Abstract
Telomere sequences at the end of chromosomes control somatic cell division; therefore, telomere length in a given cell population provides information about its replication potential. This unit describes a method for flow cytometric measurement of telomere length in subpopulations using fluorescence in situ hybridization of fluorescently-labeled probes (Flow-FISH) without prior cell separation. After cells are stained for surface immunofluorescence, antigen-antibody complexes are covalently cross-linked onto cell membranes before FISH with a telomere-specific probe. Cells with long telomeres are included as internal standards. Addition of a DNA dye permits exclusion of proliferating cells during data analysis. DNA ploidy measurements of cells of interest and internal standard are performed on separate aliquots in parallel to Flow-FISH. Telomere fluorescence of G0/1 cells of subpopulations and internal standards obtained from Flow-FISH are normalized for DNA ploidy and telomere length in subsets of interest is expressed as a fraction of the internal standard telomere length.
Keywords: fluorescence in situ hybridization, Flow-FISH, telomeres, telomere length, cell surface immunofluorescence
Introduction
Telomeres are highly conserved specialized structures at the ends of eukaryotic chromosomes. Human telomeres consist of hundreds to thousands of (T2AG3) repeats and associated telomere specific proteins, with important functions for chromosomal stability and replication (Moyzis, Buckingham et al. 1988). The length of telomere repeats varies between chromosomes and between species (Lansdorp, Verwoerd et al. 1996). In most human cells, the length of telomere repeats is remarkably heterogeneous ranging from a few to 20 kilo base pairs (kb) depending on the age of the donor, the type of tissue and the replicative history of the cells (Lansdorp, Verwoerd et al. 1996, Martens, Zijlmans et al. 1998). During cell division, cellular DNA is duplicated by the action of DNA polymerase; however, the polymerase does not completely replicate the 3′ ends of chromosomes in the so-called telomeric region (Harley, Futcher et al. 1990, Hastie, Dempster et al. 1990). As a consequence, telomeres get shortened during each cell division. Accelerated telomere shortening contributes to genetic instability of cells, replicative cellular senescence (Harley, Futcher et al. 1990, Hastie, Dempster et al. 1990), cell cycle arrest or apoptosis (Allsopp, Vaziri et al. 1992) and neoplastic transformation in vitro (Harley, Vaziri et al. 1992) and in vivo (Blasco, Lee et al. 1997). Thus, telomeres are involved in the pathogenesis and clinical progression of a variety of disorders, including congenital diseases (Vulliamy, Marrone et al. 2001), acquired disorders of the lympho-hematopoietic system (Brummendorf and Balabanov 2006), and age-related diseases (Valdes, Andrew et al. 2005). In view of the role of telomere length in biological functions ranging from aging to carcinogenesis (Stewart and Weinberg 2006), there is a need for techniques that can measure telomere length as a surrogate marker for both the proliferative history and replicative reserve of normal somatic cells.
Methods used to determine telomere length
Various methods exist for measuring telomere length. The most common are Southern blot analysis, PCR methods for measurements of average (Cawthon, Smith et al. 2003) and chromosome-specific telomere length (Baird, Rowson et al. 2003), fluorescence microscopy using directly labeled (CCCTAA)3 peptide nucleic acid (PNA) probes (Egholm, Buchardt et al. 1993) and Flow-FISH. The latter combines fluorescent in situ hybridization (FISH) with flow cytometry (Rufer, Dragowska et al. 1998). Although Southern blotting, which analyzes telomere restriction fragments (TRF), represents the widely accepted standard method for telomere length measurements, the Flow-FISH methodology, especially multicolor Flow-FISH which measures telomere length and surface markers, recently became increasingly important for several reasons: First, compared to Southern blotting and conventional Flow-FISH, magnetic bead separation of phenotypically-defined subpopulations is not required; this aspect is most important for the analysis of rare cell populations. Second, in contrast to Southern blotting, Flow-FISH does not measure sub-telomeric DNA, which overestimates the average telomere length by several kb (de Lange, Shiue et al. 1990). Furthermore, the inclusion of internal control cells in each individual tube (Baerlocher, Mak et al. 2002, Baerlocher and Lansdorp 2004) allows for sufficient correction of intra- and inter-experimental variability in hybridization efficiencies between samples. Multicolor Flow-FISH needs substantially fewer cells (in the order of 105 cells) than Southern Blotting and is less time-consuming. Finally, Flow-FISH is the only rapid way to measure telomeres in a population of cells marked by a surface-specific antigen (Rufer, Dragowska et al. 1998, Schmid, Dagarag et al. 2002). While hybridized PNA probe telomere length analysis by imaging and image cytometry tends to be more accurate than flow (due to the better control of cell orientation and quantitation of FISH spots in an imaging system (Egholm, Buchardt et al. 1993), Flow-FISH has the advantage of far faster throughput, allowing rapid analysis of thousands of cells and some amelioration of fluorescent measurement errors (Kapoor, Hakim et al. 2009). Automated multicolor flow FISH is currently the fastest and most sensitive method available to measure the average or median telomere length in granulocytes, naive T cells, memory T cells, B cells and natural killer (NK) cells in human blood (Baerlocher and Lansdorp 2003, Baerlocher and Lansdorp 2004). The introduction of new lasers and fluorophores, as well as the increasing sensitivity of instruments, have opened up new approaches and consequently have expanded the measurement parameters that can be applied to Flow-FISH. Thus, multicolor Flow-FISH allows the analysis of defined cell subpopulations in clinical scale sample sizes and might help to gain a deeper insight into the pathogenesis and disease progression of both immunological as well as hemato-oncological diseases.
Limitations and challenges with Flow-FISH
In recent years, various fluorescence in situ hybridization (FISH) protocols using telomere-specific peptide nucleic-acid (PNA) probes for the flow cytometric estimation of telomere length in individual cells (Flow-FISH) have been developed (Rufer, Dragowska et al. 1998, Baerlocher, Mak et al. 2002, Schmid, Dagarag et al. 2002, Baerlocher and Lansdorp 2003, Baerlocher, Vulto et al. 2006, Beier, Balabanov et al. 2007). DNA denaturation into single strands is achieved by heating a cell suspension to at least 80°C in a formamide-containing reaction mix and is followed by hybridization of the PNA telomere-specific probe to complementary DNA sequences (Rufer, Dragowska et al. 1998, Baerlocher, Mak et al. 2002, Schmid, Dagarag et al. 2002, Baerlocher and Lansdorp 2003). Utilization of flow cytometry’s greatest potential lies in simultaneous measurement of telomere length and cell surface antigen expression (Rufer, Dragowska et al. 1998, Baerlocher, Mak et al. 2002, Schmid, Dagarag et al. 2002, Baerlocher and Lansdorp 2003); however, development of this technique has been difficult owing to the harsh conditions required for DNA denaturation for probe hybridization which compromise the retention of the fluorescence associated with the surface-bound antibody molecules. Protocols which overcome these obstacles use heat-stable fluorochromes with suitable fluorescence emissions that are separate from the telomere fluorescence channel for antibody labeling. Antigen-antibody complexes are then cross-linked onto the cell membrane before cells undergo probe hybridization (Batliwalla, Damle et al. 2001) (Schmid, Dagarag et al. 2002). Compared with immunophenotyping of cells (in which absolute expression levels are seldom of interest), the Flow-FISH technique requires accurate, absolute measurements of relatively weak fluorescence signals. Flow-FISH measures single cells; however, it is not a valid method to identify single events with longer than the average telomere length in a population (Van Ziffle, Baerlocher et al. 2003). Reliable assessment of telomere length differences between cellular sub-populations can only be performed within populations of the same donor due to the large variation of telomere length among individuals, and a subpopulation, i.e. an identifiable peak shift in a histogram of telomere fluorescence of cells with a telomere length that is distinguishable from the major population is required (Van Ziffle, Baerlocher et al. 2003). The necessary calibrations and controls make the technique time consuming and technically demanding.
The current, newly-revised chapter describes the detailed protocol of the Flow-FISH method described in Schmid et al. (Schmid, Dagarag et al. 2002) for telomere length determination. The unit provides a primary procedure for analyzing immunophenotype, telomere length, and DNA content (see Basic Protocol), along with a supplementary method for determining DNA ploidy (see Support Protocol). Moreover, it contains detailed approaches for assay standardization, including controls to account for inter-individual telomere length variability, for experimental inconsistencies, and for instrument variations. The chapter includes extensive discussions and troubleshooting tips for the numerous critical parameters that can affect results and lists previously validated antibodies and fluorochromes and their potential combination into two different multicolor (up to 6 colors) Flow-FISH panels.
BASIC PROTOCOL: ANALYSIS OF IMMUNOPHENOTYPE, TELOMERE LENGTH, AND DNA CONTENT
This protocol describes a method for flow cytometric estimation of telomere length in cellular subsets. First, cells to be analyzed are stained for cell surface immunofluorescence. Then, antigen-antibody complexes are covalently cross-linked onto the cell surface using bis (sulfosuccinimidyl)suberate (BS3). Because Flow-FISH is a complex process that is sensitive to minor variations in experimental conditions, cells with long telomeres are added as an internal control to each reaction tube. Cell surface–labeled cells are mixed with unstained internal control cells that differ in telomere length from the cells of interest, resuspended in the reaction mix containing telomere-specific PNA probe, and heated 10 min at 80°C for DNA denaturation. Probe hybridization is performed overnight, followed by two washes at 40°C to remove probe that has bound nonspecifically. For determination of cellular autofluorescence, mixtures of the cells of interest and internal control cells are incubated with the hybridization solution without PNA probe.
