Abstract
Viral infections induce the differentiation of naïve CD4 T cells into two distinct lineages, Th1 and TFH cells. Two recent studies demonstrated that the microRNA cluster miR-17-92 selectively promotes CD4 TFH responses. However, here we show that miR-17-92 expression is required for the clonal expansion of both virus-specific Th1 and TFH cells. Upon viral infection, miR-17-92-deficient CD4 T cells showed impaired clonal expansion and subsequent memory formation. While miR-17-92 deficiency impaired the clonal expansion of both Th1 and TFH cells, the expansion of Th1 cells was more affected. Over-expression of miR-17-92 in CD4 T cells resulted in increased expansion of both virus-specific Th1 and TFH cells, but selectively enhanced the Th1 response. Taken together, our data suggest that miR-17-92 is necessary for both Th1 and TFH cells to respond efficiently to viral infections and that the Th1 response is more sensitive to the level of miR-17-92 expression.
Introduction
Effector CD4 T cells regulate immune responses through communicating with other immune cells via cytokines and direct engagement of cell surface molecules. Upon viral infection, naïve antigen-specific CD4 T cells differentiate into two subsets of effector CD4 T cells, T helper 1 (Th1) cells and T follicular helper (TFH) cells (1). Th1 cells contribute to the control of viral infections by producing cytokines such as IFN-γ and TNF-α (2). TFH cells, characterized by their expression of CXCR5, are necessary for the initiation and maintenance of the germinal center (GC) reaction, and therefore are crucial for antibody affinity maturation and the generation of memory B cells and long-lived plasma cells (3). Bcl-6, a transcriptional repressor, promotes the differentiation of TFH cell cells, while Blimp-1 antagonizes Bcl-6 and enforces the differentiation into non-TFH cells (4–6). After antigen clearance, the majority of effector CD4 T cells undergo apoptosis, while a fraction survives and differentiates into memory cells maintaining their commitment to the Th1 or TFH lineages (1, 7, 8).
MicroRNAs (miRNAs) are a family of small regulatory RNAs of ~22 nucleotides, which bind to the 3’UTR of target transcripts and suppress gene expression by blocking translation and/or degrading target transcripts (9, 10). Our group has previously shown a critical role of the miR-17-92 cluster in regulating virus-specific CD8 T cell differentiation (11). Two recent publications have shown that miR-17-92 is also necessary for TFH cell differentiation and the humoral immune response (12, 13). However, our current study shows that miR-17-92 positively regulates the expansion of both Th1 and TFH cells during viral infection. The impaired humoral response observed in miR-17-92 conditional knockout mice is caused by the defective CD4 T cell expansion rather than a selective defect in the differentiation of TFH cells. In fact, our data show that Th1 cells are more dependent on the expression of miR-17-92 for their clonal expansion as well as the production of effector cytokines than TFH cells.
Materials and methods
Mice and infection
SMARTA mice expressing a transgenic TCR specific for the GP61-80 epitope of lymphocytic choriomeningitis virus (LCMV) were backcrossed to B6.CD45.1+ strain (Jackson Laboratory) (14). C57BL/6 (CD45.1-) mice purchased from Jackson Laboratory were used as recipients of SMARTA CD4 T cells. miR-17-92 conditional knockout and miR-17-92 conditional over-expression mice were both purchased from Jackson Laboratory and were bred to a CD4-cre transgenic strain (Taconic) (15–17). For infection, mice were injected i.p. with 2×105 PFU of LCMV Armstrong. Animal experiments were conducted in accordance with Emory University IACUC protocols.
In vitro T cell proliferation assay
Purified CD4 T cells were labeled with CFSE, stimulated with plate-bound anti-CD3 and soluble anti-CD28 antibodies (BD Biosciences) for 48 hours, and cell division was determined by flow cytometry analysis of CFSE dilution.
Flow cytometry
I-AbGP66-77 specific CD4 T cells were labeled by I-AbGP66-77 tetramers (NIH Tetramer Core Facility) as previously described (1). A three-step CXCR5 staining was performed as previously described (4). Phosphorylated-ribosomal protein S6 was detected using anti-phospho-S6 antibody (Cell Signaling Technology) (11).
