN-MKI67 | 0.898 | 0.015 | |
| 1W-OM-PL | ALPL-CLEC3B | 0.881 | 0.020 |
| BGN-COL1A1 | 0.856 | 0.029 | |
| BGN-ELN | 0.902 | 0.014 | |
| BGN-RUNX2 | 0.993 | 6.60E-05 | |
| CADM1-COL1A1 | 0.869 | 0.025 | |
| COL1A1-RUNX2 | 0.820 | 0.046 | |
| ELN-RUNX2 | 0.936 | 0.006 |
(r2): coefficient of determination.
Inter-donor heterogeneity included individual examples of high gene induction (Fig 2B); e.g. BGN was only upregulated markedly in cells from donor #1 (16.7-fold with OM-FBS, p = 0.00018 and 30.4-fold with OM-PL, p = 0.00014), similarly DLX5 in cells from donor #2 (64-fold with OM-FBS, p = 0.0004 and 2106-fold with OM-PL, p = 0.0003) and ELN in cells from donor #5 using OM-FBS (258.3-fold, p = 0.03). In contrast, DCN showed significant OM-PL mediated gene induction in all donors, ranging from 3.8-fold, p = 0.0001 in cells from donor #5 to 82-fold, p = 0.0002 in cells from donor #3.
Focusing on the assay most relevant to the clinical protocol, 1W-OM-PL treatment, two-way analysis of variance indicated that the donor-specific influence on gene upregulation was significant (p<0.001) accounting for over 70% of the total variance in seven of the twelve osteogenic biomarker genes, namely BGN, CADM1, CLEC3B COL1A1, COL1A2, DCN and RUNX2. Overall, there was considerable variation in the extent of gene upregulation during differentiation and most importantly, osteogenic biomarker gene expression measured after only one week of OM-PL treatment was suitable for identification of inter-donor differences.
Inter-donor heterogeneity for OM-PL-induced ALZ+, VK+, MKI67+ ex-vivo phenotypes
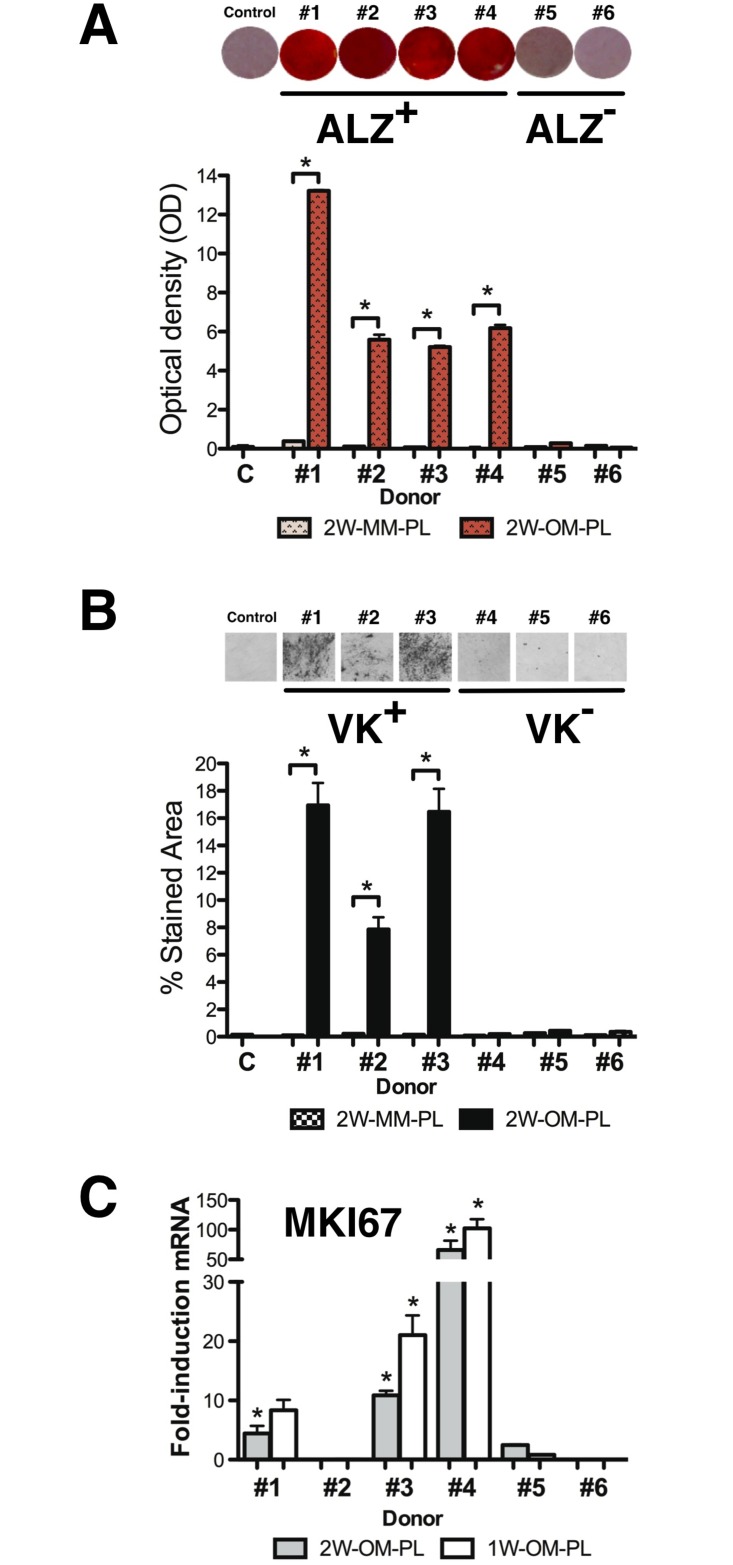
There was relatively little ALZ+ staining observed after OM treatment for one week, but after two weeks, two main donor-specific phenotypes were found. Cells from four donors (#1, #2, #3, #4) exhibited extensive matrix mineralization and were characterized as ALZ+ with OD values ranging between 5 and 13, significantly greater than the negative control samples (p = 0.0003) (Fig 3A). In contrast, cells derived from donors #5 and #6 exhibited weak ALZ+ staining, resembling that of negative control cells and were characterized as ALZ- with a background level OD less than 0.5 (p<0.015).
Fig 3. Inter-donor heterogeneity for OM-PL-induced ALZ+, VK+, MKI67+ ex-vivo phenotypes.
After two weeks of osteogenic induction in OM-PL, the hBM-MSCs were stained for matrix mineralization. (A) Representative photomicrographs (10X) of donor-specific cGMP-hBM-MSC populations positive for Alizarin red S stain (ALZ+) and histogram of eluted dye staining intensity measurement at 562 nm (*p < 0.05). (B) Representative photomicrographs (10X) of donor-specific cGMP-hBM-MSC populations positive for Von Kossa stain (VK+) and histogram of positively stained area quantified using Image J software (*p < 0.05). (C) Histogram showing the extent of gene upregulation of MKI67 determined after induction with OM-PL for two weeks (dark columns) or one week (light columns). Measurements from triplicate determinations that were statistically significant are indicated (*p < 0.05).
Over the same two-week treatment period, only three of the four ALZ+ donors, namely #1, #2, #3 were also VK+, showing diffuse areas of calcium phosphate stained by black silver ion particles covering 8 to 16% of the surface area evaluated by Image J software (Fig 3B). In contrast, cells from donors #4, #5 and #6 had no significant VK staining and resembled the non-OM treated negative control cells, data in S2 Table.
Given proposals that proliferation plays an important role in the early stages of differentiation, we analyzed MKI67 expression in OM-PL treated cells. The inter-donor heterogeneity for MKI67 upregulation did not correspond directly with the mineralization phenotypes. After 2W-OM-PL treatment the low MKI67 expression of donors #2, #5, #6 remained low, whereas donors #1, #3 and #4 showed 4 to 66-fold MKI67 upregulation. After 1W-OM-PL treatment, the significant MKI67 upregulation for donors #1, #3 and #4 ranged from 8 to 102-fold (p<0.05) (Fig 3C), data in S1 Table. Thus both time points confirmed inter-donor heterogeneity for MKI67+ expression with greater upregulation of the active cell cycle biomarker during early phases of osteogenic differentiation.
Inter-donor heterogeneity for the extent of bone formation in-vivo
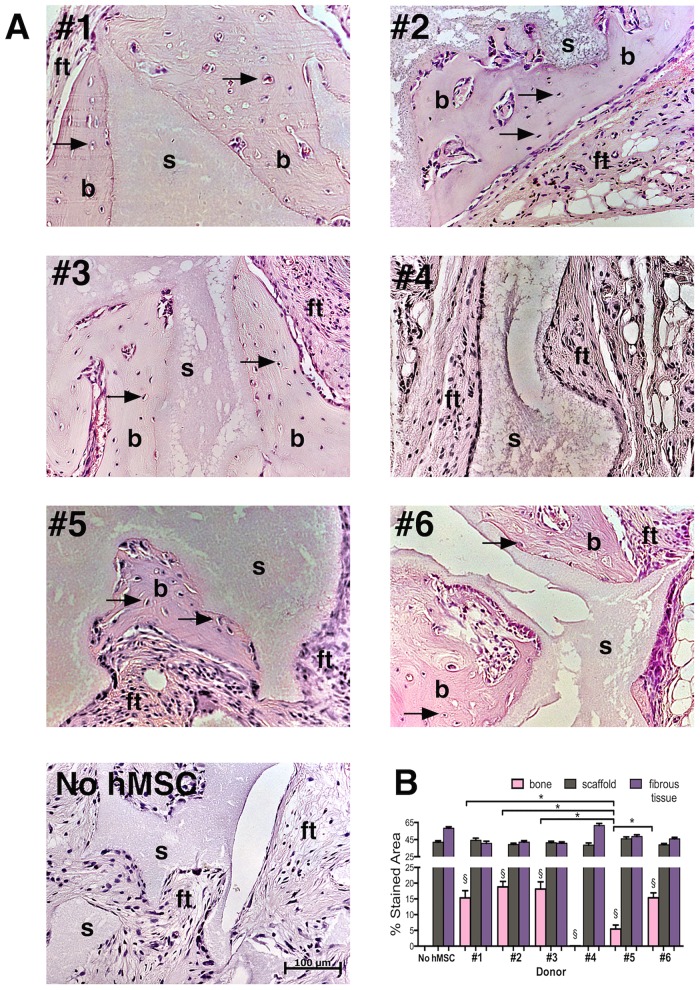
All NOD/SCID mice subjected to xenograft implants were healthy after the experimental procedures and the small implant incisions healed within 7 days without infection or complications. H&E stained sections of hBM-MSCs and HA-βTCP xenografts six weeks post-implantation revealed donor-specific heterogeneity for the extent of ectopic bone formation among fields of view capturing equivalent areas of scaffold. cGMP-hBM-MSC derived from donors #1, #2, #3 and #6 had a good positive bone-forming phenotype (BF+) forming new bone comprising 15–18% of the total tissue section area (Fig 4A and 4B). In contrast, xenografts specific to donor #4 showed cells aligned along the scaffold surface and vascularization of the tissue but no significant bone formation (Fig 4A). Unlike the other bone-forming xenografts, donor #5 cells failed to show any evidence for hematopoietic territories adjacent to the newly formed bone and the amount of bone formed by donor #5 cells (5.4%) was significantly less (p<0.05) than the average bone formation among the other bone-forming donors (16.91%) (Fig 4B), thus it was defined as poor bone-forming (BF-), data in S3 Table.