Accurate assessment of telomere length requires that only singlet cells (not cell aggregates) containing one genome be considered. Thus, a nucleic-acid dye with a fluorescence emission separate from the emission of the telomere-specific probe is added to the Flow-FISH samples before sample acquisition to permit exclusion of proliferating cells and cell aggregates during data analysis. Duplicate samples are analyzed on the cytometer and the resulting telomere-specific mean fluorescence of G0/1 cells of the cell subset is then compared to the mean background fluorescence of G0/1 subset cells. At the same time, telomere-specific mean fluorescence of the G0/1 internal control cells over background fluorescence is determined.
The amount of DNA per cell influences telomere fluorescence, because it directly correlates with the number of chromosomes and telomere ends. Consequently, relative DNA contents (DNA ploidy) of the cells of interest and internal control cells need to be reliably assessed. To improve the precision of DNA ploidy measurements, separate sample aliquots are processed in parallel to Flow-FISH using an optimized nucleic-acid staining and analysis protocol as outlined below (see Support Protocol).
As the final step, telomere fluorescence values obtained from Flow-FISH are then normalized for DNA content, and telomere length of the cell subset in the sample is expressed as a fraction of the telomere length of the internal control cells.
Materials
PBS-washed cells
Staining solution (see recipe)
PBS (APPENDIX 2A), room temperature and 40°C
Cross-linking solution (see recipe)
1 M Tris·Cl, pH 8.0 (APPENDIX 2A)
Internal control cells, cryopreserved (see recipe)
Hybridization buffer (see recipe) with and without fluorescently labeled, telomere-specific PNA probe, (C3TA2)3(PerSeptive Biosystems or equivalent)
Nucleic-acid stain compatible with other fluorochromes used (e.g., PI, 7-AAD, Hoechst 33342), in PBS
12 × 75–mm polystyrene tubes
1.5-ml polypropylene tubes
80°C shaking water bath
40°C water bath or heating block
Flow cytometer with 488-nm blue excitation, 633-nm red excitation, and optional UV or violet excitation depending on fluorochrome choices made during strategic planning of the experiments
Additional reagents and equipment for staining cells (UNIT 6.2), counting cells (APPENDIX 3A), and standardizing flow cytometers (UNIT 1.3)
Stain cells for cell surface immunofluorescence
-
1
Place ~2.5–3 × 106 PBS-washed cells into a 12 × 75–mm polystyrene tube and resuspend in 250 μl staining solution. Perform cell-surface staining procedure (either direct or indirect) appropriate for the antigen(s) to be studied (see Support Protocol and UNIT 6.2).
If cells are peripheral blood mononuclear cells (PBMCs) separated by Ficoll-Hypaque density gradient, make sure that there is no visible red-cell contamination, as it has been reported that erythrocytes interfere with FITC telomere fluorescence (see Critical Parameters and Troubleshooting). Residual red cells can be lysed by an ~2 min treatment with ammonium chloride lysing solution (APPENDIX 2A) at 20° to 25°C.Starting cell numbers can be lower than the numbers indicated above, as cell recovery will vary depending whether a direct or an indirect staining method is used. However, it is necessary to start with sufficient cells, taking into account that 0.25 × 106 stained cells per reaction are required for Flow-FISH. Initially, follow manufacturer’s recommendation for the appropriate antibody concentration but optimize staining conditions to obtain the maximal signal separation between background and specific staining for Flow-FISH (see UNIT 4.1 for performing antibody titrations). Preparation of a sample stained with the appropriate isotype-matched control(s) is essential for determination of background staining. -
2
After the last washing step, resuspend the cell pellet in 200 μl PBS. Slowly add 200 μl cross-linking solution dropwise. Incubate 30 min at 2° to 8°C protected from light, then add 8 μl of 1 M Tris·Cl, pH 8.0. Mix well and incubate 15 min at 20° to 25°C.
BS3 is a highly reactive cross-linker that easily hydrolyzes; therefore, it is critical that the BS3 solution be prepared immediately before use (see Critical Parameters and Troubleshooting). The reaction buffers must not contain any primary amines because they compete with the desired cross-linking action on cell surface antigen–antibody complexes. Tris buffer added at the end of the first incubation period provides an excess of primary amines to stop cross-linking. -
3
Add 1 ml PBS, centrifuge cells 5 min at 300 × g, 2° to 8°C, and remove supernatant. Add 1 ml PBS. Count cells (APPENDIX 3A).
Cells have to be counted carefully before fluorescence in situ hybridization (FISH) to account for cell losses during staining and cross-linking, because accurate cell numbers are critical for reliable hybridization reactions.
Denature DNA and perform FISH
-
4
Thaw an aliquot of cryopreserved cells from the batch to be used as an internal standard and perform a cell count (APPENDIX 3A).
-
5
Mix 1 × 106 cell surface–stained cells with 1 × 106 thawed internal control cells. Place 0.5 × 106 cells of mixture into a 1.5-ml polypropylene tube. Add PBS to 1 ml, mix well, and centrifuge cells 5 min at 300 × g, 2° to 8°C. Completely remove supernatant.
Internal control cells are added to the reaction tubes in equal numbers compared to the cells to be studied to compensate for variations in the numerous factors that influence FISH of the telomere probe to DNA (see Critical Parameters and Troubleshooting). Samples are prepared in duplicate to reduce assay variability. Two tubes are needed for hybridization without probe, and two for hybridization with telomere-specific probe.Note that tubes have to be made of polypropylene to withstand the high temperature required for DNA denaturation. Tubes larger than 1.5 ml can be used, but as reaction conditions (i.e., evaporation of reaction mix) are influenced by tube airspace, scatter parameters will be altered. -
6
Add 300 μl hybridization buffer with or without fluorescently labeled, telomere-specific probe. Mix well and incubate 10 min at 80°C in a shaking water bath protected from light.
CAUTION: Note that formamide is a toxic substance. Observe proper precautions and perform reactions in a fume hood.Tubes containing hybridization mix without probe are needed for determination of background fluorescence. The appropriate telomere probe concentration has to be determined by titration. Concentrations of 4, 10, and 55 nM have been reported as optimal depending on the type of Flow-FISH procedure used (see Critical Parameters and Troubleshooting).Temperature needs to be maintained with great precision, because 80°C is considered the minimum temperature for proper DNA denaturation. Alternatively, tubes can be heated in a heating block; however, temperature differences between individual tubes will be more pronounced than in a shaking water bath (also see Critical Parameters and Troubleshooting). -
7
Take the tubes out of the water bath and incubate overnight at 20° to 25°C, protected from light.
Alternate protocols use shorter incubation times for probe hybridization (see Critical Parameters and Troubleshooting, Flow-FISH). -
8
The next day, add 1 ml PBS, 40°C, to the tubes which were hybridized overnight. Mix well and incubate 10 min at 40°C. Wash by centrifugation 5 min at 500 × g, 20° to 25°C. Repeat once.
Stain for DNA content
-
9
Add 0.5 ml appropriate nucleic acid stain compatible with other fluorochromes used in PBS to all tubes. Mix well and incubate samples at least 2 to 3 hr at 20° to 25°C protected from light.
Which dye will be most suitable for any given experiment will depend on the other fluorochromes used for phenotyping and for detection of telomere fluorescence (see Strategic Planning, Critical Parameters and Troubleshooting, and Anticipated Results).
Analyze samples on the flow cytometer
-
10
Run samples on a flow cytometer with light sources and filters appropriate for the fluorochromes used. Standardize the flow cytometer for each experiment using standard particles and procedures as described in UNIT 1.3.
-
11
Obtain data using the following parameters:
-
Collect cell-surface fluorescence and telomere fluorescence with log amplification, and DNA fluorescence with linear amplification using area and width signals for doublet discrimination.
Log amplification of the telomere-specific signal is needed for simultaneous measurement of sample and internal standard. Use a low sample differential during sample acquisition to reduce the coefficients of variation of DNA fluorescence measurements.
-
-
12
Analyze samples using plots and gating strategies as described (see Anticipated Results).
SUPPORT PROTOCOL: DNA PLOIDY DETERMINATION
The protocol describes a method for assessment of relative DNA contents of cells of interest and internal control cells using a nucleic-acid staining procedure that has been optimized for high-resolution DNA analysis.
Materials
Unstained cells
Thawed internal control cells
PBS (APPENDIX 2A)
PI staining solution
Prepare and analyze samples
On the day of the original experiment, place 0.5 × 106 unstained cells to be studied and 0.5 × 106 freshly thawed internal control cells into two separate tubes.
-
Wash once with 1 ml PBS by centrifugation 5 min at 300 × g, 2° to 8°C. Add 0.5 ml propidium iodide staining buffer to the cell pellets. Mix and incubate 15 min at 2° to 8°C protected from light.