ELISA and ELISPOT assays
ELISA and ELISPOT assays to detect LCMV specific antibodies and antibody-secreting cells respectively were performed as previously (18).
Retroviral transduction
MSCV-PGK-GFP plasmid with miR-17-92 insert has been previously described (11). Activated SMARTA CD4 T cells were purified, and infected by retroviruses with or without miR-17-92 insert. Cells were cultured with 10ng/ml IL-2, and GFP+ cells were sorted 2 days later. 2×104 GFP+ cells were transferred into each recipient.
Statistical analysis
All data analysis was performed using GraphPad Prism v5. P values were determined by a two-tailed, unpaired Student’s t test and considered significant if p<0.05. *p<0.05, ** p<0.01, ***p<0.001.
Results and Discussion
miR-17-92 is critical for proliferation of virus-specific CD4 T cells
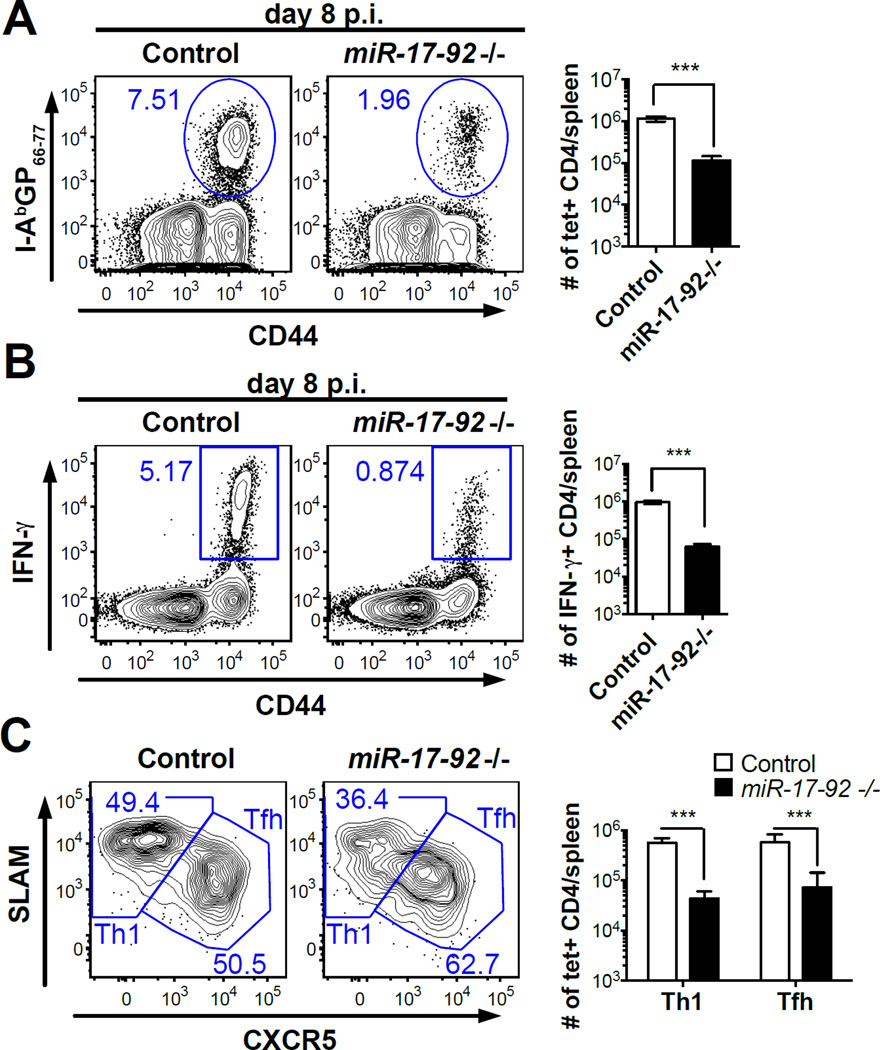
To study the role of miR-17-92 in the CD4 T cell response to viral infections, we bred miR-17-92 loxP/loxP mice to a CD4-cre transgenic strain, resulting in a T-cell specific miR-17-92 deficiency (17). miR-17-92−/− (miR-17-92 loxP/loxP; CD4-cre) mice and littermate controls (miR-17-92 loxP/loxP) were infected with LCMV Armstrong. On day 8 post-infection (p.i.), there was a ~10 fold decrease in the number of I-AbGP66-77 tetramer+ CD4 T cells as well as a reduction in the total CD44high effector CD4 T cell population in miR-17-92−/− mice compared to littermate controls (Fig. 1A). Moreover, CFSE labeled miR-17-92−/− CD4 T cells divided less than their WT counterparts after in vitro stimulation with anti-CD3 and anti-CD28 antibodies (Supplemental Fig. 1A). Therefore, the diminished clonal expansion of virus-specific miR-17-92−/− effector cells was likely caused by the reduced proliferative capacity of these cells after antigenic stimulation.