Fig 4. Inter-donor heterogeneity for bone formation in-vivo.
Photomicrographs of H&E stained sections of decalcified paraffin-embedded Xenografts under bright field illumination. The Xenografts consisted of hydroxyapatite ß-tricalcium scaffold granules seeded with cGMP-hBM-MSC derived from (A) donors #1 to donor #6 respectively. Regions adjacent to the scaffold (s) contained newly formed osteoid bone (b) stained more homogeneously pink relative to the surrounding fibrous tissue (ft) and contained numerous osteocytes within lacunae (arrows). A representative section of the control implant of hydroxyapatite ß-tricalcium scaffold granules without cells revealed scaffold (s) and fibrous tissue (ft) only. (B) Histogram of the histological section area governed by scaffold (grey column), stromal fibrous tissue (purple column) or bone osteoid matrix (pink column) showing significant bone formation (§p<0.05). Donor heterogeneity with regard to the relative amount of bone formed showed statistically significant differences (*p < 0.05). Scale bar = 100 μm.
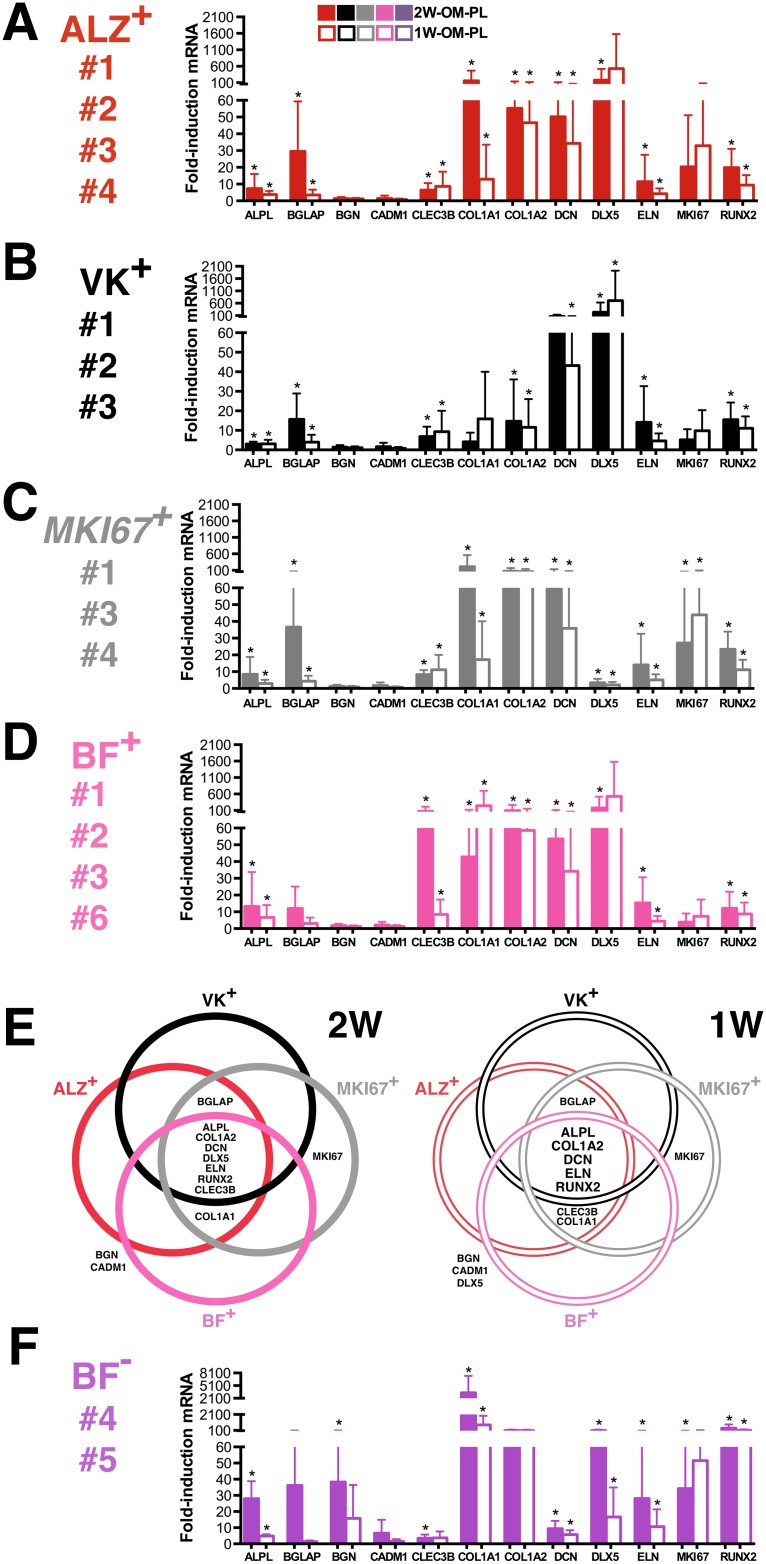
Biomarker characterization of ALZ+, VK+, MKI67+, BF+ cell populations
To accommodate inter-donor heterogeneity, we focused on measuring biomarker expression after OM-PL treatment in phenotypically functional cGMP-hBM-MSC populations. Thus, the representative average biomarker expression associated with the ALZ+ phenotype represented donors #1, #2, #3, #4 (Fig 5A); the VK+ phenotype represented donors #1, #2, #3 (Fig 5B); the proliferation related MKI67+ phenotype represented donors #1, #3, #4 (Fig 5C) and the BF+ phenotype represented donors #1, #2, #3, #6 (Fig 5D). For each phenotype, a specific subset of the biomarker genes showed statistically significant upregulation among all the relevant donor-specific cGMP-hBM-MSC populations. Statistics for pooled expression patterns for 2W-OM-PL showed seven biomarker genes were significantly upregulated in association with all ex vivo osteogenic phenotypes and in vivo bone formation (Fig 5E, 2W Venn diagram). However, more relevant to the need for a prompt assay within cGMP cell expansion timelines, after 1W-OM-PL treatment five biomarkers ALPL, COL1A2, DCN, ELN, RUNX2 represented the osteogenic “signature genes” significantly upregulated in all phenotypic contexts (Fig 5E, 1W Venn diagram).
Fig 5. Biomarker characterisation of ALZ+, VK+, MKI67+, BF+ and BF- cell populations.
Histograms of the average extent of osteogenic biomarker gene upregulation in cGMP-hBM-MSC derived from (A) donors #1, #2, #3, #4 with cells positive for Alizarin red S (ALZ+) when treated with OM-PL for two weeks (red column) or one week (light column). (B) donors #1, #2, #3 with cells positive for Von Kossa staining (VK+) when treated with OM-PL for two weeks (black column) or one week (light column). (C) donors #1, #3, #4 with significant MKI67+ upregulation when cells were treated with OM-PL for two weeks (grey column) or one week (light column). (D) donors #1, #2, #3, #6 with good bone formation (BF+) when cells were treated with OM-PL for two weeks (pink column) or one week (light column). (E) Venn diagrams show the relation between osteogenic function and significantly upregulated biomarkers after (left hand side) OM-PL treatment for two weeks (2W) or (right hand side) OM-PL treatment for one week. (1W). (F) donors #4, #5, incapable of good bone formation (BF-) when cells were treated with OM-PL for two weeks (purple column) or one week (light column). Error bars indicate S.D. of means. (*) Constituent mean values were statistically significant (p<0.05).
The signature gene induction measurements following OM treatment in the non-bone forming (BF-) cGMP-hBM-MSC from donors #4 and #5 (Fig 5F), data in S1 Table, remained statistically significant with one exception. Specifically, COL1A2 was upregulated ≈177-fold in donor #4 cells yet negligibly (1.1-fold) in cells from donor #5. Another difference was that at two weeks of OM-PL treatment upregulation of MKI67 was significant for BF- cells (≈13 fold) but negligible for the BF+ population, suggesting that cells that did not differentiate had more persistent cell cycle activity. Notably, the two genes constituting the type I collagen protein behaved differently, in contrast to COL1A2, COL1A1 mRNA was not included as a signature gene since it was not significantly upregulated in cells with the VK+ phenotype. Of note, COL1A1 expression at 1W-OM-PL was significantly greater in BF- than in BF+ cells (p < 0.001).
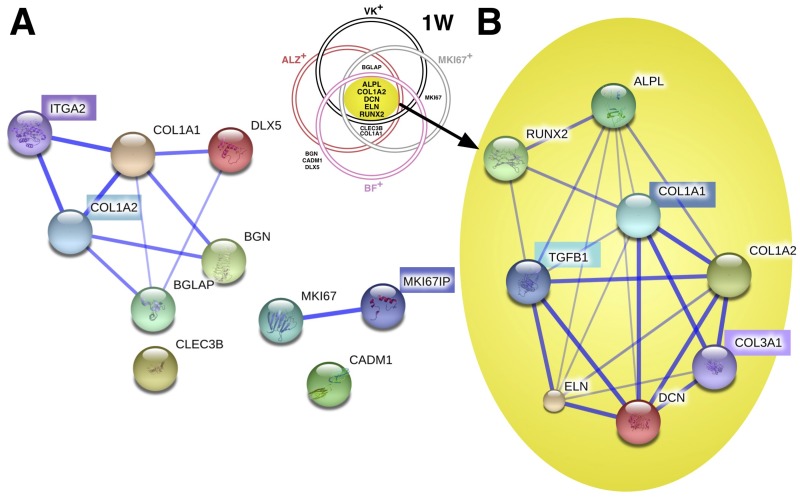
Bioinformatic analysis of signature gene products implicated TGF-ß1 interactions
The Bioinformatic software STRING was used to explore known and predicted interactions for the protein products of the osteogenic biomarker genes. Asking whether the five 1W-OM-PL osteogenic signature genes were preferentially implicated with a particular signaling pathway, we compared their STRING data independently from that of the seven excluded biomarker genes; BGN, BGLAP, CADM1, CLEC3B, COL1A1, DLX5, MKI67, whose significant upregulation was only found in a subset of phenotypes.