Alternately, perform a staining protocol for flow cytometric DNA analysis as described in Basic Protocol 1 of UNIT 7.5. Whenever analyzing malignant cell samples that may be aneuploid (e.g., samples from leukemic patients), it is advisable to include an internal DNA ploidy standard consisting of a mixture of chicken and trout erythrocytes (see Basic Protocol 2 in UNIT 7.5). -
Acquire samples for DNA ploidy on a flow cytometer using the following parameters:
Blue excitation
Detection of PI emission at linear amplification at orange-red wavelength
Constant detector voltage and gain between sample runs
PI area and width signal processing for doublet discrimination
-
Low sample differential setting.
The last parameter is used to obtain low coefficients of variation on DNA fluorescence measurements.
Analyze the resulting DNA histograms with appropriate DNA histogram deconvolution software for an accurate determination of the relative positions of the G0/1 peaks.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX.
Cross-linking solution
Store bis(sulfosuccinimidyl)suberate (BS3; Pierce) desiccated at −20°C in powder aliquots suitable for the experiment sizes planned. Prepare BS3 solution immediately before use by taking out one vial of BS3 powder. Wait until the vial has warmed to ambient room temperature before opening. Add 5 mM sodium citrate buffer, pH 5, to a final concentration of 4 mM (w/v) BS3.
Hybridization buffer
70% (v/v) deionized formamide (APPENDIX 2A)
1% (w/v) BSA
10 mM Tris·Cl, pH 7.2 (APPENDIX 2A)
Store 6 months in a tightly sealed container protected from light at 2° to 8°C
Internal control cells
Use cells with long telomeres—e.g., 1301 cells (European Collection of Cell Cultures, https://www.phe-culturecollections.org.UK; Sigma-Aldrich), calf thymocytes or mouse spleen cells (see also Critical Parameters and Troubleshooting, Assay Standardization). Generate a large batch of cells and cryopreserve by liquid-nitrogen storage of aliquots suitable for the typical experiment size planned (APPENDIX 3B).
PI staining solution
0.1 g sodium citrate
0.3 ml Triton X-100
10 mg propidium iodide (PI)
2 mg DNase-free ribonuclease A (APPENDIX 2A)
Adjust volume to 100 ml with H2O
Store 6 months in a tightly sealed container protected from light at 2° to 8°C
Staining solution
PBS without Ca2+ and Mg2+ (APPENDIX 2A)
0.1% (w/v) sodium azide
2% (w/v) newborn calf serum (e.g., Omega Scientific)
Adjust pH to 7.2 with 0.1 M NaOH or 0.1 M HCl
Store up to 6 months at 20° to 25°C
COMMENTARY
Background Information
Significance of telomere length measurement
Telomeres represent important regulatory elements for controlling how often somatic cells can divide. Although telomere shortening is counteracted by the activity of the enzyme telomerase and an alternative lengthening of telomeres (ALT) pathway (Cech 2004), a slow but inexorable decrease in a cell’s replicative potential accompanies cell division. Thus, the average telomere length of a given cell population relates to its replication potential and is linked to cellular senescence (Effros, Allsopp et al. 1996, Effros, Dagarag et al. 2003). Owing to the pivotal role of telomere dynamics in tumor formation, aging, and immune exhaustion pertinent to infectious disease control, interest in telomere biology has increased in recent years and has spurred the development of novel technologies for assessing telomere length.
Measuring telomere length
The traditional method of measuring telomere length is the determination of the mean terminal restriction fragment (TRF) size of genomic DNA by Southern blot analysis. This technique is robust but labor intensive and time consuming. In addition, the restriction enzymes used for TRF analysis cut the DNA in subtelomeric regions; as a consequence, non-telomeric sequences are included in the DNA fragments and make the measurement less accurate.
Fluorescently labeled peptide nucleic-acid (PNA) probes hybridize with complementary oligonucleotide sequences at the low ionic strength unfavorable for re-annealing of the target strands and form very stable duplexes with DNA. Availability of these PNA probes facilitated the development of novel fluorescence in situ hybridization (FISH) protocols for quantitation of telomeric repeat sequences (Lansdorp, Verwoerd et al. 1996). FISH measurements of telomere length correlate well with TRF analysis; however, a linear regression line comparing the two methods is not expected to pass through the origin, because subtelomeric sequences do not interfere with telomere-specific FISH as hexameric T2AG3 repeats are rare except at the ends of chromosomes. Rather, the calculated line should cross the Southern blot axis at a point corresponding to the approximate mean of the subtelomeric DNA length of the samples tested (Hultdin, Gronlund et al. 1998, Law and Lau 2001, Roos and Hultdin 2001, Schmid, Dagarag et al. 2002). The FISH method for estimation of telomere length was first performed by digital imaging microscopy on metaphase spreads of chromosomes and was termed Q-FISH, but sample preparation is complicated, lengthy, and requires a high degree of technical expertise. It is the method of choice, however, to analyze telomeric sequences on specific chromosomes.
In contrast, subsequent Flow-FISH protocols (Hultdin, Gronlund et al. 1998, Rufer, Dragowska et al. 1998) that utilize rapid detection of fluorescence signals of individual cells in suspension, although complex, are less technically demanding, use standard analytic flow cytometers, are able to provide results within 24 hr, and can be combined with phenotypic analysis of cells. Nevertheless, because the conditions required for DNA denaturation during FISH are detrimental to standard staining of cell surface antigens, until recently, analysis of telomere length in cell subpopulations had to be performed on cells that were separated prior to Flow-FISH either by cell sorting (Rufer, Brummendorf et al. 1999, Batliwalla, Rufer et al. 2000) or by magnetic bead separations (Rufer, Dragowska et al. 1998, Son, Murray et al. 2000). Isolating cell subpopulations by these means is time consuming and can be costly and difficult, particularly if the frequency of the cells of interest is low and when large numbers of samples have to be separated. Klapper et al. and Kurosaka et al. have used immunomagnetic separation of cell subsets and cell sorting followed by either Flow-FISH or Southern blotting to determine telomere length in cell subsets, but these studies were limited by small sample sizes (Klapper, Moosig et al. 2004, Kurosaka, Yasuda et al. 2006). In contrast, methods that are able to stabilize antigen-antibody complexes on the cell surface either by covalent cross-linking (Batliwalla, Damle et al. 2001, Schmid, Dagarag et al. 2002) or by fixation (Plunkett, Soares et al. 2001) permit simultaneous measurement of telomere length and cell surface markers without the need for cell separation.
Factors affecting telomere length
Inter-individual heterogeneity of telomere length has been shown to be largely based on genetic factors (Slagboom, Droog et al. 1994, Andrew, Aviv et al. 2006); however, this variability may be also influenced by socio-environmental factors (Valdes, Andrew et al. 2005, Cherkas, Aviv et al. 2006), although this aspect remains controversial (Lansdorp 2006). The great variation in telomere length of cells with the same surface phenotype between individual donors, especially in hematopoietic stem cells (Van Ziffle, Baerlocher et al. 2003) prevents direct donor-to-donor comparisons of telomere length data from a particular cell population. Donor-to-donor variation in the telomere length highlights the need to determine the telomere length in subpopulations from the same donor in order to establish a reference point for telomere length comparisons (Van Ziffle, Baerlocher et al. 2003). In addition, clinical parameters such as age, disease duration, grade of lymphopenia, and treatment, may have an effect on telomere length in lymphocytes (Wu, Hsieh et al. 2007). Monocyte populations have been shown to have a weaker diurnal rhythm than lymphocytes and other PBMC subpopulations and may be more sensitive to intra-individual factors affecting telomere length (Ackermann, Revell et al. 2012).
Both telomere length and rate of telomere shortening in lymphocytes are typically different from myeloid cells (i.e., monocytes and granulocytes) and CD4+, CD8+, and B cells have shorter telomeres then CD14+ monocytes (Rufer, Brummendorf et al. 1999, Beier, Balabanov et al. 2007). Kapoor et al. found that the telomere length for the CD14+ cell population was on average 17.7% higher than that of the CD3+ cells (Kapoor, Hakim et al. 2009). B cells, especially germinal center B cells, and certain memory B cells have longer telomeres than T cells and NK cells from the same donor (Weng, Granger et al. 1997, Martens, Brass et al. 2002, Baerlocher, Mak et al. 2003, Ouyang, Baerlocher et al. 2007). NK cells, T cells, and granulocytes have similar telomere length dynamics and cell subpopulations with a mature phenotype and with an effector phenotype or expressing activating receptors have shorter telomeres compared to those with an immature phenotype (Ouyang, Baerlocher et al. 2007).
Data on telomere length analysis of subpopulations of immune and hematopoietic cells supports the hypothesis that cells with the greatest proliferative potential have the longest telomeres (Van Ziffle, Baerlocher et al. 2003, Ouyang, Baerlocher et al. 2007). Borisov et al. studied in vitro telomere length changes after T lymphocyte activation in CD4+ and CD8+ cells (Borisov, Korolkova et al. 2014). Before activation, telomeres in CD4+ T cells were 3.7% longer than in CD8+ T cells, but after one week in culture, the ratio of telomere length changed, and telomeres in CD4+ cells became 2.2% shorter. This probably occurs due to the faster proliferative response of CD8+ cells. In summary, due to the numerous factors that can influence telomere length in vivo and in vitro, careful standardization of the Flow-FISH method is needed.