Figure 1. miR-17-92 is critical to the CD4 T cell response during LCMV infection but is not required for the TFH cell differentiation.
Splenocytes of miR-17-92−/− mice and littermate controls were collected on day 8 p.i. (A) FACS plots of I-AbGP66-77 tetramer staining (gated on CD4 T cells) and number of I-AbGP66-77 tetramer+ CD4 T cells per spleen. (B) FACS plots of intracellular IFN-γ staining (gated on CD4 T cells) and number of IFN-γ+ CD4 T cells per spleen. (C) FACS analysis of CXCR5 and SLAM expression on I-AbGP66-77 tetramer+ CD4 T cells. (D) Number of Th1 or TFH I-AbGP66-77 tetramer+ CD4 T cells per spleen. Results are representative of at least three independent experiments with n≥3.
miR-17-92 is required for the generation of virus-specific Th1 as well as TFH cells
Like many antiviral CD4 T cell responses, the CD4 T cell response to LCMV infection exhibits a strong Th1 polarization and expansion of effector CD4 T cells producing the Th1 signature cytokine IFN-γ (2, 19). A recent study has suggested that the Th1 compartment and IFN-γ producing CD4 T cells are largely unaffected by the loss of miR-17-92 after LCMV infection (13). However, considering the substantial reduction in the number of LCMV-specific CD4 effectors in miR-17-92−/− mice, as indicated by both tetramer and CD44 staining, it is highly unlikely that the number of Th1 cells remains unchanged. To examine whether miR-17-92 is necessary for the generation of LCMV-specific IFN-γ producing CD4 T cells, we re-stimulated splenocytes from miR-17-92−/− and control mice with GP61-80 peptide and quantified IFN-γ+ CD4 T cells on day 8 p.i.. Strikingly, miR-17-92−/− mice showed a ~16 fold decrease in the number of IFN-γ+ CD4 T cells as well as a reduction in the IFN-γ protein level among IFN-γ+ cells compared to littermate controls (Fig. 1B and Supplemental Fig. 1B). In fact, the decrease in IFN-γ+ CD4 T cells was larger than the decrease in tetramer+ cells, which suggests that the Th1 response is more susceptible to the loss of miR-17-92.
An important function of effector CD4 T cells is to support the germinal center (GC) reaction, which is mediated by CD4 TFH cells (3). To determine whether miR-17-92 regulates Th1/TFH cell differentiation, we analyzed the expression of the TFH marker CXCR5 and the Th1 marker SLAM on LCMV-specific CD4 T cells (4). On day 8 p.i., the frequency of TFH (CXCR5+SLAMint) cells within I-AbGP66-77 tetramer+ CD4 T cells in miR-17-92−/− mice was not reduced compared to control mice (Fig. 1C). We quantified I-AbGP66-77 tetramer+ Th1 and TFH cells and found that both populations were severely reduced in the knockout mice (Fig. 1D). The reduced numbers of TFH cells in miR-17-92−/− mice coincided with reduced numbers of GC B cells and anti-LCMV IgG secreting cells (Supplemental Fig. 1D). Taken together, our results suggest that the loss of miR-17-92, which is up-regulated upon T cell activation in virus-specific Th1 and TFH cells, causes a general defect in the expansion of virus-specific CD4 T cells but does not specifically impact the development of TFH cells (Supplemental Fig. 1H).