For non-signature gene products (Fig 6A), STRING software text-mining evidence indicated associations between COL1A1, DLX5 and BGLAP proteins with co-expression and experimental data also supporting a BGN-COL1A1 interaction, however CADM1, CLEC3B and MKI67 proteins did not show associations. STRING predicted high score functional partners to be COL1A2 (score 0.999), MKI67IP (score 0.996) and ITGA2 (score 0.987). The protein products of two genes CADM1 and CLEC3B remained unassociated.
Fig 6. Bioinformatic analysis of osteogenic biomarker gene product interactions.
STRING (v9.05) software confidence view, with stronger associations represented by thicker lines, for (A) the osteogenic biomarkers not common to all osteogenic functional phenotypes, constituted a disconnected network. The software predicted three closest functional partners (highlighted acronym) and relative scores were: type I collagen alpha 2 (COL1A2; 0.999); MKI67 interacting nucleolar phosphoprotein (MKI67IP; 0.996) and integrin alpha 2/CD49b; 0.987). (B) the osteogenic signature gene biomarkers common to all osteogenic functional ex vivo and in vivo phenotypes constituted a closely connected network. The software predicted three closest functional partners (highlighted) and relative scores were: type I collagen alpha 1 (COL1A1; 0.999); Transforming growth factor, beta 1 (TGFB1; 0.999); and type III collagen alpha 1 (COL3A1; 0.993).
In contrast, signature gene STRING analysis (Fig 6B) revealed a web of multiple interactions for all five gene-products. Experimental evidence supported interactions between COL1A2, DCN and ELN, all interacting with ALPL that was in turn associated with RUNX2 by text-mining evidence. This implied an interdependent functional network. The three highest score predicted functional partners were COL1A1 (score 0.999), Transforming growth factor beta-1 (TGFß1) (score 0.999) and collagen type III alpha 1 (COL3A1) (score 0.993). STRING experimental evidence supported molecular interactions between ELN, DCN, TGFß1, COL1A1 and COL1A2. Like ALPL and DCN, TGFß1 was associated with six other proteins in the network, highlighting the likelihood of it having a key role in our signature gene network for osteogenesis.
Osteogenic signature gene expression correlated with bone formation
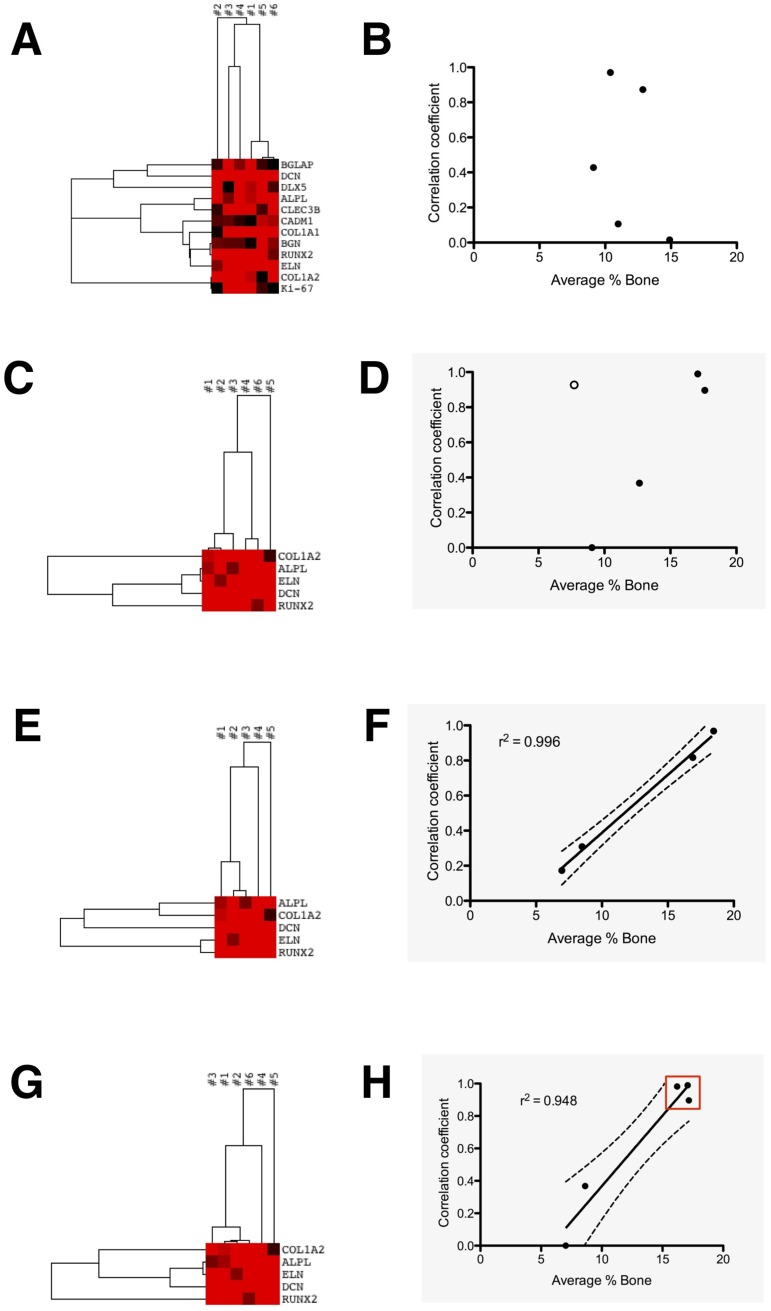
The results supported our initial strategic assumption that correlating ex vivo gene expression with in vivo bone formation would require identification of a subset of biomarker genes significant for all contexts. Simply using all twelve significantly expressed biomarker genes for agglomerative cluster analysis at 1W-OM-PL (Fig 7A) led to donor correlations unrelated to their respective amount of bone formed in vivo, even when using the Pearson centroid linkage that minimized the effect of outlier values, resulting in a very scattered plot (Fig 7B). In contrast, with the five osteogenic signature genes the cluster dendrogram showed more relevant hBM-MSC donor-specific associations (Fig 7C). Close association between bone-forming donors #1 and #2 (correlation coefficient 0.99) led the agglomerative hierarchy. Next, bone-forming hBM-MSC from donor #6 were paradoxically paired with those of non bone-forming donor #4 (correlation coefficient 0.93). Subsequently, hBM-MSC from donor #3 were grouped as similar to the other bone-forming hBM-MSC from donors #1 and #2 (correlation coefficient 0.90). Relatively high correlation coefficients between bone-forming hBM-MSC donors #1, #2, #3 contrasted sharply to the lack of any correlation with donor #5, whose hBM-MSC did not show ex vivo osteogenic differentiation or bone formation. Providing a potentially important clue, the dendrogram also highlighted COL1A2 as the most dissimilarly expressed gene among the six donors. Plotting the dendrogram data (Fig 7D) resulted in slope-aligned donor associations, with a clear outlier exception reflecting the close correlation between donors #4 and #6. Uniquely, these two cell populations both had inconsistent ex vivo and in vivo phenotypes; the donor #4 hBM-MSCs were positive for ex vivo mineralization but did not form bone, however conversely, donor #6 cells had negative mineralization assays yet their xenografts did form bone.
Fig 7. Ex-vivo osteogenic signature gene expression could cluster donor-specific cGMP-hBM-MSC populations according to bone forming potential.
The gene upregulation profiles of the six donor-specific populations induced for osteogenic differentiation with 1W-OM-PL were subjected to cluster analysis: (A) The dendrogram derived using all twelve osteogenic biomarker genes without prior selection for significance in osteogenic function led to (B) a plot of correlation coefficients between donors and their bone forming potential which revealed no significant relationship (r2 = 0.104, p = 0.596). (C) Dendrogram from cluster analysis restricted to the five osteogenic signature genes. Using a Euclidian distance, single linkage algorithm the dendogram indicated closest similarity between bone-forming donors #1 and #2. (D) The plot of correlation coefficients versus bone-forming potential suggested that most associations appeared to constitute a regression slope (closed circles), but there was an outlier association between donors #4 and #6 (open circle) preventing overall correlation (r2 = 0.225, p = 0.420). The above cluster analysis was repeated but with modification to accommodate outlying observations. (E) Dendrogram from cluster analysis excluding data from outlying donor #6, restricted to the five osteogenic signature genes, using a Pearson centroid linkage and uncentered correlation distance. (F) The resulting plot of donor correlation coefficients versus bone forming potential confirmed a presumed regression line relationship between bone-forming potential and gene expression for cells from the five congruent donors (r2 = 0.996, p = 0.0169). (G) The dendrogram resulting from a Euclidian distance, single linkage algorithm with donor#6 COL1A2 induction adjusted to 6.63-fold enhanced similarity between bone-forming donors. (H) The corresponding plot of donor correlation coefficients versus bone-forming potential showed a strong linear relationship between ex vivo gene expression and in vivo bone-forming potential in cells from all donors (r2 = 0.948, p = 0.0051) with closely clustered bone-forming donors #1, #2, #3, #6 (red box).
Removing the outlier donor #6 data before centroid-linkage clustering resulted in a dendrogram that prioritised correlations between bone-forming donors (Fig 7E). Plotting the data highlighting a marked linear regression among the remaining five donors (r2 = 0.996, p = 0.0022). Thus, bar one exception, the signature gene expression patterns measured in culture allowed discrimination according to the amount of bone formed, placing bone-forming and non-bone-forming hBM-MSC populations at opposite ends of a straight slope (Fig 7F).
Regarding the exceptional association between cells that did not and did form bone, 1W-OM-PL treated donor #4 and #6 cells both showed unusually high COL1A2 upregulation (152-fold and 199-fold, respectively), in contrast to much more modest COL1A2 upregulation in the remaining bone forming-donors (#1, 2.63-fold; #2, 3.93-fold; #3, 28.18-fold). Hypothetical mathematical adjustment of donor#6 COL1A2 upregulation to the central tendency geometric mean value of the other bone-forming donors (6.63-fold) resulted in a new Euclidian distance single linkage cluster dendrogram (Fig 7G). With this mathematical adjustment according to data, the close relationship between osteogenic signature gene expression and bone formation was extended to all six donors (r2 = 0.948, p = 0.0051) (Fig 7H), data in S4 Table. Thus, in the context of our signature gene clustering algorithm the elevated ex vivo COL1A2 expression of BF+ donor #6 sufficed to explain its inconsistent outlier status.