Alternate Protocols and Modifications of Flow-FISH
As with most techniques, changes to the original, published protocols were described in a series of papers (Rufer, Dragowska et al. 1998, Rufer, Brummendorf et al. 1999, Baerlocher, Mak et al. 2002, Baerlocher and Lansdorp 2003, Baerlocher and Lansdorp 2004). Flow-FISH has been adopted by a number of different laboratories for various applications. For example, to measure the average telomere length in whole kidney epithelial cells, it was proposed to use nuclei rather than whole cells in order to detect the weak fluorescence signals of PNA bound to telomere target sequences above the background resulting from cytoplasmic auto-fluorescence and non-specific binding of the probe (Wieser, Stadler et al. 2006). Another modified Flow-FISH protocol has been developed for the dynamic measurement of telomere length in cell division (Potter and Wener 2005, Borisov, Korolkova et al. 2014). For this method, three different procedures were performed one after another: staining of cells with a vital dye -CFSE Cell Tracker-followed by cell surface staining, and then FISH with a telomere-specific probe (Potter and Wener 2005, Borisov, Korolkova et al. 2014). A major modification from the protocol for simultaneous assessment of phenotype and telomere length described in the current chapter involves performing FISH before immunophenotyping and using specific antibodies that are able to bind to antigens (epitopes) that have not been affected by the harsh experimental conditions (See Table 1 for comparison of the methods) (Baerlocher, Vulto et al. 2006). Variations also exist in the duration of the hybridization reaction, in the concentration of the PNA probe, in DNA staining, in method standardization (Beier, Balabanov et al. 2007), and data analysis (see Table 1 for details). Finally, in some protocols the telomere fluorescence is expressed on a linear scale (Rufer, Dragowska et al. 1998) rather than a logarithmic scale (see Basic Protocol) (Hultdin, Gronlund et al. 1998). Because the ability to detect small biologically relevant differences relies largely on method accuracy and reproducibility, the effect of any modification from established procedures regarding telomere length measurement must be carefully validated.
Table 1.
Comparison of two major, validated variations of the Flow-FISH method
| Step |
Immunophenotyping → FISH (Flow-FISH) Hultdin et al. (Hultdin, Gronlund et al. 1998) and modifications (Schmid, Dagarag et al. 2002, Kapoor, Hakim et al. 2009) |
FISH → Immunophenotyping (FISH-Flow) Rufer et al. 1998 (Rufer, Dragowska et al. 1998) and modifications (Baerlocher, Vulto et al. 2006) |
| Calibration of instrument | DNA calibration beads | Calibration (MESF) beads |
| Cells | PBMC and 1301 control cells | PBMC and bovine thymocytes |
| Immunophenotyping | Few fluorochromes retain their fluorescence after the harsh conditions required for DNA denaturation in Flow-FISH (heat affects the fluorochrome) Stable fluorescent probes: Alexa Fluor 488 (Schmid, Dagarag et al. 2002), Alexa Fluor 546 (Schmid, Dagarag et al. 2002), Alexa Fluor 647 (Kapoor, Hakim et al. 2009), Qdot 605 (Kapoor, Hakim et al. 2009), Qdot 655(Kapoor, Hakim et al. 2009), Qdot 705 (Kapoor, Hakim et al. 2009) for cell surface staining Antibodies: anti-CD4, anti-CD3, anti-CD8, anti-CD14, anti-CD28 BS3 cross-linking to stabilize fluorochrome-antigen-antibody complexes |
Very few antibodies recognize epitopes that survive the harsh conditions required for DNA denaturation in Flow-FISH (heat affects the epitope) Antibodies: CD45RA–Cy5 (clone 8d2) and CD20– (PE) (clone L26), Biotinylated anti-CD57 (clone HNK-1) No use of cross-linking fixatives (Baerlocher and Lansdorp 2003) |
| Flow-FISH | Covalent cross-linking DNA denaturation: 10 min at 80°C in a shaking water bath Hybridization: overnight in the dark. |
No fixation; permeabilization only with formamide and heat DNA denaturation: 15 min at 87°C in a shaking water bath Hybridization: 2 h at RT in the dark. |
| DNA staining | Propidium iodide (PI), Hoechst 33342, LDS751 | LDS751 |
| Data analysis | Relative telomere length determination (TLR method) | Absolute telomere length determination (MESF/Kb) |
Critical Parameters and Troubleshooting
The procedure described in the primary procedure (see Basic Protocol) is complex and requires basic understanding of cellular staining principles and flow cytometry. In addition, previous experience in setting up a flow cytometer for multicolor experiments will facilitate success. In planning a combined phenotype and telomere-length assay, it is necessary for the experimenter to consider the fluorochrome choices and the capabilities of the flow cytometer to be used for data acquisition and to optimize the cell-surface and DNA staining protocol prior to performing the assay on samples to be studied. Critical parameters that may affect experimental variability including sample preparation, factors related to immunophenotyping, flow-FISH and DNA staining, assay and instrument standardization as well as data reporting and assay validation.
Sample Preparation
The Flow-FISH method for measuring telomere length is a complex protocol with various aspects needing careful consideration for a successful experiment (Lauzon, Sanchez Dardon et al. 2000). The primary method (see Basic Protocol) has been successfully used on PBMC isolated by Ficoll-Hypaque density centrifugation. Whole-blood samples cannot be readily used as it has been shown that the hemoglobin contained in erythrocytes can interfere with measuring telomere length when using a FITC-labeled probe, possibly by quenching probe fluorescence (Baerlocher, Mak et al. 2002); therefore, complete removal of red cells is important, and sample hematocrit should not exceed 2%. Preparation of a fresh NH4CL solution with the appropriate pH and the addition of washing steps may aid in achieving efficient red cell lysis (Baerlocher, Vulto et al. 2006).
Fixation and cryopreservation protocols can affect subset analysis by flow cytometry (Pinto, Trivett et al. 2005). Although the lymphocyte subset percentages were shown to be comparable between different procedures for cryopreservation and fixation, the fluorescence intensity of staining can diminish for specific markers (Pinto, Trivett et al. 2005). Cryopreservation may also affect monocytes more than lymphocytes (Wang, Hsu et al. 1998, Wiley, Ashok et al. 2014) while limited evidence suggests that thawing has minimal effects on monocytes (Hori, Heike et al. 2004). To our knowledge the effect of cryopreservation and freeze-thawing on telomere length measurement has not been validated in Flow-FISH procedures against fresh samples. However, frozen, banked PBMC samples have reliably been used to determine telomere length with Flow-FISH (Plunkett, Soares et al. 2001, Van Ziffle, Baerlocher et al. 2003, Potter and Wener 2005, Rufer, Reichenbach et al. 2005, Beier, Balabanov et al. 2007, Kapoor, Hakim et al. 2009). Reduction of the time interval of storing samples prior to freezing, checking the viability of cells to be frozen, thawing cells quickly, and replacing freezing solutions with buffers that support cell viability will help in obtaining cells suitable for Flow-FISH assays. The addition of DNAse (1 ug/per mL) can reduce loss of leukocytes after thawing by preventing the formation of cell aggregates caused by dead cells present in the samples (Baerlocher, Vulto et al. 2006).
Immunophenotyping
Stability of fluorochromes used for immunophenotyping of membrane antigens during Flow-FISH
The harsh experimental conditions required for proper DNA denaturation for binding of the telomere-specific probe provided a major obstacle for the generation of reliable protocols for Flow-FISH assays with concomitant cell surface staining as the thermal stability of most fluorophore-antibody conjugates has been shown to be poor in the 80 to 87°C temperature range required for probe hybridization (Schmid, Dagarag et al. 2002, Baerlocher, Vulto et al. 2006). Traditional bright fluorochromes such as phycoerythrin (PE), allophycocyanin (APC), or tandem fluorochromes containing PE or APC are protein based and therefore destroyed by heat, and commonly used low molecular weight fluorochromes such as Pacific Blue do not retain sufficient brightness to allow detection of antibody-labeled cells (Kapoor, Hakim et al. 2009). Thus, only heat-stable fluorochromes can be utilized in telomere Flow-FISH assays. Possible fluorochrome choices include fluorescein isothiocyanate (FITC), cyanine dyes (e.g., Cy3, Cy5), Alexa Fluor reagents (e.g., Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 647, and Qdots (e.g., Qdots 605, 655, 705). Fluorochrome brightness is important because after FISH, cell-surface fluorescence intensity still decreases markedly despite BS3 cross-linking, while cellular autofluorescence increases. Furthermore, maintenance of cell-surface fluorescence varies between different fluorochromes and antigen-antibody complexes, but choosing an antigen with a high expression level can facilitate success, because after the hybridization procedure, discrimination of positive from negative cells for less abundant antigens may not be possible (Borisov, Korolkova et al. 2014).
Cyanine dyes are heat stable, and Batliwalla et al. (Batliwalla, Damle et al. 2001) have used the Cy5 as the fluorochrome for detection of cell surface antigens. Availability of several heat-stable, bright Alexa Fluor–type fluorochromes has opened up the possibility to combine two cell surface markers with telomere Flow-FISH (Schmid, Dagarag et al. 2002). In the published protocol (Schmid, Dagarag et al. 2002), which provides the basis for the Basic Protocol described in this unit, the authors used Alexa Fluor 488 and Alexa Fluor 546 for cell surface staining and employed an indirect staining method that, although time-consuming, permitted the detection of positive cells above background with frequency similar to that for samples that were not subjected to Flow-FISH. Nevertheless, it is possible that for staining of other antigens, Alexa Fluor monoclonal antibodies that are directly conjugated are suitable. For instance, Kapoor et al. showed that CD4 Alexa Fluor 647 fluorescence is preserved well after heating to 82°C, retaining more than 80% of its pre-hybridization fluorescence (Kapoor, Hakim et al. 2009).