miR-17-92 deficiency reduces the number of virus-specific memory CD4 T cells
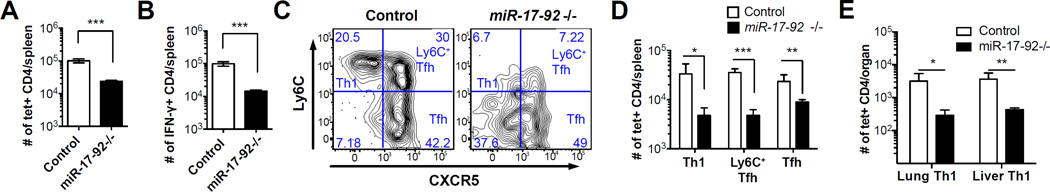
To investigate how miR-17-92 deficiency impacts CD4 memory differentiation, we analyzed knockout and control mice on day 108 p.i.. There were ~4 fold fewer I-AbGP66-77 tetramer+ CD4 T cells and ~7 fold fewer IFN-γ+ CD4 T cells after GP61-80 peptide stimulation in the knockout mice than in the littermate controls (Fig. 2A, 2B). Consistent with our results at the effector time points, the more profound loss of IFN-γ+ cells in miR-17-92−/− mice confirms that the generation of IFN-γ+ Th1 cells is more dependent on miR-17-92. Moreover, production of IFN-γ was significantly reduced in miR-17-92−/− memory CD4 T cells, suggesting that miR-17-92 is important for the functionality of virus-specific CD4 T cells (Supplemental Fig. 1C). We have recently shown that LCMV-specific memory CD4 T cells can be segregated into three populations: CXCR5-Ly6C+ memory Th1 cells, CXCR5+Ly6C+ memory TFH cells and CXCR5+Ly6C- memory TFH cells (1). Both Ly6C+ memory CD4 T cell subsets were strikingly reduced in miR-17-92−/− mice compared to littermate controls (Fig. 2C, 2D). In addition, miR-17-92−/− mice had significantly lower numbers of I-AbGP66-77 tetramer+ memory CD4 T cells in liver and lung (Fig. 2E), which are primarily occupied by Th1 rather than TFH memory cells (1). Therefore, miR-17-92−/− mice not only mounted compromised effector CD4 T cell responses but also showed significant defects in the number of virus-specific memory CD4 T cells.
Figure 2. miR-17-92 deficiency reduces the number of virus-specific memory CD4 T cells generated after the primary immune response.
Splenocytes of miR-17-92−/− mice and littermate controls were collected on day 108 p.i. (A) Number of I-AbGP66-77 tetramer+ CD4 T cells and (B) number of IFN-γ+ CD4 T cells per spleen. (C,D) FACS plots of CXCR5 and Ly6C and number of I-AbGP66-77 specific Th1, Ly6C+ TFH, and TFH memory CD4 T cells. (E) Number of I-AbGP66-77 tetramer+ CD4 T cells in liver and lung on day 244 p.i.. Results are representative of at least three independent experiments with n≥3.