COL1A2 down-regulation via TGF-ß1 antagonists restored ex-vivo matrix mineralization
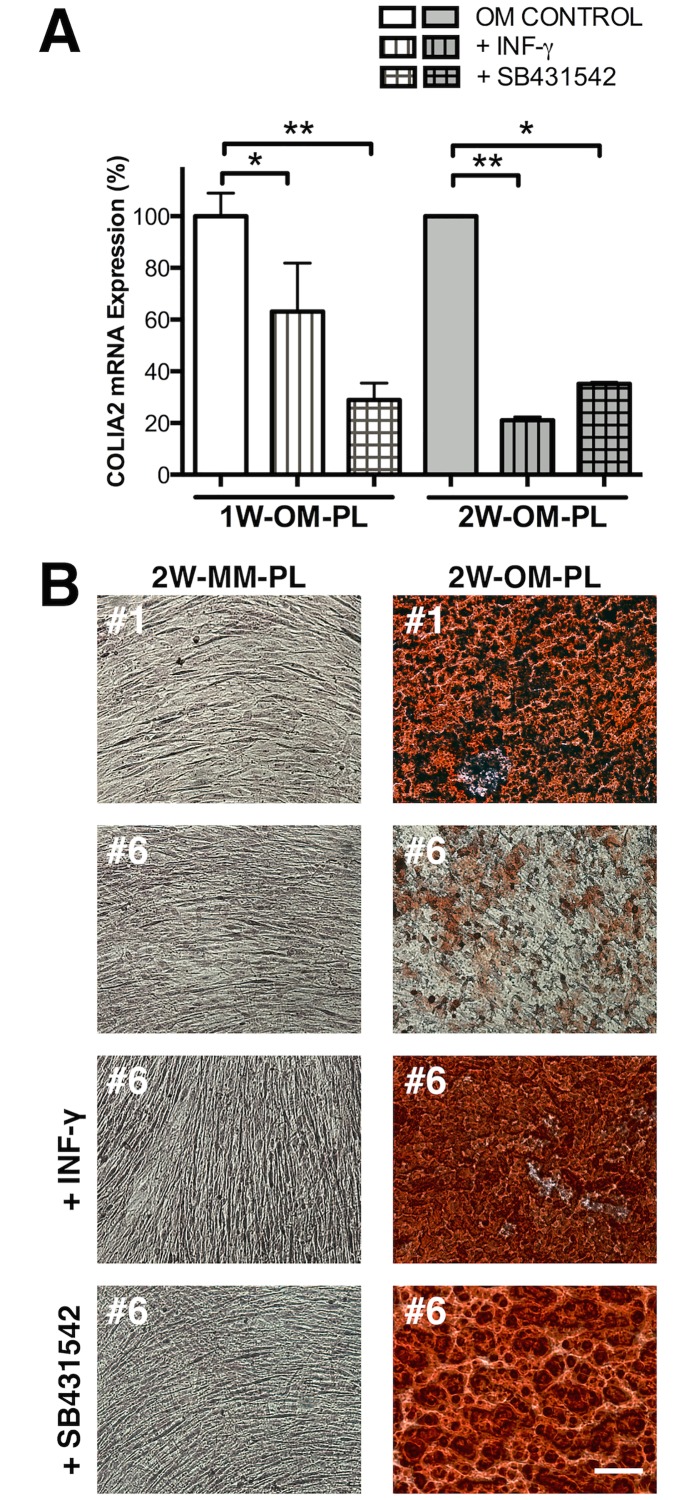
According to our mathematical adjustments, reducing COL1A2 mRNA levels in donor #6 cGMP-hBM-MSC to 23% of the measured value sufficed to maintain a significant correlation (p≤0.05) between signature gene data and bone formation. We explored whether we could obtain corroborative experimental evidence by a brief three-day treatment of the cells with the physiological factor INF-γ, a cytokine known to antagonise TGF-ß1 mediated induction of collagen expression. Donor #6 cGMP-hBM-MSC treated with either MM alone, MM plus IFN-γ, or MM plus the TGF-beta type I receptor inhibitor SB-431542, grew for three days with similar rates to near-confluence before osteogenic differentiation was initiated with OM-PL. RT-PCR analysis (Fig 8A) showed reduced COL1A2 induction by 1W-OM-PL after pretreatment with either INF-γ (≈63% of control) or SB431542 (≈29% of control). Moreover these responses were persistent with long-term consequences; the relative level of COL1A2 induction in donor#6 cells after two weeks of OM-PL treatment remained reduced in cells pre-treated with INF-γ (≈21% of control) or SB431542 (≈35% of control) (Fig 8A), data in S5 Table. With regard to Alizarin Red staining after differentiation, control cells from donor #1 or donor #6 kept in MM-PL, reached a high cell density without mineralization. Following 2W-OM-PL treatment, the positive control donor #1 cells stained strongly with Alizarin Red, but donor #6 cells much less so (Fig 8B). However, donor #6 cells pre-treated with INF-γ or SB431542 for just three days before 2W-OM-PL treatment achieved strong Alizarin red staining.
Fig 8. Downregulation of type I collagen restored ex-vivo matrix mineralization.
(A) Histogram of RT-PCR determined COL1A2 mRNA downregulation following treatment of donor #6 cells with Interferon gamma (IFN-γ) or TGF-ß1 signaling inhibitor SB431542 for three days before treatment with osteogenic medium for one or two weeks. *p <0.05, **p <0.005. (B) Representative photomicrographs of Alizarin Red S staining after two-week treatment of cells with maintenance medium (MM) or osteogenic medium (OM) using donor #1 cells as a positive control compared to donor #6 cells and donor #6 cells pre-treated with MM supplemented with 40U/mL INF-γ or 2 μM SB431542 for three days. Bar = 100 μM.
Discussion
It is increasingly appreciated that among important rigorous requirements for translation of science into effective therapies, potency assays giving a priori indication of administered cell functionality are key for properly conducted ethical trials. Nonetheless, the development of an accurate osteogenic potency assay is particularly challenging. Diverse isolation and culture methods, donor-specific characteristics and methods used to characterise the differentiated phenotype can all contribute to the functional heterogeneity reported for primary cultures of bone marrow derived mesenchymal stem cells. Considering the above we devised a strategy to test whether gene expression of primary cGMP-hBM-MSC ex vivo could predict bone forming potential.
The isolation, cell characterisation and culture methods adopted for this study are those currently being employed by the Reborne EU FP7 consortium in phase I clinical trials [40], ensuring that our data should be relevant to clinical contexts. A history of different osteogenic medium (OM) formulations in the literature complicates selection of candidate osteogenic biomarker genes. For example, collagen fibril synthesis and assembly by ascorbic acid-2-phosphate, the long-acting vitamin C derivative is concentration dependent [41]. Beta-glycerophosphate, a substrate for alkaline phosphatase and source of inorganic phosphate is important for the formation of hydroxyapatite Ca10(PO4)6(OH)2, yet intracellular incorporation of inorganic phosphate also affects cell function and gene expression [42]. Dexamethasone, a synthetic agonist for the glucocorticoid receptor induces osteogenic differentiation through activation of the Wnt/beta-catenin signaling pathway with variable context and concentration dependent effects on cell proliferation, metabolism and differentiation [43]. We used 10 nM as opposed to 100 nM Dexamethasone to enhance our potential to see osteogenic differences since 100 nM Dexamethasone treatment could artefactually drive matrix mineralization in skin-derived cells [44], co-stimulate expression of adipogenic genes [45] and obscure donor-specific variations in osteoblastic differentiation [46].
Although ascorbic acid, beta-glycerophosphate and dexamethasone sufficed to induce osteogenic differentiation of MSC [47], we added the physiologically relevant supplement BMP-2, that can potentiate differentiation in preosteoblast cells [48, 49]. BMP-2 has a predominant role in the early phase of hMSC differentiation and preserves cell-type specificity since it did not enhance osteogenic differentiation of adipose derived stem cells [50] or periodontal ligament cells [51]. Co-administration of BMP-2 with hMSC increased bone formation in immune deficient mice [52]. It is crucially relevant to graft healing [53] and despite a short half-life [54] BMP-2 is under consideration for clinical applications [55–57]. Our ex vivo dose of BMP-2 (100 ng/mL) fell within the range of 25 ng/mL to 25 μg/mL shown to yield near-equivalent amounts of bone in vivo [58]. In comparison to some other BMP family members, BMP-2 showed modest improvement in inducing ex vivo osteogenic differentiation [59]. Thus BMP-2 was well suited for a discriminatory assay, it wouldn’t necessarily undermine the contribution of a cell’s innate osteogenic potential, as might an alternative excessively dominant inducer of osteogenic differentiation.
The choice of human PL or FBS, both modulators of osteogenic inducers, was considered a predominant characteristic distinguishing OM-PL from OM-FBS. In previous reports, individual hMSC populations had a marginally improved propensity to form bone when grown in PL (9/9; 100%) versus FBS (6/9; 67%), whilst ex vivo osteogenic assays were equivalent [60]. Even without osteogenic medium supplements, PL helped prime hMSC towards an osteogenic phenotype by raising levels of ALPL mRNA, and matrix proteins integrin-binding sialoprotein IBSP and OPN/SPP1 [61]. The latter secreted protein can bind hydroxyapatite avidly and also act as a cytokine upregulating expression of IFN-γ [62]. Our results supported the view that osteogenic biomarker expression must be considered and interpreted within the context of the specific osteogenic inducers used. For example, serum may be suboptimal for BMP-2 responsiveness [63] and under our conditions CADM1 expression was not a predictor of hMSC function [64]. Previous predictive biomarkers from model systems using FBS need not necessarily remain relevant when hMSC are derived and cultured with PL. For example, although ELN expression may be governed by dexamethasone acting on several glucocorticoid-responsive elements in its promoter [65], its expression was significantly higher in OM-FBS versus OM-PL (p<0–001). This likely reflected complex interactive growth factor networks; e.g. a possible inhibitory effect of basic FGF [66] a significant component of PL [67].
Pooling data from all donors, most genes (7/12; 58%) were significantly induced by either OM-FBS or OM-PL after one week, implying this principally reflected fundamental features of differentiation rather than particular responses to unique growth factors. We confirmed several osteogenic biomarker gene expression patterns reported in an immortalised hMSC-TERT cell model [68] extending their relevance for osteogenic differentiation to primary hMSC with PL-based OM [69]. We also confirmed that early events in the differentiation process were linked to end-stage phenotypic expression such as mineral deposition [70]. Meeting the first of our main aims, prompt differentiation with more robust osteogenic biomarker induction in 1W-OM-PL could facilitate performing the osteogenic potency assay within a cGMP cell expansion time frame.