Quantum dots (Qdot nanoparticles) are inorganic semiconductor complexes that are highly fluorescent and are excited by the short-wavelength lasers (ultraviolet or violet) which have become more prevalent on flow analyzers (Jamieson, Bakhshi et al. 2007). Qdots were shown to possess greater thermal stability than traditional low molecular weight fluorophores (Kapoor, Hakim et al. 2009). Qdot antibody conjugates directed against monocyte and T cell antigens were found to retain most of their fluorescence following the high-temperature annealing step, allowing ready immunophenotyping and telomere length measurement. Since Qdots have very narrow emission bandwidths, several Qdot-antibody conjugates (Qdot 605, 655, and 705) can be analyzed simultaneously in a Flow-FISH assay permitting complex cell subset-specific analysis. For multicolor cell-surface staining the emission spectra of fluorochromes have to be separate, but can overlap as long as fluorescence compensation can be applied; however, neither can overlap into the channel used for detection of telomere fluorescence, as compensation of the overlap would alter the position of the telomere-specific fluorescent peak. Setting proper fluorescence compensation is critical and it has to be performed using single color controls, e.g., using cells labeled with antibodies conjugated to the same fluorochromes than the ones used for antibody and probe labeling (Borisov, Korolkova et al. 2014). Table 2 shows previously validated antibodies and fluorochromes and their potential combination into two different multicolor (up to 6 colors) Flow-FISH panels. Note that the successful modification and expansion of panels (as shown in Table 2) beyond the reagent combinations that have been published, i.e., the ones developed by Schmid et al. (Schmid, Dagarag et al. 2002) and Kapoor et al. (Kapoor, Hakim et al. 2009), will depend on the instrument configuration, laser options, and filter sets and will require careful establishment of instrument settings and fluorescence compensation.
Table 2.
Potential multicolor panels for Flow-FISH using previously validated antibodies/fluorochromes and BS3 cross-linkinga
| Fluorochrome | Target/Antigen (reference) | Company | Excitation (nm) (laser) | Emission |
|---|---|---|---|---|
| Panel 1 | ||||
| Hoechst 33342 (HO342) | DNA stain (Schmid, Dagarag et al. 2002) | Life Technologies | 361 (355) | 497 |
| Alexa Fluor 488 | CD8 (Schmid, Dagarag et al. 2002) or CD28 (Schmid, Dagarag et al. 2002) | BD Biosciences | 494 (488) | 518 |
| PE or Alexa Fluor 546 | CD57 PE (Baerlocher, Vulto et al. 2006) – used after FISH or CD8 labeled with GAM Alexa Fluor 546 (Schmid, Dagarag et al. 2002) |
BD Biosciences BD Biosciences/Life Technologies (GAM Fab’2 Alexa Fluor 546) |
496 (488, 561) 556 (488, 561) |
578 573 |
| Qdot 605 | CD14 (Kapoor, Hakim et al. 2009) | Thermo Scientific | Broad UV and violet excitation (350, 405) | 605 |
| Cy5 | PNA probe (Schmid, Dagarag et al. 2002) | Panagene | 647 (633) | 665 |
| Qdot 705 | CD4 (Kapoor, Hakim et al. 2009) | Thermo Scientific | Broad UV and violet excitation (350, 405) | 705 |
| Panel 2 | ||||
| Alexa Fluor 488/FITC | PNA probe (Kapoor, Hakim et al. 2009) | Dako, Panagene | 494 (488) | 518 |
| Alexa Fluor 546 | CD8 (Schmid, Dagarag et al. 2002) | BD Biosciences/Life Technologies (GAM Fab’2 Alexa Fluor 546) | 556 (561b) | 573 |
| Qdot 605 | CD14 (Kapoor, Hakim et al. 2009) | Thermo Scientific | Broad UV and violet excitation (350, 405) | 605 |
| Qdot 655 | CD3 (Kapoor, Hakim et al. 2009) | Thermo Scientific | Broad UV and violet excitation (350, 405) | 655 |
| Qdot 705 | CD4 (Kapoor, Hakim et al. 2009) | Thermo Scientific | Broad UV and violet excitation (350, 405) | 705 |
| LDS-751 | DNA stain (Baerlocher, Vulto et al. 2006, Kapoor, Hakim et al. 2009) | Thermo Scientific | 543 (488, 561, 532) | 712 |
Note that successful implementation of these expanded panels that extend beyond the reagent combinations that have been published, i.e., by Schmid et al. (Schmid, Dagarag et al. 2002) and Kapoor et al. (Kapoor, Hakim et al. 2009), will depend on the instrument configuration, laser options, and filter sets and will require careful establishment of instrument settings and fluorescence compensation.
Although Alexa Fluor 546 can also be excited by a 488nm laser (see first panel), for the second panel, exciting it with a 561nm laser is preferable.
Covalent crosslinking of the fluorophore-antibody complex to the cell surface
Standard fixation with formaldehyde or commercial reagent systems such as Fix & Perm (Caltag Medsystems) or FACS Lysing solution (BD Biosciences) is insufficient to maintain surface fluorescence in Flow-FISH protocols. Batliwalla et al. (Batliwalla, Damle et al. 2001) developed a novel strategy for covalently cross-linking antigen-antibody complexes onto the cell surface membrane with bis(sulfosuccinimidyl)suberate (BS3). BS3, the water-soluble, noncleavable cross-linker utilized in the Basic Protocol, is a homobifunctional N-hydroxysuccinimide ester which predominantly reacts with the ε-amino group of lysine. BS3 is more stable in solutions with low pH, as hydrolysis increases under alkaline conditions. Nevertheless, because of its inherent instability, the generation of BS3 stock solutions is not recommended; rather, aliquots of the powder need to be stored at −20°C in a desiccator for retention of cross-linking activity, and BS3 solutions should be prepared just before use; otherwise, the crosslinker will not work properly and surface labeling with antibodies will be unsuccessful (Borisov, Korolkova et al. 2014). BS3 is excluded from intact cells and most importantly, does not hinder probe access to the DNA.
Considerations for immunophenotyping of membrane antigens during Flow-FISH
It is critical for the reliability of the telomere length measurements that there be no fluorescence overlap of other fluorochromes into the channel used for detection of telomere fluorescence. Thus, the choice of label for the telomere-specific probe should be made before selecting the other colors. Because the conditions during FISH increase cellular autofluorescence and at the same time reduce the fluorescence intensity of the antibody labels used for cell surface staining, maximization of the specific fluorescence signal is critical. As monoclonal antibody clones directed against the same antigen can differ in their stability during Flow-FISH, it may be necessary to compare reagents from various sources. In addition, variations in reagent concentration, staining time and temperature, and the number of washes need to be investigated to optimize the brightness of the specific signal above background. To achieve sufficient signal amplification for reliable discrimination of cell subpopulations in the Flow-FISH assay, it may be necessary to utilize two-layer staining protocols that use either the streptavidin-biotin system or a species-specific second-step reagent. In addition, indirect staining protocols can result in better stabilization of cell surface antigen–antibody complexes, but they increase the staining time and potential cell losses from multiple washing steps.
Flow-FISH
Denaturation of DNA
While Hultdin et al. (Hultdin, Gronlund et al. 1998) exposed cells to a fixation and permeabilization step before Flow-FISH, the protocol by Rufer et al. (Rufer, Dragowska et al. 1998) showed that sufficient access of the telomere-specific probe can be achieved without a fixation step. The small size of the probe and the reaction conditions used for Flow-FISH (i.e., formamide and heat) appear to cause adequate cell membrane permeability for probe access to DNA. One central experimental condition for Flow-FISH is the complete denaturation of double-stranded DNA into single strands by heat and deionized formamide for proper telomere-specific PNA probe hybridization. Early protocols exposed cell preparations to a temperature of 80°C and a concentration of formamide of 70% for 10 min (Hultdin, Gronlund et al. 1998, Rufer, Dragowska et al. 1998). These conditions have subsequently been used successfully by others (Son, Murray et al. 2000, Law and Lau 2001, Plunkett, Soares et al. 2001, Schmid, Dagarag et al. 2002). Recently, however, it has been indicated that temperatures of 85° to 87°C may provide further optimization of the DNA denaturation step (Baerlocher, Mak et al. 2002). Nevertheless, as higher temperatures create extremely unfavorable conditions for cells, these considerations have to be balanced against maintenance of cellular integrity and preservation of cell surface antigens in cases where subset analysis of cell populations is desired. Furthermore, tubes placed into a shaking water bath will reach the desired temperature faster and with less time and temperature difference between individual tubes than tubes placed into a heating block and therefore will require less overall heat exposure.
Hybridization with telomere-specific fluorescent probe
The proper concentration of the PNA telomere-specific probe and hybridization conditions are important experimental considerations. The use of a 96-well automated dispenser, such as the Hydra system, can assist with the many pipetting steps in the procedure (Baerlocher, Vulto et al. 2006). Active vortexing or intense shaking of the hybridization solution should be avoided because it contains 10% FBS, and can easily foam. On the other hand, after the hybridization procedure, during washing the cells tend to aggregate, so vigorous vortexing is recommended (Borisov, Korolkova et al. 2014).