miR-17-92 over-expression selectively enhances the Th1 response
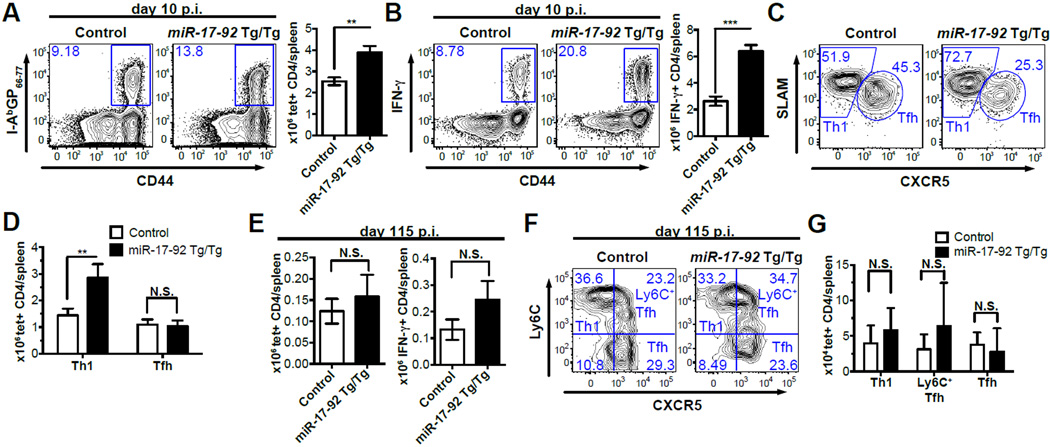
We next sought to determine whether increasing miR-17-92 expression can enhance CD4 T cell responses and generate more IFN-γ+ effectors. We bred a previously described conditional miR-17-92 over-expression strain to CD4-cre transgenic mice, resulting in a T-cell specific over-expression of miR-17-92 (15). Cre+ mice bearing two copies of the miR-17-92 transgene (Tg) are designated as miR-17-92 Tg/Tg, and their cre- littermates were used as controls. TG/TG mice and their littermate controls were infected with LCMV Arm and analyzed on day 10 p.i.. We found that miR-17-92 Tg/Tg mice had almost twice as many I-AbGP66-77 tetramer+ CD4 T cells as control mice, indicating an enhanced CD4 T cell response (Fig. 3A). Moreover, there were ~3 fold more IFN-γ+ CD4 T cells and more IFN-γ expression on a per cell basis in miR-17-92 Tg/Tg mice than in control mice on day 10 p.i. (Fig. 3B and Supplemental Fig. 1E). Therefore, these results confirm that miR-17-92 positively regulates the antiviral immune response mediated by CD4 T cells and exerts a stronger effect on IFN-γ+ CD4 T cells. The frequency of CXCR5+SLAMint TFH cells was almost reduced by half in miR-17-92 Tg/Tg tetramer+ CD4 T cells relative to their WT counterparts (Fig. 3C). Accordingly, the frequencies of CXCR5-SLAMhigh Th1 cells increased upon over-expression of miR-17-92. However, the decrease in TFH cell frequency was due to an increased Th1 response rather than a decreased TFH response (Fig. 3D). Accordingly, the number of GC B cells in Tg/Tg mice was largely unaffected (Supplemental Fig. 1G). Thus, miR-17-92 over-expression enhances the generation of IFN-γ+ CD4 T cells and promotes the expansion of Th1 cells.
Figure 3. miR-17-92 over-expression preferentially enhances the Th1 effector response.
(A-D) Splenocytes from miR-17-92 Tg/Tg and control mice were collected on day 10 p.i.. (A) FACS plots of I-AbGP66-77 tetramer staining (gated on CD4 T cells) and number of I-AbGP66-77 tetramer+ CD4 T cells per spleen. (B) FACS plots of intracellular IFN-γ staining (gated on CD4 T cells), the number of IFN-γ+ CD4 T cells per spleen, and IFN-γ MFI. (C) FACS plots of CXCR5 and SLAM staining (gated on I-AbGP66-77 tetramer+ CD4 T cells). (D) Number of I-AbGP66-77 tetramer+ Th1 and TFH CD4 T cells per spleen. (E-G) Splenocytes of miR-17-92 Tg/Tg and control mice were collected on day 115 p.i.. (E) Number of I-AbGP66-77 tetramer+ CD4 T cells and IFN-γ+ CD4 T cells per spleen. (F,G) FACS plots of CXCR5 and Ly6C and number of I-AbGP66-77 specific Th1, Ly6C+ TFH, and TFH memory CD4 T cells. Results are representative of at least three independent experiments with n≥3.