To categorise heterogeneous donor-specific cultures we focused on significantly expressed genes in cells that functioned positively in multiple osteogenic assays. Alizarin Red and Von Kossa staining for matrix mineralization were complementary staining methods, each reacting with the target calcium molecule differently [32]. Donor #4 cells with their ALZ+ yet VK- phenotype highlighted the multifactorial complexity of the matrix deposition and mineralization process. RT-PCR based analysis of MKI67 expression, a measure for cell cycle activity equivalent to the positive Ki67 index from immunocytochemical staining [71], advantageously provided a phenotypic end-point that could be measured concurrently with the osteogenic biomarker genes at both 1W and 2W time points. In contrast to ALZ+ or VK+ cells, MKI67+ cells expressed BGLAP, COL1A1, COL1A2 and RUNX2 at relatively high levels, consistent with observations that collagen type I can enhance hMSC growth rate [72]. hMSC proliferation may be associated open chromatin structures broadening the gene expression repertoire [73, 74] and indicative of multipotency [75] but we could not confirm that proliferation was a reliable correlate with bone formation [25].
The ectopic bone formation assay cleared showed inter-donor heterogeneity and despite a small sample size of six donors, our % incidence of donors with good bone formation (4/6; 67%) was very representative of heterogeneity for successful bone formation in studies with larger sample sizes: (20/34; 59%) [76], (74/120; 62%) [77], (8/14; 57%) [20] and (11/20; 55%) [25]. Parameters such as age of donor, CFU efficiency of primary cultures, or any single gene did not correlate directly with bone formation. Although BGLAP was associated with ex vivo matrix mineralization it was not strongly associated with the bone forming phenotype, confirming its distinct significance for ex-vivo and in-vivo osteogenic differentiation pathways [26]. Despite such caveats, seeking promptly expressed osteogenic biomarkers relevant for both ex vivo and in vivo contexts in 1W-OM-PL data we did identify five genes; ALPL, COL1A2, DCN, ELN and RUNX2 fitting the “signature” of being significantly expressed in all contexts. Providing just five signature genes was advantageous for donor discrimination, since too many clustering variables might reduce the probability of finding clear dissimilarity. Fortunately, only the RUNX2 and ELN genes were co-induced in a parallel manner among the six donors, reducing concern for biased overrepresentation from too many highly correlated variables.
Previous publications aiming to draw correlations between gene expression and bone forming potency have also been based on relatively small sample sizes, yet there is need for caution when interpreting the regression coefficient as an indicator of the relative effectiveness of our signature gene biomarkers. We ensured careful and significant measurement for all our signature genes, because when the variance of individual observations is small, accurate prediction may still be possible despite a small sample size. Given many potential differences between immortalized hMSC-TERT versus primary hMSC expanded and differentiated in different media, the consistent identification of a predictive quality for COL1A2, DCN and ELN expression was striking. Correlation need not imply causation, nonetheless, the five signature genes have well recognised roles in bone formation. Mice null for tissue-nonspecific alkaline phosphatase (ALPL) suffer bone abnormalities and its overexpression can increase skeletal mineralization [78]. Mutations in COL1A2 or its partner COL1A1 that encode type I collagen can cause dominant inheritance of osteogenesis imperfecta [79]. A strong correlation between the steady-state mRNA levels of type I collagen and periostal bone formation highlights the in vivo relevance of its gene expression level [80]. Decorin (DCN) a major matrix proteoglycan in bone helps regulate matrix mineralization by influencing collagen assembly [81]. Elastin (ELN) degradation products can synergise with TGF-ß1 to promote osteogenic differentiation in fibroblasts [82] and the multifunctional runt related transcription factor 2 (RUNX2) is essential for osteoblast development and bone mineralization [83]. It has been suggested that ex vivo induction of ALPL mRNA levels and ALPL activity could predict the bone forming capacity of human bone marrow stromal cells [84]. We confirmed that testing the response to osteogenic induction can be informative and ALPL was one of our five signature genes, however no single genetic biomarker sufficed to reliably predict osteogenic performance among our six donor specific hBM-MSC populations. Rather our data indicated that the measurement of induced levels of expression among a cohort of osteogenic genes was required to reliably correlate ex vivo and in vivo osteogenesis.
Seeking further clues regarding the role of our five signature genes, a STRING database bioinformatic search identified the three most highly scored associations from known and predicted proteins. As might be expected, COL1A1, the collagen type I triple helix partner of signature gene COL1A2 had the highest interaction score. The STRING database prioritised interactions with morphogenic cytokine TGF-ß1 over the inducing agent BMP-2, a particularly compelling result since counterbalanced BMP-2 and TGF-ß1 signalling pathways converge at the RUNX2 gene to control hMSC differentiation with a coordinated activity that is critical for skeleton formation [85]. TGF-ß1 can enhance BMP-2 ectopic bone formation [86] but in excess can be responsible for inhibition of osteogenesis that can in turn be counteracted with BMP-2 treatment [87]. For the third interactor highlighted by STRING, transgenic mouse studies have confirmed that COL3A1 regulates osteoblastogenesis and the quantity of trabecular bone [88]. The high in vivo relevance of these influential signalling pathways [89] may help explain why expression of our osteogenic potency signature genes ex vivo could overcome contextual differences to remain relevant for in vivo bone formation.
Notably, the most highly upregulated gene for all the osteogenic phenotypes we tested, decorin, was less upregulated in cells from our BF- donors and has been reported to be essential for maintaining mature osteoblasts through an ability to sequester and modulate the activity of TGF-ß1 [90]. A role for decorin in neutralizing the activity of TGF-ß1 would be consistent with observations that neutralizing TGF-ß with specific antibodies could induce an 11% increase in the mineral-to-collagen ratio in murine bone [91] and that excessive TGF-ß1 activity can underlie bone disease [92]. In summary, bioinformatic analysis highlighted that the signature genes form part of a network involving TGF-ß1, which was consistent with this cytokine’s central role in bone remodeling [93].
We also shed light on an apparent anomaly from earlier studies. Despite Collagen type I’s key role in bone formation [94], its ex vivo expression has been correlated with bone forming potential [95], but sometimes not [96]. Notably, measurement of baseline COL1A1 expression was not related to the bone forming potential of hMSC-TERT clones [26], whereas in a follow-up study using the same cells, OM-induced COL1A2 expression did predict bone formation [27]. The often used, term “collagen type I gene” is ambiguous since two independently regulated COL1A1 and COL1A2 genes found on different chromosomes contribute to the two α1(I) chains and one α2(I) chain forming the heterotrimer Type I collagen molecule. These two collagen genes share common [97] and distinct DNA promoter elements [98]. Our data indicated COL1A2 rather than COL1A1 gene induction was more precisely correlated with subsequent bone formation. True to the iterative nature of hierarchical cluster analysis, discriminating between the cGMP-hBM-MSC from different donors using the five signature genes required optimization and compensation for apparent outliers. Exclusion of just one outlier, the donor #6 cell population, sufficed to reveal a remarkably linear correlation between the ex vivo expression of osteogenic signature genes and bone formation among the remaining five donors (coefficient of determination, r2 = 0.996).
Rather than be ignored, the outlier donor #6 was a useful source of hypothesis; since cluster analysis highlighted COL1A2 as the most disparate signature gene, very highly induced in donor #6 cells. Given that donor #6 cells ultimately did form bone, we conceptualized substituting the donor #6 COL1A2 high measured value with the lower geometric mean value of the other bone-forming donors. This sufficed to assimilate donor #6 cells with the other bone-forming populations and greatly improve the linear correlation between signature gene expression and bone formation for all donors. The closest clustering of the bone-forming donors with greatest separation from non-bone-forming donors was achieved by the more quantatively discriminatory Euclidian distance and single linkage cluster analysis (r2 = 0.948, p = 0.0051). Notably, the phenotype of donor #6 cGMP-hBM-MSC resembled the impaired mineralization and reduced Alizarin red staining in human osteoarthritic (OA) osteoblasts with abnormally high expression of the type I collagen genes [36]. Couchourel et al., rescued Alizarin staining by correcting an abnormal COL1A1-to-COL1A2 expression ratio with inhibition of TGF-ß1 signalling in OA osteoblasts. Given the paradox that donor #6 cells had contrasting ex vivo and in vivo phenotypes we explored the effects of IFN-γ, a physiologically relevant inhibitor of COL1A1 and COL1A2 expression. This cytokine can cross-talk antagonistically with the TGF-ß1 pathway [99] and is produced locally in the bone microenvironment by inflammatory cells as well as hMSC [100] with an anabolic role in bone formation [101]. Brief initial treatment of donor #6 cells with either IFN-γ or an inhibitor of the TGF-ß1 pathway receptor ALK5, significantly reduced subsequent OM induced COL1A2 gene expression and evoked strong Alizarin red staining. Thus mathematical simulation was qualitatively and quantitatively supported by experimental evidence to confirm that correlations drawn between the levels of signature gene expression and bone formation were consistent with hypothetical and real outcomes.
Additional complementary early parameters of freshly isolated hBM-MSC have recently been proposed for predicting cell growth potential from CFU-F morphology [102] and osteogenic function from monolayer cell morphology [103, 104] or mitochondrial function [105]. However, a key advantage of our RT-PCR based analysis includes the possibility to provide a stored cDNA resource that can be subsequently explored further in relation to patient outcomes and/or as new biomarkers are discovered. Further studies will be needed to help verify our assay and determine whether it may be further improved to meet specific requirements by inclusion of further genes.
In summary, we have provided proof of principal that a promptly performed ex vivo potency assay measuring OM induced gene expression of five biomarker “signature genes” could discriminate donor-specific cGMP-hBM-MSC populations according to their bone forming potential in vivo. That such linear correlations could be drawn from relatively few samples encourages consideration that the osteogenic potency assay is applicable to individual cases. As cardinal predictive biomarkers, the five signature genes showed very coherent biological qualities consistent with a governing influence of TGF-ß1 signaling in early phases of hMSC osteogenic differentiation. Having demonstrated feasibility, we propose that ex vivo functional potency assays will be very valuable for the therapeutic application of adult stem cells.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This work was supported in parts by the European Commission Seventh Framework Programme (FP7/2007–2013) (grant no. 241879), through the REBORNE project. We are thankful to Flavia Parise and Renata Battini for their constant support in pre-clinical investigation studies. We also thank Markus T. Rojewski and Hubert Schrezenmeier from the Institute of Clinical Transfusion Medicine and Immungenetics Ulm, German Red Cross Blood Transfusions Service and University of Ulm, Germany as well as Pierre Layrolle, INSERM U957, Laboratory of Pathophysiology of Bone Resorption, Faculty of Medicine, University of Nantes, France and Nathalie Chevallier, Cell Therapy Facility, EFS Ile de France, 94010 Creteil, France for the provision of cGMP-hBM-MSC.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in parts by the European Commission Seventh Framework Programme (FP7/2007–2013) (grant no. 241879), through the REBORNE project.