Methods that were published by the two independent laboratories which have pioneered the Flow-FISH technique use markedly divergent probe concentrations and hybridization times for their individual protocols. Rufer et al. (Rufer, Dragowska et al. 1998) used 55 nM telomere-specific probe and 2 hr of hybridization, while Hultdin et al. (Hultdin, Gronlund et al. 1998) found that overnight hybridization with a probe concentration of 4 nM was optimal for their assay. Some of these differences may be due to the cell fixation-permeabilization step and the addition of a cell line with long telomeres as an internal standard to each reaction tube by Hultdin et al. (Hultdin, Gronlund et al. 1998). In their assay, and in the procedure described in the Basic Protocol, telomere probe is also taken up by the standard. Thus, conditions that provide optimal separation of telomere fluorescence between sample and standard may differ from those most suited for reactions without an internal standard. In a published protocol for dual-color cell surface staining and telomere length measurements Schmid, Dagarag et al. 2002) that also utilizes an internal standard, the authors selected (a concentration of 10 nM for a Cy5-labeled telomere-specific probe as optimal for Flow-FISH. For a given probe concentration, cell numbers have to be kept extremely constant between sample tubes, because specific binding is highly dependent on cell concentration, and therefore reliable telomere length assessment of rare cell populations can be problematic, because the ratio of the cell concentration to the PNA probe concentration can have a dramatic effect on labeling (Kapoor, Hakim et al. 2009).
Baerlocher et al. (Baerlocher, Mak et al. 2002) showed that the hybridization reaction can reach a plateau after ~2 hr, but Hultdin et al. (Hultdin, Gronlund et al. 1998) and Schmid et al. (Schmid, Dagarag et al. 2002) have successfully used overnight hybridization. Baerlocher et al. further reduced the hybridization time to 10 minutes at 82°C in a temperature-controlled water bath (Baerlocher, Vulto et al. 2006). Kapoor et al found that varying the hybridization time from 10 to14 minutes had no significant effect on probe hybridization (Kapoor, Hakim et al. 2009). The samples were then stored overnight at room temperature (22–24°C). Heating in formamide significantly changes the size of the cells; thus, forward scatter and side scatter signals of cells will decrease compared to cells analyzed before the hybridization procedure (Baerlocher, Mak et al. 2002) requiring different instrument settings.
Hybridization must be followed by sufficient washing steps (at least two) to remove unbound and nonspecifically bound PNA probe. While most published procedures include formamide in their washing solutions, a commercially available kit for Flow-FISH (Dako, Agilent Technologies) uses formamide-free washing steps at 40°C, and the authors have adopted this in the published procedure ((Schmid, Dagarag et al. 2002); also see Basic Protocol). Thus, cells can be washed twice with wash buffer provided with the Telomere PNA Kit/FITC (Dako) according to the kit directions while the third wash can be performed with PBS + 1% BSA (Kapoor, Hakim et al. 2009).
For each test sample two samples are analyzed: one in which the cells were hybridized to the peptide nucleic acid (PNA) probe and one that was treated identically but without the PNA probe. The latter is required to measure the level of autofluorescence in cells of interest and to enable telomere length to be calculated from specific PNA hybridization (Baerlocher, Vulto et al. 2006). The type of fluorochrome used for telomere-specific probe labeling can influence the ability to detect specific probe fluorescence above the high green autofluorescence generated by the Flow-FISH procedure (Mosiman, Patterson et al. 1997). Using Cy5, which emits red fluorescence away from the parts of the spectrum with the highest contributions to autofluorescence, improves measurement of telomere fluorescence (Schmid, Dagarag et al. 2002).
DNA Staining
Cells tend to form aggregates during the Flow-FISH procedure. Clumped cells interfere with accurate determination of telomere fluorescence and they need to be gated out on plots of DNA fluorescence width versus area during data analysis. Furthermore, only cells in G0/1 which have not started to increase their DNA content can be used for an accurate determination of telomere length, as Hultdin et al. (Hultdin, Gronlund et al. 1998) have shown that replication of telomere sequences can already begin in the early S-phase of the cell cycle. The addition of a nucleic-acid dye before sample acquisition on the cytometer can address both these problems. The selection of the appropriate nucleic-acid dye for staining will depend on its compatibility with the other fluorochromes that are used for detection of telomere-specific probe and cell surface antigens. Combinations of fluorochromes that have been used successfully for simultaneous measurement of phenotype, telomere length, and DNA content include Cy5, FITC, and propidium iodide (PI) for single-color cell surface staining (Batliwalla, Damle et al. 2001, Plunkett, Soares et al. 2001), and Alexa Fluor 488, Alexa Fluor 546, Cy5, and Hoechst 33342 (HO342) for measuring two cell-surface markers (Schmid, Dagarag et al. 2002), and Quantum dot (Qdot) 605, 655, and 705 and LDS 751 for simultaneous detection of three surface markers (Kapoor, Hakim et al. 2009). Dyes are added at much lower concentrations than the ones commonly used for DNA staining to minimize their interference with telomere fluorescence due to quenching or energy transfer (Baerlocher, Mak et al. 2002). Titrations to find the lowest possible dye concentration that still produces adequate DNA histograms are advisable. Concentrations that have been used previously are 0.06 to 0.2 μg/ml for PI (Hultdin, Gronlund et al. 1998, Rufer, Dragowska et al. 1998), 0.06 μg/ml for 7-aminoactinomycin D (7-AAD; (Rufer, Dragowska et al. 1998)), 0.3 μg/ml for HO342 (Schmid, Dagarag et al. 2002), and 200 μg/ml for LDS751 (Baerlocher, Vulto et al. 2006).
Longer than usual incubation times are necessary to reach adequate staining; nevertheless, coefficients of variation of DNA distributions are not as low as with standard staining methods and dye concentrations. Dyes are selected depending on the fluorochrome used for the detection of telomere fluorescence and cannot have notable fluorescence emissions overlapping into the telomere fluorescence channel to avoid interference with the correct determination of the mean telomere fluorescence. Many protocols, including ours, have used propidium iodide (PI) for DNA staining (Schmid, Dagarag et al. 2002, Beier, Balabanov et al. 2007) whereas others have used LDS751 (Baerlocher, Vulto et al. 2006). However the LDS dye has limitations when used in telomere length analysis since it cannot allow precise discrimination of 2N versus >2N DNA content (Van Ziffle, Baerlocher et al. 2003). A DNA dye that provides more precise DNA distributions such as DAPI (Harley, Futcher et al. 1990), is required for improved cell cycle determination. Ideally, samples should be acquired immediately after the completion of DNA staining, although Kapoor et al. found that no significant variations in the measurements were observed up to 24 hours (Kapoor, Hakim et al. 2009).
Assay Standardization
Controls to account for Interindividual telomere length variability
In order to correct for the variability in telomere length between individuals of the same age (see section on Factors Affecting Telomere Length for details), studies that investigated the telomere length in leukemic cells by Flow-FISH have used CD3 T cells from the same individual (which can be expected to be unaffected by the disease process) as an internal standard (Brummendorf, Holyoake et al. 2000, Drummond, Lennard et al. 2004). Similarly, to better describe disease specific changes in telomere biology in immune cell subsets in Lupus patients, Beier et al. utilized CD14+ monocytes as a subject-specific internal control (Beier, Balabanov et al. 2007). Vigorous statistical approaches such as multiple linear regression and appropriate matching of experimental groups can be used to reduce the influence of clinical parameters such as age, disease duration, grade of lymphopenia, and treatment on the telomere length assessment (Beier, Balabanov et al. 2007).
Controls to account for experimental variability in determination of telomere length
Flow-FISH of telomere length depends on the reliability of the generation and measurement of a fluorescent signal for which minor alterations or erratic or systematic errors in the procedure can lead to relatively large changes in the readout. Thus, for improvement of the accuracy of the assay, addition of a stable internal standard that controls for the variations between individual reaction tubes is critical as it limits statistical errors from tube-to-tube and day-to-day analysis. Hultdin et al. (Hultdin, Gronlund et al. 1998) have introduced the use of the 1301 cell line, a subline of CCRF-CEM, as an internal standard. 1301 cells have extremely long telomeres (>25 kb), are near tetraploid, and therefore can be easily distinguished from any human sample on the flow cytometer. In principle, any cells that exhibit these features are suitable. For instance, mouse spleen cells that are known to have long telomeres could also be a useful standard. Other reference controls suitable for standardization of the Flow-FISH assay include calf thymocytes (CT, a long telomere control), Jurkat T cells (a short telomere control), and standardized mononuclear cells (MNC) from a middle-aged donor (Kapoor, Hakim et al. 2009). CTs which have telomeres (~15–20 kb) that are about 2–3 times longer than human cells and can be easily distinguished from human test cells can be prepared from freshly harvested calf thymus (U.S. Department of Agriculture) using the method of Baerlocher et al. (Baerlocher, Vulto et al. 2006). To further improve their usefulness, the bovine thymocytes are mildly fixed in 0.2% formaldehyde and aliquoted and frozen for later use as a convenient internal reference. Fixation results in decreased staining with the LDS751 dye (and decreased PNA fluorescence) but increased stability of the cells during the many steps in the Flow-FISH protocol. Alternatively, a single passage of Jurkat T cells can be expanded in large quantity, cryopreserved in DMSO, and used as reference control for the duration of the study (Kapoor, Hakim et al. 2009).