We next sought to determine whether excessive miR-17-92 expression disrupts memory CD4 T cell differentiation. On day 115 p.i., Tg/Tg mice showed comparable or even slightly more I-AbGP66-77 tetramer+ CD4 T cells as well as IFN-γ+ CD4 T than their WT counterparts, indicating that miR-17-92 over-expression does not compromise the generation of memory CD4 T cells (Fig. 3E, Supplemental Fig. 1F). Consistent with our observation that miR-17-92 deficiency causes a reduction in the Ly6C+ memory CD4 T cells, miR-17-92 over-expression seems to favor the formation of Ly6C+ memory CD4 T cell subsets (Fig. 3F,3G).
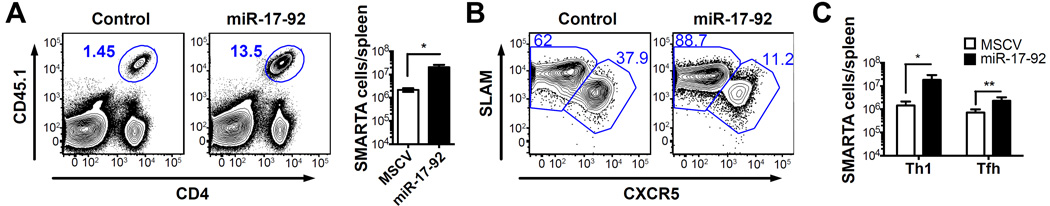
To confirm that the effect of miR-17-92 over-expression on effector CD4 T cell differentiation and expansion is cell autonomous, we adoptively transferred SMARTA CD4 T cells retrovirally transduced with a MSCV construct with or without a miR-17-92 insert into WT recipients and infected the chimeras with LCMV (Supplemental Fig. 2A). On day 8 p.i., SMARTA cells over-expressing miR-17-92 expanded significantly more and produced more IFN-γ than those transduced with the empty MSCV (Fig. 4A and Supplemental Fig. 2B), which supports that miR-17-92 enhances Th1 differentiation in a CD4 T cell intrinsic manner. While the expansion of both subsets increased after miR-17-92 over-expression, the extent was much greater in Th1 cells than in TFH cells (~12 fold vs ~3 fold) (Fig. 4B,4C). Comparable to our previous results in LCMV-specific CD8 T cells, miR-17-92 over-expressing SMARTA cells contained more phosphorylated ribosomal protein S6 (Supplemental Fig. 2C), suggesting elevated mTOR signaling (11).
Figure 4. The effect of miR-17-92 over-expression on LCMV-specific CD4 T cell expansion and differentiation is cell autonomous.
SMARTA cells were transduced with MSCV construct with or without miR-17-92 insert and transferred to C57BL/6 recipients. On day 8 p.i., splenocytes were collected for analysis. (A) FACS plots of SMARTA cells (gated on total lymphocytes) and number of SMARTA cells per spleen. (B) FACS plots of CXCR5 and SLAM staining (gated on SMARTA cells). (C) Number of Th1 and TFH SMARTA cells per spleen. Results are representative of at least three independent experiments with n≥3.
In summary, by employing both loss-of-function and gain-of-function approaches, we have demonstrated an essential role of miR-17-92 in regulating the CD4 T cell response to viral infection. miR-17-92 is critical for the clonal expansion of CD4 T cells during acute LCMV infection. Moreover, the Th1 polarization and secretion of effector cytokine IFN-γ also requires miR-17-92 expression. While both Th1 and TFH responses are positively regulated by miR-17-92, the magnitude of Th1 expansion is more sensitive to the level of miR-17-92 expression.
Our results show that miR-17-92 enhances mTOR signaling and is required for antiviral Th1 and TFH responses. It is likely that miR-17-92 suppresses the expression of PTEN (Phosphatase and tensin homolog), a negative regulator of the PI3K-mTOR signaling pathway and well-documented target of miR-17-92, in both subsets to achieve optimal PI3K-mTOR signaling for their clonal expansion (15). Over-expression of miR-17-92 in Tg/Tg mice increased the Th1 response without affecting the TFH response, while retroviral over-expression of miR-17-92 enhanced the Th1 response and, to a lesser extent, the TFH response. We cannot exclude that excessive expression of miR-17-92 driven by the retroviral construct might cause potential off-target effects, but given that the retroviral LTR promoter drove a more robust expression of miR-17-92 than the CAG promoter in the Tg/Tg mice, our data suggest that while miR-17-92 positively regulates both Th1 and TFH cell expansion, Th1 cells are more sensitive to the dose of miR-17-92. The higher sensitivity of Th1 expansion could be due to lineage-specific differences in the expression of miR-17-92 targets.