References
- 1.Quarto R, Mastrogiacomo M, Cancedda R, Kutepov SM, Mukhachev V, Lavroukov A et al. Repair of large bone defects with the use of autologous bone marrow stromal cells. N Engl J Med. 2001; 344(5):385–386. 10.1056/NEJM200102013440516 [DOI] [PubMed] [Google Scholar]
- 2.Bhumiratana S, Vunjak-Novakovic G. Concise review: personalized human bone grafts for reconstructing head and face. Stem Cells Transl Med. 2012; 1(1):64–69. 10.5966/sctm.2011-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen BJ, Gilissen C, Roelofs H, Schaap-Oziemlak A, Veltman JA, Raymakers RA et al. Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 2010; 19(4):481–490. 10.1089/scd.2009.0288 [DOI] [PubMed] [Google Scholar]
- 4.Al-Nbaheen M, Vishnubalaji R, Ali D, Bouslimi A, Al-Jassir F, Megges M et al. Human stromal (mesenchymal) stem cells from bone marrow, adipose tissue and skin exhibit differences in molecular phenotype and differentiation potential. Stem Cell Rev. 2013; 9(1):32–43. 10.1007/s12015-012-9365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sensebe L, Bourin P, Tarte K. Good manufacturing practices production of mesenchymal stem/stromal cells. Hum Gene Ther. 2011; 22(1):19–26. 10.1089/hum.2010.197 [DOI] [PubMed] [Google Scholar]
- 6.Aldahmash A, Haack-Sorensen M, Al-Nbaheen M, Harkness L, Abdallah BM, Kassem M et al. Human serum is as efficient as fetal bovine serum in supporting proliferation and differentiation of human multipotent stromal (mesenchymal) stem cells in vitro and in vivo. Stem Cell Rev. 2011; 7(4):860–868. 10.1007/s12015-011-9274-2 [DOI] [PubMed] [Google Scholar]
- 7.Ben Azouna N, Jenhani F, Regaya Z, Berraeis L, Ben Othman T, Ducrocq E et al. Phenotypical and functional characteristics of mesenchymal stem cells from bone marrow: comparison of culture using different media supplemented with human platelet lysate or fetal bovine serum. Stem Cell Res Ther. 2012; 3(1):6 10.1186/scrt97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung S, Panchalingam KM, Wuerth RD, Rosenberg L, Behie LA. Large-scale production of human mesenchymal stem cells for clinical applications. Biotechnol Appl Biochem. 2012; 59(2):106–120. 10.1002/bab.1006 [DOI] [PubMed] [Google Scholar]
- 9.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008; 3(5):e2213 10.1371/journal.pone.0002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenderup K, Justesen J, Eriksen EF, Rattan SI, Kassem M. Number and proliferative capacity of osteogenic stem cells are maintained during aging and in patients with osteoporosis. J Bone Miner Res. 2001; 16(6):1120–1129. 10.1359/jbmr.2001.16.6.1120 [DOI] [PubMed] [Google Scholar]
- 11.Shi S, Gronthos S, Chen S, Reddi A, Counter CM, Robey PG et al. Bone formation by human postnatal bone marrow stromal stem cells is enhanced by telomerase expression. Nat Biotechnol. 2002; 20(6):587–591. 10.1038/nbt0602-587 [DOI] [PubMed] [Google Scholar]
- 12.Simonsen JL, Rosada C, Serakinci N, Justesen J, Stenderup K, Rattan SI et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002; 20(6):592–596. 10.1038/nbt0602-592 [DOI] [PubMed] [Google Scholar]
- 13.Bernardo ME, Cometa AM, Pagliara D, Vinti L, Rossi F, Cristantielli R et al. Ex vivo expansion of mesenchymal stromal cells. Best Pract Res Clin Haematol. 2011; 24(1):73–81. 10.1016/j.beha.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Phinney DG. Functional heterogeneity of mesenchymal stem cells: implications for cell therapy. J Cell Biochem. 2012; 113(9):2806–2812. 10.1002/jcb.24166 [DOI] [PubMed] [Google Scholar]
- 15.Tohma Y, Dohi Y, Ohgushi H, Tadokoro M, Akahane M, Tanaka Y et al. Osteogenic activity of bone marrow-derived mesenchymal stem cells (BMSCs) seeded on irradiated allogenic bone. J Tissue Eng Regen Med. 2012; 6(2):96–102. 10.1002/term.401 [DOI] [PubMed] [Google Scholar]
- 16.Lian JB, Stein GS. Development of the osteoblast phenotype: molecular mechanisms mediating osteoblast growth and differentiation. Iowa Orthop J. 1995; 15 118–140. [PMC free article] [PubMed] [Google Scholar]
- 17.Madras N, Gibbs AL, Zhou Y, Zandstra PW, Aubin JE. Modeling stem cell development by retrospective analysis of gene expression profiles in single progenitor-derived colonies. Stem Cells. 2002; 20(3):230–240. 10.1634/stemcells.20-3-230 [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Sotome S, Wang J, Orii H, Uemura T, Shinomiya K et al. Correlation of in vivo bone formation capability and in vitro differentiation of human bone marrow stromal cells. J Med Dent Sci. 2005; 52(1):27–34. [PubMed] [Google Scholar]
- 19.Jaquiery C, Schaeren S, Farhadi J, Mainil-Varlet P, Kunz C, Zeilhofer HF et al. In vitro osteogenic differentiation and in vivo bone-forming capacity of human isogenic jaw periosteal cells and bone marrow stromal cells. Ann Surg. 2005; 242(6):859–67, discussion 867. 10.1097/01.sla.0000189572.02554.2c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes SC, Tibbe JM, Veenhof M, Both S, Oner FC, van Blitterswijk CA et al. Relation between in vitro and in vivo osteogenic potential of cultured human bone marrow stromal cells. J Mater Sci Mater Med. 2004; 15(10):1123–1128. 10.1023/B:JMSM.0000046394.53153.21 [DOI] [PubMed] [Google Scholar]
- 21.Castano-Izquierdo H, Alvarez-Barreto J, van den Dolder J, Jansen JA, Mikos AG, Sikavitsas VI et al. Pre-culture period of mesenchymal stem cells in osteogenic media influences their in vivo bone forming potential. J Biomed Mater Res A. 2007; 82(1):129–138. 10.1002/jbm.a.31082 [DOI] [PubMed] [Google Scholar]
- 22.Sabatini F, Petecchia L, Tavian M, Jodon de Villeroche V, Rossi GA, Brouty-Boye D et al. Human bronchial fibroblasts exhibit a mesenchymal stem cell phenotype and multilineage differentiating potentialities. Lab Invest. 2005; 85(8):962–971. 10.1038/labinvest.3700300 [DOI] [PubMed] [Google Scholar]
- 23.Sudo K, Kanno M, Miharada K, Ogawa S, Hiroyama T, Saijo K et al. Mesenchymal progenitors able to differentiate into osteogenic, chondrogenic, and/or adipogenic cells in vitro are present in most primary fibroblast-like cell populations. Stem Cells. 2007; 25(7):1610–1617. 10.1634/stemcells.2006-0504 [DOI] [PubMed] [Google Scholar]
- 24.Cordonnier T, Langonne A, Corre P, Renaud A, Sensebe L, Rosset P et al. Osteoblastic differentiation and potent osteogenicity of three-dimensional hBMSC-BCP particle constructs. J Tissue Eng Regen Med. 2012; 10.1002/term.1529 [DOI] [PubMed] [Google Scholar]
- 25.Janicki P, Boeuf S, Steck E, Egermann M, Kasten P, Richter W et al. Prediction of in vivo bone forming potency of bone marrow-derived human mesenchymal stem cells. Eur Cell Mater. 2011; 21 488–507. [DOI] [PubMed] [Google Scholar]
- 26.Larsen KH, Frederiksen CM, Burns JS, Abdallah BM, Kassem M. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J Bone Miner Res. 2010; 25(4):796–808. 10.1359/jbmr.091018 [DOI] [PubMed] [Google Scholar]
- 27.Burns JS, Rasmussen PL, Larsen KH, Schroder HD, Kassem M. Parameters in three-dimensional osteospheroids of telomerized human mesenchymal (stromal) stem cells grown on osteoconductive scaffolds that predict in vivo bone-forming potential. Tissue Eng Part A. 2010; 16(7):2331–2342. 10.1089/ten.TEA.2009.0735 [DOI] [PubMed] [Google Scholar]
- 28.Stein GS, Lian JB, Gerstenfeld LG, Shalhoub V, Aronow M, Owen T et al. The onset and progression of osteoblast differentiation is functionally related to cellular proliferation. Connect Tissue Res. 1989; 20(1–4):3–13. 10.3109/03008208909023869 [DOI] [PubMed] [Google Scholar]
- 29.Isaksson H, van Donkelaar CC, Huiskes R, Yao J, Ito K. Determining the most important cellular characteristics for fracture healing using design of experiments methods. J Theor Biol. 2008; 255(1):26–39. 10.1016/j.jtbi.2008.07.037 [DOI] [PubMed] [Google Scholar]
- 30.Fekete N, Rojewski MT, Furst D, Kreja L, Ignatius A, Dausend J et al. GMP-compliant isolation and large-scale expansion of bone marrow-derived MSC. PLoS One. 2012; 7(8):e43255 10.1371/journal.pone.0043255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrezenmeier H, Seifried E. Buffy-coat-derived pooled platelet concentrates and apheresis platelet concentrates: which product type should be preferred? Vox Sang. 2010; 99(1):1–15. 10.1111/j.1423-0410.2009.01295.x [DOI] [PubMed] [Google Scholar]
- 32.Veronesi E, Murgia A, Caselli A, Grisendi G, Piccinno MS, Rasini V et al. Transportation Conditions for Prompt Use of Ex Vivo Expanded and Freshly Harvested Clinical-Grade Bone Marrow Mesenchymal Stromal/Stem Cells for Bone Regeneration. Tissue Eng Part C Methods. 2013; 10.1089/ten.TEC.2013.0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puchtler H, Meloan SN, Terry MS. On the history and mechanism of alizarin and alizarin red S stains for calcium. J Histochem Cytochem. 1969; 17(2):110–124. 10.1177/17.2.110 [DOI] [PubMed] [Google Scholar]
- 34.Dennis JE, Konstantakos EK, Arm D, Caplan AI. In vivo osteogenesis assay: a rapid method for quantitative analysis. Biomaterials. 1998; 19(15):1323–1328. 10.1016/S0142-9612(97)00170-1 [DOI] [PubMed] [Google Scholar]