Instrument Standardization
The reliability of the fluorescent signal from Flow-FISH is also highly dependent on flow cytometer quality control and standardization (UNIT 1.3). Sensitive detection and calibration of the channel used for the detection of the hybridized PNA probe (typically fluorescein or Cy5) is particularly important to distinguish low levels of specific fluorescence from background or auto-fluorescence.
When the telomere signal is processed linearly as described by Rufer et al. (Rufer, Dragowska et al. 1998) and for DNA ploidy determinations, it is advisable to check the linearity of the fluorescent scale with appropriate calibration beads, such as DNA check beads available from BD Biosciences. When the telomere fluorescence is processed logarithmically (Hultdin, Gronlund et al. 1998), the correct behavior of the log amplifier must be verified by using commercially available beads and methods (UNIT 1.3), or published methods (Schmid, Schmid et al. 1988). The use of calibration beads enables instrument problems to be rapidly identified, indicates instrument sensitivity, and provides a record of instrument settings and day-to-day variations in instrument performance. Marked differences in the ability of different instruments to distinguish beads with low levels of fluorescence (corresponding to less than 10,000 Molecules of Equivalent Soluble Fluorochrome (MESF) units (see also section below on Data Reporting and Assay Validation) from unlabeled beads, have been described (Baerlocher and Lansdorp 2003). If the instrument shows deviations from expected results, cleaning the instrument (following the procedures recommended by the manufacturer) may resolve the issues. If the problems persist, optical realignment, a new flow cell, or replacement of a photomultiplier tube by service personnel may increase sensitivity of detection (Baerlocher, Vulto et al. 2006). FITC MESF beads, e.g., from Bangs Laboratories, Inc., can be used for both measurement of absolute telomere length and for calibration of the flow cytometer to adjust the FITC channel voltage (Kapoor, Hakim et al. 2009). The optimal PMT voltage can be found by using methods previously described for determining voltage ranges with both maximum sensitivity and a good linear response. The PMT setting is subsequently left constant and used for all MESF microsphere and FITC PNA telomere measurements (Kapoor, Hakim et al. 2009) to improve the reproducibility of Flow-FISH.
Data Reporting and Assay Validation
Telomere length can be quantified either as a relative or absolute telomere length. Hultdin et al. (Hultdin, Gronlund et al. 1998) expressed the telomere length of the samples of interest as a fraction of the telomere length of the internal standard to allow for a direct comparison between samples. To compensate for differing DNA content between the samples and the 1301 cells (i.e., diploid versus tetraploid), they measured their DNA indices and normalized the telomere values for their DNA content. This is critical, because DNA content directly correlates to the number of chromosomes and telomere ends ((Hultdin, Gronlund et al. 1998, Law 2001, Roos and Hultdin 2001). DNA histograms obtained with the nucleic-acid counterstain in Flow-FISH samples are suboptimal owing to the low concentration of the stain; therefore, it is advisable to perform the determination of DNA indices using a method optimized for the determination of DNA content (Hultdin, Gronlund et al. 1998, Schmid, Dagarag et al. 2002) such as the one published by Fried et al. (Fried, Perez et al. 1980) see Support Protocol and also protocols described in UNIT 7.5).
For a ready comparison of data obtained in different laboratories, however, it is necessary to generate a linear regression line between the Flow-FISH technique and the TRF fragment size in kilo bases as measured by Southern blotting (Hultdin, Gronlund et al. 1998, Law and Lau 2001, Schmid, Dagarag et al. 2002). This can be achieved by parallel analysis of samples of various telomere lengths, e.g., human samples from individuals that differ in age or cell lines that have long telomeres. After establishing the correlation between TRF values and Flow-FISH data, the correlation equation can then be applied to subsequent samples that are processed with the same Flow-FISH method and analyzed on the same flow cytometer.
Reporting of relative telomere length only allows the comparison of the results of experiments carried out with the same standard; however, it is also possible to express telomere lengths in absolute units i.e., base pairs. Rufer et al. (Rufer, Dragowska et al. 1998) reported their data in terms of arbitrary fluorescence units or Molecules of Equivalents of Soluble fluorochrome (MESF) units using as the Quantum™ MESF beads from Bangs Laboratories Inc. (Rufer, Brummendorf et al. 1999). The FITC-labeled beads contain five different populations, each labeled with a known number of FITC molecules. The mean fluorescence intensity (MFI) value for each bead peak corresponds to the approximate number of fluorescein molecules; thus, a standard curve for MESF values, and by extension FITC molar concentration, can be generated. By doing side-by-side Southern blotting and Flow-FISH using a PNA probe conjugated with a known molar amount of FITC and using MESF standards, a mathematical formula can be empirically derived relating relative fluorescence to telomere length in kilo base pairs (Kb) (Rufer, Dragowska et al. 1998, Kapoor and Telford 2004). The average FITC MFI for the unlabeled PNA probe samples (run in duplicate) is subtracted from the average FITC MFI for the FITC-labeled PNA probe sample (also run in duplicate) resulting in the equivalent MESF value for the sample (Rufer, Brummendorf et al. 1999, Baerlocher, Mak et al. 2002, Kapoor and Telford 2004, Baerlocher, Vulto et al. 2006). This can be calculated using both the Quantum™ FITC MESF software (Bangs Laboratory) and a linear regression equation relating the MESF values to the FITC histogram scale.
Anticipated Results
Figures 7.26.1 and 7.26.2 show the application of the procedure described in the Basic Protocol to the determination of telomere length in CD28 subsets of CD8+ cells in human PBMCs. CD28 is a molecule expressed on the cell surface of most T cells. It is known to be transiently downregulated after activation (Azuma, Phillips et al. 1993), and an irreversible loss of CD28 on CD8+ cells has been associated with aging (Valenzuela 2000). CD8+CD28− cells have a diminished capacity for in vitro proliferation, raising the possibility that some of these cells may have reached a state of replicative senescence (Effros, Allsopp et al. 1996, Monteiro, Batliwalla et al. 1996). Shortening of telomeres in the CD8+CD28− subset has been previously reported using various methods (Effros, Allsopp et al. 1996, Monteiro, Batliwalla et al. 1996, Batliwalla, Rufer et al. 2000). Thus, application of the method described here (see Basic Protocol) is expected to demonstrate differences in telomere length between CD8+CD28+ and CD8+CD28− human lymphocytes.
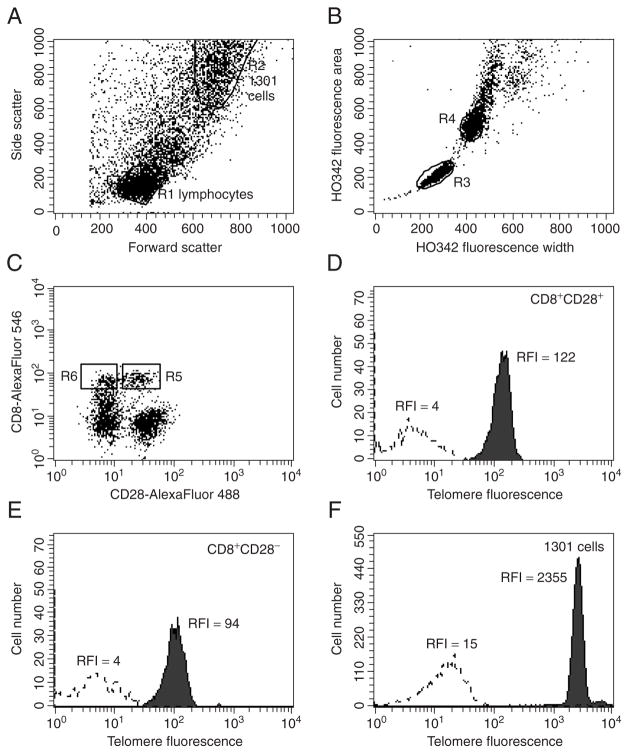
Figure 7.26.1.
Fresh human PBMCs were processed as described (see Basic Protocol) and detailed (see Anticipated Results). (A) Forward-versus side-scatter dot plot, ungated; (B) HO342 width versus area dot plot, gated on R1 or R2; (C) CD28 Alexa Fluor 488 versus Alexa Fluor 546 dot plot, gated on R1 and R3; (D) histogram overlay of Cy5 telomere-specific fluorescence of cells falling within gate R1, gate R3, and gate R5 over background fluorescence; (E) histogram overlay of Cy5 telomere-specific fluorescence of cells falling within gate R1, gate R3, and gate R6 over background fluorescence; (F) histogram overlay of 1301 cells falling within gate R2 and gate R4 over background fluorescence. RFI: relative mean fluorescence intensity.