Two recent studies have concluded that miR-17-92 selectively regulates TFH cell differentiation after LCMV infection (12, 13). In contrast, our results demonstrate that miR-17-92 is required for both Th1 and TFH cells to respond effectively to viral infections. These studies identified effector TFH cells by staining total CD4 T cells for CXCR5 and PD-1. While this staining strategy can be used as a surrogate to quantitate antigen-specific TFH effector cells, it does not provide any information regarding the number of Th1 effector cells. It is difficult to judge whether miR-17-92 deficiency selectively affects TFH cell differentiation without comparing the numbers of antigen-specific TFH and Th1 cells between miR-17-92 knockout and control mice. We used MHC-II tetramer staining to identify antigen-specific CD4 T cells and distinguished TFH and Th1 lineages within the antigen-specific CD4 T cells based on the differential expression of various surface markers such as CXCR5 and SLAM. This approach demonstrates that the numbers of virus-specific TFH and Th1 cells are both substantially reduced in miR-17-92 knockout mice after infection. Moreover, given that the decrease in the GC response was comparable to, if not less than, the decrease in the effector CD4 T cell response (Fig. 1A,1F), a compromised B cell helper function of miR-17-92−/− CD4 T cells seems implausible. Our results also show a significant reduction of IFN-γ producing CD4 T cells in the absence of miR-17-92, which is in contrast to a recent study (13). This discrepancy might be due to different promoters controlling Cre expression. In our study naïve CD4 T cells already lacked miR-17-92 (CD4-cre), whereas in the other study miR-17-92 deficiency was only achieved about three days after T cell activation (OX40-cre). These results suggest that miR-17-92 may function at different stages of the Th1 and TFH response: miR-17-92 might be required in Th1 cells during initial activation, while it might be required in TFH cells after initial activation. Our study thus underlines the importance of miR-17-92 to the antiviral immune response of both CD4 Th1 and TFH cells. A better understanding of the regulatory pathways involved in CD4 T cell differentiation will provide the framework for the design of rational vaccination strategies.
Supplementary Material
Footnotes
This work was supported by the National Institutes of Health (Grant R01-AI030048 to R.A.), and by the Cancer Research Insitute’s Irvington Institute Fellowship Program (to J.-H.H.).
References
- 1.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nature reviews. Immunology. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 4.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Stockinger B, Bourgeois C, Kassiotis G. CD4+ memory T cells: functional differentiation and homeostasis. Immunol Rev. 2006;211:39–48. doi: 10.1111/j.0105-2896.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- 8.Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Wu T, Wieland A, Araki K, Davis CW, Ye L, Hale JS, Ahmed R. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9965–9970. doi: 10.1073/pnas.1207327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nature immunology. 2013;14:840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, Fremgen D, Verdin E, Oldstone MB, Qi H, Teijaro JR, Xiao C. MicroRNAs of the miR-17 approximately 92 family are critical regulators of T(FH) differentiation. Nature immunology. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 15.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature immunology. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 18.Rasheed MA, Latner DR, Aubert RD, Gourley T, Spolski R, Davis CW, Langley WA, Ha SJ, Ye L, Sarkar S, Kalia V, Konieczny BT, Leonard WJ, Ahmed R. Interleukin-21 is a critical cytokine for the generation of virus-specific long-lived plasma cells. Journal of virology. 2013;87:7737–7746. doi: 10.1128/JVI.00063-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varga SM, Welsh RM. High frequency of virus-specific interleukin-2-producing CD4(+) T cells and Th1 dominance during lymphocytic choriomeningitis virus infection. Journal of virology. 2000;74:4429–4432. doi: 10.1128/jvi.74.9.4429-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.