- 35.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013; 41(Database issue):D808–15. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couchourel D, Aubry I, Delalandre A, Lavigne M, Martel-Pelletier J, Pelletier JP et al. Altered mineralization of human osteoarthritic osteoblasts is attributable to abnormal type I collagen production. Arthritis Rheum. 2009; 60(5):1438–1450. 10.1002/art.24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C et al. Inhibition of transforming growth factor (TGF)-β1–induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542. Molecular pharmacology. 2002; 62(1):58–64. 10.1124/mol.62.1.58 [DOI] [PubMed] [Google Scholar]
- 38.Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I et al. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J Biol Chem. 2003; 278(44):43470–43479. 10.1074/jbc.M302339200 [DOI] [PubMed] [Google Scholar]
- 39.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002; 62(1):65–74. 10.1124/mol.62.1.65 [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Barrena E, Rosset P, Lozano D, Stanovici J, Ermthaller C, Gerbhard F et al. Bone fracture healing: cell therapy in delayed unions and nonunions. Bone. 2015; 70 93–101. 10.1016/j.bone.2014.07.033 [DOI] [PubMed] [Google Scholar]
- 41.Choi KM, Seo YK, Yoon HH, Song KY, Kwon SY, Lee HS et al. Effect of ascorbic acid on bone marrow-derived mesenchymal stem cell proliferation and differentiation. J Biosci Bioeng. 2008; 105(6):586–594. 10.1263/jbb.105.586 [DOI] [PubMed] [Google Scholar]
- 42.Beck GRJ, Moran E, Knecht N. Inorganic phosphate regulates multiple genes during osteoblast differentiation, including Nrf2. Exp Cell Res. 2003; 288(2):288–300. 10.1016/S0014-4827(03)00213-1 [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Pang B, Li Y, Zhu D, Pang T, Liu Y et al. Dexamethasone has variable effects on mesenchymal stromal cells. Cytotherapy. 2012; 14(4):423–430. 10.3109/14653249.2011.652735 [DOI] [PubMed] [Google Scholar]
- 44.Hee CK, Nicoll SB. Differential surface antigen expression and 1alpha,25-dihydroxyvitamin D3 responsiveness distinguish human dermal fibroblasts with age-dependent osteogenic differentiation potential from marrow-derived stromal cells in vitro. Cytotherapy. 2011; 13(5):528–538. 10.3109/14653249.2010.542454 [DOI] [PubMed] [Google Scholar]
- 45.Mostafa NZ, Fitzsimmons R, Major PW, Adesida A, Jomha N, Jiang H et al. Osteogenic differentiation of human mesenchymal stem cells cultured with dexamethasone, vitamin D3, basic fibroblast growth factor, and bone morphogenetic protein-2. Connect Tissue Res. 2012; 53(2):117–131. 10.3109/03008207.2011.611601 [DOI] [PubMed] [Google Scholar]
- 46.Alm JJ, Heino TJ, Hentunen TA, Vaananen HK, Aro HT. Transient 100 nM dexamethasone treatment reduces inter- and intraindividual variations in osteoblastic differentiation of bone marrow-derived human mesenchymal stem cells. Tissue Eng Part C Methods. 2012; 18(9):658–666. 10.1089/ten.TEC.2011.0675 [DOI] [PubMed] [Google Scholar]
- 47.Fiorentini E, Granchi D, Leonardi E, Baldini N, Ciapetti G. Effects of osteogenic differentiation inducers on in vitro expanded adult mesenchymal stromal cells. Int J Artif Organs. 2011; 34(10):998–1011. 10.5301/ijao.5000001 [DOI] [PubMed] [Google Scholar]
- 48.Lecanda F, Avioli LV, Cheng SL. Regulation of bone matrix protein expression and induction of differentiation of human osteoblasts and human bone marrow stromal cells by bone morphogenetic protein-2. J Cell Biochem. 1997; 67(3):386–396. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen NR, Henriksen Z, Sorensen OH, Civitelli R. Dexamethasone, BMP-2, and 1,25-dihydroxyvitamin D enhance a more differentiated osteoblast phenotype: validation of an in vitro model for human bone marrow-derived primary osteoblasts. Steroids. 2004; 69(4):219–226. 10.1016/j.steroids.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 50.Cruz AC, Silva ML, Caon T, Simoes CM. Addition of bone morphogenetic protein type 2 to ascorbate and beta-glycerophosphate supplementation did not enhance osteogenic differentiation of human adipose-derived stem cells. J Appl Oral Sci. 2012; 20(6):628–635. 10.1590/S1678-77572012000600007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khanna-Jain R, Agata H, Vuorinen A, Sandor GK, Suuronen R, Miettinen S et al. Addition of BMP-2 or BMP-6 to dexamethasone, ascorbic acid, and beta-glycerophosphate may not enhance osteogenic differentiation of human periodontal ligament cells. Growth Factors. 2010; 28(6):437–446. 10.3109/08977194.2010.495719 [DOI] [PubMed] [Google Scholar]
- 52.Yamagiwa H, Endo N, Tokunaga K, Hayami T, Hatano H, Takahashi HE et al. In vivo bone-forming capacity of human bone marrow-derived stromal cells is stimulated by recombinant human bone morphogenetic protein-2. J Bone Miner Metab. 2001; 19(1):20–28. [DOI] [PubMed] [Google Scholar]
- 53.Chappuis V, Gamer L, Cox K, Lowery JW, Bosshardt DD, Rosen V et al. Periosteal BMP2 activity drives bone graft healing. Bone. 2012; 51(4):800–809. 10.1016/j.bone.2012.07.017 [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, Wang S, Bao J, Sun X, Zhang X, Zhang X et al. Trehalose maintains bioactivity and promotes sustained release of BMP-2 from lyophilized CDHA scaffolds for enhanced osteogenesis in vitro and in vivo. PLoS One. 2013; 8(1):e54645 10.1371/journal.pone.0054645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kisiel M, Ventura M, Oommen OP, George A, Walboomers XF, Hilborn J et al. Critical assessment of rhBMP-2 mediated bone induction: an in vitro and in vivo evaluation. J Control Release. 2012; 162(3):646–653. 10.1016/j.jconrel.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 56.Razzouk S, Sarkis R. BMP-2: biological challenges to its clinical use. N Y State Dent J. 2012; 78(5):37–39. [PubMed] [Google Scholar]
- 57.Zara JN, Siu RK, Zhang X, Shen J, Ngo R, Lee M et al. High doses of bone morphogenetic protein 2 induce structurally abnormal bone and inflammation in vivo. Tissue Eng Part A. 2011; 17(9–10):1389–1399. 10.1089/ten.TEA.2010.0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wegman F, van der Helm Y, Oner FC, Dhert W, Alblas J. BMP-2 plasmid DNA as a substitute for BMP-2 protein in bone tissue engineering. Tissue Eng Part A. 2013; 10.1089/ten.TEA.2012.0569 [DOI] [PubMed] [Google Scholar]
- 59.Açil Y, Ghoniem AA, Wiltfang J, Gierloff M. Optimizing the osteogenic differentiation of human mesenchymal stromal cells by the synergistic action of growth factors. J Craniomaxillofac Surg. 2014; 42(8):2002–2009. 10.1016/j.jcms.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 60.Prins HJ, Rozemuller H, Vonk-Griffioen S, Verweij VG, Dhert WJ, Slaper-Cortenbach IC et al. Bone-forming capacity of mesenchymal stromal cells when cultured in the presence of human platelet lysate as substitute for fetal bovine serum. Tissue Eng Part A. 2009; 15(12):3741–3751. 10.1089/ten.TEA.2008.0666 [DOI] [PubMed] [Google Scholar]
- 61.Chevallier N, Anagnostou F, Zilber S, Bodivit G, Maurin S, Barrault A et al. Osteoblastic differentiation of human mesenchymal stem cells with platelet lysate. Biomaterials. 2010; 31(2):270–278. 10.1016/j.biomaterials.2009.09.043 [DOI] [PubMed] [Google Scholar]
- 62.O’Regan AW, Hayden JM, Berman JS. Osteopontin augments CD3-mediated interferon-gamma and CD40 ligand expression by T cells, which results in IL-12 production from peripheral blood mononuclear cells. J Leukoc Biol. 2000; 68(4):495–502. [PubMed] [Google Scholar]
- 63.Mizuno D, Agata H, Furue H, Kimura A, Narita Y, Watanabe N et al. Limited but heterogeneous osteogenic response of human bone marrow mesenchymal stem cells to bone morphogenetic protein-2 and serum. Growth Factors. 2010; 28(1):34–43. 10.3109/08977190903326362 [DOI] [PubMed] [Google Scholar]
- 64.Mentink A, Hulsman M, Groen N, Licht R, Dechering KJ, van der Stok J et al. Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials. 2013; 10.1016/j.biomaterials.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 65.Del Monaco M, Covello SP, Kennedy SH, Gilinger G, Litwack G, Uitto J et al. Identification of novel glucocorticoid-response elements in human elastin promoter and demonstration of nucleotide sequence specificity of the receptor binding. J Invest Dermatol. 1997; 108(6):938–942. 10.1111/1523-1747.ep12295241 [DOI] [PubMed] [Google Scholar]
- 66.Rich CB, Fontanilla MR, Nugent M, Foster JA. Basic fibroblast growth factor decreases elastin gene transcription through an AP1/cAMP-response element hybrid site in the distal promoter. J Biol Chem. 1999; 274(47):33433–33439. 10.1074/jbc.274.47.33433 [DOI] [PubMed] [Google Scholar]
- 67.Fekete N, Gadelorge M, Furst D, Maurer C, Dausend J, Fleury-Cappellesso S et al. Platelet lysate from whole blood-derived pooled platelet concentrates and apheresis-derived platelet concentrates for the isolation and expansion of human bone marrow mesenchymal stromal cells: production process, content and identification of active components. Cytotherapy. 2012; 14(5):540–554. 10.3109/14653249.2012.655420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdallah BM, Haack-Sorensen M, Burns JS, Elsnab B, Jakob F, Hokland P et al. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite [corrected] extensive proliferation. Biochem Biophys Res Commun. 2005; 326(3):527–538. 10.1016/j.bbrc.2004.11.059 [DOI] [PubMed] [Google Scholar]
- 69.Xia W, Li H, Wang Z, Xu R, Fu Y, Zhang X et al. Human platelet lysate supports ex vivo expansion and enhances osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Cell Biol Int. 2011; 35(6):639–643. 10.1042/CBI20100361 [DOI] [PubMed] [Google Scholar]
- 70.Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem. 1997; 64(2):295–312. [DOI] [PubMed] [Google Scholar]
- 71.Niikura N, Iwamoto T, Masuda S, Kumaki N, Xiaoyan T, Shirane M et al. Immunohistochemical Ki67 labeling index has similar proliferation predictive power to various gene signatures in breast cancer. Cancer Sci. 2012; 103(8):1508–1512. 10.1111/j.1349-7006.2012.02319.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathews S, Bhonde R, Gupta PK, Totey S. Extracellular matrix protein mediated regulation of the osteoblast differentiation of bone marrow derived human mesenchymal stem cells. Differentiation. 2012; 84(2):185–192. 10.1016/j.diff.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 73.Quivy V, Calomme C, Dekoninck A, Demonte D, Bex F, Lamsoul I et al. Gene activation and gene silencing: a subtle equilibrium. Cloning Stem Cells. 2004; 6(2):140–149. 10.1089/1536230041372454 [DOI] [PubMed] [Google Scholar]
- 74.Achille V, Mantelli M, Arrigo G, Novara F, Avanzini MA, Bernardo ME et al. Cell-cycle phases and genetic profile of bone marrow-derived mesenchymal stromal cells expanded in vitro from healthy donors. J Cell Biochem. 2011; 112(7):1817–1821. 10.1002/jcb.23100 [DOI] [PubMed] [Google Scholar]
- 75.Russell KC, Lacey MR, Gilliam JK, Tucker HA, Phinney DG, O’Connor KC et al. Clonal analysis of the proliferation potential of human bone marrow mesenchymal stem cells as a function of potency. Biotechnol Bioeng. 2011; 108(11):2716–2726. 10.1002/bit.23193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuznetsov SA, Krebsbach PH, Satomura K, Kerr J, Riminucci M, Benayahu D et al. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J Bone Miner Res. 1997; 12(9):1335–1347. 10.1359/jbmr.1997.12.9.1335 [DOI] [PubMed] [Google Scholar]
- 77.Mankani MH, Kuznetsov SA, Avila NA, Kingman A, Robey PG. Bone formation in transplants of human bone marrow stromal cells and hydroxyapatite-tricalcium phosphate: prediction with quantitative CT in mice. Radiology. 2004; 230(2):369–376. 10.1148/radiol.2302011529 [DOI] [PubMed] [Google Scholar]
- 78.Narisawa S, Yadav MC, Millan JL. In vivo overexpression of tissue-nonspecific alkaline phosphatase increases skeletal mineralization and affects the phosphorylation status of osteopontin. J Bone Miner Res. 2013; 10.1002/jbmr.1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang ZL, Zhang H, Ke YH, Yue H, Xiao WJ, Yu JB et al. The identification of novel mutations in COL1A1, COL1A2, and LEPRE1 genes in Chinese patients with osteogenesis imperfecta. J Bone Miner Metab. 2011; 10.1007/s00774-011-0284-6 [DOI] [PubMed] [Google Scholar]
- 80.Turner RT, Spelsberg TC. Correlation between mRNA levels for bone cell proteins and bone formation in long bones of maturing rats. Am J Physiol. 1991; 261(3 Pt 1):E348–53. [DOI] [PubMed] [Google Scholar]
- 81.Mochida Y, Parisuthiman D, Pornprasertsuk-Damrongsri S, Atsawasuwan P, Sricholpech M, Boskey AL et al. Decorin modulates collagen matrix assembly and mineralization. Matrix Biol. 2009; 28(1):44–52. 10.1016/j.matbio.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simionescu A, Simionescu DT, Vyavahare NR. Osteogenic responses in fibroblasts activated by elastin degradation products and transforming growth factor-beta1: role of myofibroblasts in vascular calcification. Am J Pathol. 2007; 171(1):116–123. 10.2353/ajpath.2007.060930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010; 339(1):189–195. 10.1007/s00441-009-0832-8 [DOI] [PubMed] [Google Scholar]
- 84.Prins HJ, Braat AK, Gawlitta D, Dhert WJ, Egan DA, Tijssen-Slump E et al. In vitro induction of alkaline phosphatase levels predicts in vivo bone forming capacity of human bone marrow stromal cells. Stem Cell Res. 2014; 12(2):428–440. 10.1016/j.scr.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 85.Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012; 8(2):272–288. 10.7150/ijbs.2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tachi K, Takami M, Sato H, Mochizuki A, Zhao B, Miyamoto Y et al. Enhancement of bone morphogenetic protein-2-induced ectopic bone formation by transforming growth factor-beta1. Tissue Eng Part A. 2011; 17(5–6):597–606. 10.1089/ten.TEA.2010.0094 [DOI] [PubMed] [Google Scholar]
- 87.Quarto N, Li S, Renda A, Longaker MT. Exogenous activation of BMP-2 signaling overcomes TGFbeta-mediated inhibition of osteogenesis in Marfan embryonic stem cells and Marfan patient-specific induced pluripotent stem cells. Stem Cells. 2012; 30(12):2709–2719. 10.1002/stem.1250 [DOI] [PubMed] [Google Scholar]
- 88.Volk SW, Shah SR, Cohen AJ, Wang Y, Brisson BK, Vogel LK et al. Type III collagen regulates osteoblastogenesis and the quantity of trabecular bone. Calcif Tissue Int. 2014; 94(6):621–631. 10.1007/s00223-014-9843-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sibiya SJ, Olivier EI, Duneas N. High yield isolation of BMP-2 from bone and in vivo activity of a combination of BMP-2/TGF-beta1. J Biomed Mater Res A. 2013; 101(3):641–646. 10.1002/jbm.a.34365 [DOI] [PubMed] [Google Scholar]
- 90.Bi Y, Stuelten CH, Kilts T, Wadhwa S, Iozzo RV, Robey PG et al. Extracellular matrix proteoglycans control the fate of bone marrow stromal cells. J Biol Chem. 2005; 280(34):30481–30489. 10.1074/jbc.M500573200 [DOI] [PubMed] [Google Scholar]
- 91.Edwards JR, Nyman JS, Lwin ST, Moore MM, Esparza J, O’Quinn EC et al. Inhibition of TGF-beta signaling by 1D11 antibody treatment increases bone mass and quality in vivo. J Bone Miner Res. 2010; 25(11):2419–2426. 10.1002/jbmr.139 [DOI] [PubMed] [Google Scholar]
- 92.Grafe I, Yang T, Alexander S, Homan EP, Lietman C, Jiang MM et al. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat Med. 2014; 20(6):670–675. 10.1038/nm.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crane JL, Cao X. Bone marrow mesenchymal stem cells and TGF-beta signaling in bone remodeling. J Clin Invest. 2014; 124(2):466–472. 10.1172/JCI70050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Linsley C, Wu B, Tawil B. The Effect of Fibrinogen, Collagen Type I, and Fibronectin on Mesenchymal Stem Cell Growth and Differentiation into Osteoblasts. Tissue Eng Part A. 2013; 10.1089/ten.TEA.2012.0523 [DOI] [PubMed] [Google Scholar]
- 95.De Bari C, Dell’Accio F, Karystinou A, Guillot PV, Fisk NM, Jones EA et al. A biomarker-based mathematical model to predict bone-forming potency of human synovial and periosteal mesenchymal stem cells. Arthritis Rheum. 2008; 58(1):240–250. 10.1002/art.23143 [DOI] [PubMed] [Google Scholar]
- 96.Manferdini C, Gabusi E, Grassi F, Piacentini A, Cattini L, Zini N et al. Evidence of specific characteristics and osteogenic potentiality in bone cells from tibia. J Cell Physiol. 2011; 226(10):2675–2682. 10.1002/jcp.22618 [DOI] [PubMed] [Google Scholar]
- 97.Karsenty G, Park RW. Regulation of type I collagen genes expression. Int Rev Immunol. 1995; 12(2–4):177–185. 10.3109/08830189509056711 [DOI] [PubMed] [Google Scholar]
- 98.Ramirez F, Tanaka S, Bou-Gharios G. Transcriptional regulation of the human alpha2(I) collagen gene (COL1A2), an informative model system to study fibrotic diseases. Matrix Biol. 2006; 25(6):365–372. 10.1016/j.matbio.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 99.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009; 19(1):71–88. 10.1038/cr.2008.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Y, Wang L, Kikuiri T, Akiyama K, Chen C, Xu X et al. Mesenchymal stem cell-based tissue regeneration is governed by recipient T lymphocytes via IFN-gamma and TNF-alpha. Nat Med. 2011; 17(12):1594–1601. 10.1038/nm.2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duque G, Huang DC, Dion N, Macoritto M, Rivas D, Li W et al. Interferon-gamma plays a role in bone formation in vivo and rescues osteoporosis in ovariectomized mice. J Bone Miner Res. 2011; 26(7):1472–1483. 10.1002/jbmr.350 [DOI] [PubMed] [Google Scholar]
- 102.Gothard D, Dawson JI, Oreffo RO. Assessing the potential of colony morphology for dissecting the CFU-F population from human bone marrow stromal cells. Cell Tissue Res. 2013; 10.1007/s00441-013-1564-3 [DOI] [PubMed] [Google Scholar]
- 103.Seiler C, Gazdhar A, Reyes M, Benneker LM, Geiser T, Siebenrock KA et al. Time-lapse microscopy and classification of 2D human mesenchymal stem cells based on cell shape picks up myogenic from osteogenic and adipogenic differentiation. J Tissue Eng Regen Med. 2012; 10.1002/term.1575 [DOI] [PubMed] [Google Scholar]
- 104.Matsuoka F, Takeuchi I, Agata H, Kagami H, Shiono H, Kiyota Y et al. Morphology-based prediction of osteogenic differentiation potential of human mesenchymal stem cells. PLoS One. 2013; 8(2):e55082 10.1371/journal.pone.0055082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pietila M, Lehtonen S, Narhi M, Hassinen IE, Leskela HV, Aranko K et al. Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng Part C Methods. 2010; 16(3):435–445. 10.1089/ten.tec.2009.0247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.