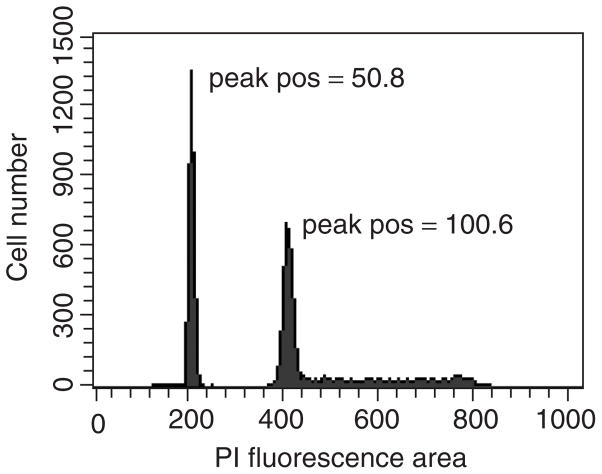
Figure 7.26.2.
Data shown are derived from parallel processing of the same human PBMCs and 1301 control cells that were used for the determination of telomere length for determination of the DNA indices as described in the Support Protocol. Relative positions of the singlet G0/1 peaks indicated are derived from histogram deconvolution using Modfit LT software, which utilizes a scale with 256-channel resolution.
In the example shown in Figure 7.26.1 an indirect staining strategy was used for dual-color cell-surface labeling utilizing purified CD8 antibody followed by goat anti-mouse Fab’2 Alexa Fluor 546 and biotinylated CD28 followed by streptavidin Alexa Fluor 488. Cy5-labeled telomere-specific probe for FISH requires red excitation but emits around 660 nm, separate from both cell-surface fluorochromes, and was used at a concentration of 10 nM. Ultraviolet-excitable HO342 was selected for counterstaining DNA, because its blue emission does not interfere with Cy5 emission or with the emissions from the Alexa Fluor fluorochromes. The 1301 cell line was used as internal control. 1301 cells have more forward and side scatter; thus, they appear in the upper right corner of the scatter plot (Fig. 7.26.1A, gate R2), and are gated separately from lymphocytes in the lower left corner (Fig. 7.26.1A, gate R1). Next, singlet resting lymphocytes are discriminated from doublets and proliferating cells on a plot of HO342 fluorescence width versus area (Fig. 7.26.1B, gate R3) and are displayed on a plot of Alexa Fluor 488 versus Alexa Fluor 546 fluorescence for identification of CD8bright+CD28+ and CD8bright+CD28− subsets (Fig. 7.26.1C). Cy5 telomere fluorescence above background for each CD8+ subset is shown in plots D and E, respectively. Telomere fluorescence of singlet 1301 control cells within gates R2 and R4 is displayed in plot F. The geometric mean of the telomere fluorescence is lower in CD8bright+CD28− cells than in CD8bright+CD28+ cells, indicating that in the sample analyzed the average telomere length in CD28− cells is indeed shorter. DNA histograms to be used for DNA ploidy analysis are shown in Figure 7.26.2 and were generated following the procedure outlined above (see Support Protocol). The DNA index of the 1301 line is calculated by dividing the mean of the G0/1 peak of the cell line by the mean of the sample peak. The value obtained is used as the correction factor for the differences in DNA ploidy of the control cell line 1301 compared to the sample (see Critical Parameters and Troubleshooting). Calculation of sample telomere length as a percentage of the telomere length of 1301 control cells (%TL) then follows the equation:
| Equation 7.26.1 |
where tel. fluor. is the geometric mean of the telomere fluorescence of the sample, bkg is the geometric mean of the sample background, DI1301 is the DNA index of 1301 cells, tel. fluor.1301 is the geometric mean of the telomere fluorescence of 1301 cells, bkg1301 is the geometric mean of the 1301 background, and DI is the DNA index of the sample.
When this equation is applied to the current example, CD8bright+CD28− cells have a mean telomere length of 7.7% compared to 10.1% for the CD8bright+CD28+ cells. Conversion of these results into TRF values requires that each laboratory determine the linear regression line by parallel measurement of samples by Flow-FISH and TRF analysis as discussed under Critical Parameters and Troubleshooting. In the authors’ laboratory the correlation equation is determined as follows (Schmid, Dagarag et al. 2002): TRF size (kb) = % TL × 0.77 + 2.02. Application of this equation to the results from data shown in Figure 7.26.1 indicates that on average, telomeres in CD8bright+CD28− cells are 2 kb shorter than those of CD8bright+CD28+ cells.
When other internal controls such as CT are used the CT telomere length can be initially calculated for a large number of samples (greater than 6) and then this value can be taken as a fixed internal value per experiment. The calculated sample telomere length can then be normalized using the measured telomere length calculated for the CTs present in the sample tube and multiplied by the expected telomere length for the CTs (Kapoor, Hakim et al. 2009).
Time Considerations
Ficoll-Hypaque separation of PBMCs will take ~2 hr, thawing of frozen samples ~1 hr. The time required for cell surface staining will depend on the number of antigens to be labeled, whether a direct or an indirect staining protocol is used, and how many samples are processed. Thus, expect staining to take from <1 hr up to 3 hr, and cross-linking ~1 hr. Preparing and counting the samples and the internal control cells for Flow-FISH, including DNA denaturation, takes ~45 min. Determination of the DNA indices requires ~30 min. The washing steps performed the next day take ~45 min and DNA staining will be completed in 2 to 3 hr. Instrument standardization and setup will vary according to the protocols and flow cytometer used. At a minimum, expect sample acquisition to take at least 5 min per sample, as a low sample differential has to be used to optimize DNA content measurements.
Acknowledgments
This work was performed in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility and was supported by National Institutes of Health awards CA-16042 and AI-28697.
Contributor Information
Theodoros Kelesidis, Email: tkelesidis@mednet.ucla.edu.
Ingrid Schmid, Email: schmid@mednet.ucla.edu.
References
- Ackermann K, V, Revell L, Lao O, Rombouts EJ, Skene DJ, Kayser M. Diurnal rhythms in blood cell populations and the effect of acute sleep deprivation in healthy young men. Sleep. 2012;35:933–940. doi: 10.5665/sleep.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew T, Aviv A, Falchi M, Surdulescu GL, Gardner JP, Lu X, Kimura M, Kato BS, Valdes AM, Spector TD. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am J Hum Genet. 2006;78:480–486. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma M, Phillips JH, Lanier LL. CD28- T lymphocytes. Antigenic and functional properties. J Immunol. 1993;150:1147–1159. [PubMed] [Google Scholar]
- Baerlocher GM, Lansdorp PM. Telomere length measurements in leukocyte subsets by automated multicolor flow-FISH. Cytometry A. 2003;55:1–6. doi: 10.1002/cyto.a.10064. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Lansdorp PM. Telomere length measurements using fluorescence in situ hybridization and flow cytometry. Methods Cell Biol. 2004;75:719–750. doi: 10.1016/s0091-679x(04)75031-1. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Mak J, Roth A, Rice KS, Lansdorp PM. Telomere shortening in leukocyte subpopulations from baboons. J Leukoc Biol. 2003;73:289–296. doi: 10.1189/jlb.0702361. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Mak J, Tien T, Lansdorp PM. Telomere length measurement by fluorescence in situ hybridization and flow cytometry: tips and pitfalls. Cytometry. 2002;47:89–99. doi: 10.1002/cyto.10053. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- Batliwalla FM, Damle RN, Metz C, Chiorazzi N, Gregersen PK. Simultaneous flow cytometric analysis of cell surface markers and telomere length: analysis of human tonsilar B cells. J Immunol Methods. 2001;247:103–109. doi: 10.1016/s0022-1759(00)00297-0. [DOI] [PubMed] [Google Scholar]
- Batliwalla FM, Rufer N, Lansdorp PM, Gregersen PK. Oligoclonal expansions in the CD8(+)CD28(−) T cells largely explain the shorter telomeres detected in this subset: analysis by flow FISH. Hum Immunol. 2000;61:951–958. doi: 10.1016/s0198-8859(00)00157-9. [DOI] [PubMed] [Google Scholar]
- Beier F, Balabanov S, Amberger CC, Hartmann U, Manger K, Dietz K, Kotter I, Brummendorf TH. Telomere length analysis in monocytes and lymphocytes from patients with systemic lupus erythematosus using multi-color flow-FISH. Lupus. 2007;16:955–962. doi: 10.1177/0961203307084299. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande P, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Borisov VI, Korolkova OY, Kozhevnikov VS. Application of flow-FISH for dynamic measurement of telomere length in cell division. Curr Protoc Cytom. 2014;69:8.14.11–18.14.10. doi: 10.1002/0471142956.cy0814s69. [DOI] [PubMed] [Google Scholar]
- Brummendorf TH, Balabanov S. Telomere length dynamics in normal hematopoiesis and in disease states characterized by increased stem cell turnover. Leukemia. 2006;20:1706–1716. doi: 10.1038/sj.leu.2404339. [DOI] [PubMed] [Google Scholar]
- Brummendorf TH, Holyoake TL, Rufer N, Barnett MJ, Schulzer M, Eaves CJ, Eaves AC, Lansdorp PM. Prognostic implications of differences in telomere length between normal and malignant cells from patients with chronic myeloid leukemia measured by flow cytometry. Blood. 2000;95:1883–1890. [PubMed] [Google Scholar]
- Cawthon RM, Smith KR, O’Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- Cherkas LF, Aviv A, Valdes AM, Hunkin JL, Gardner JP, Surdulescu GL, Kimura M, Spector TD. The effects of social status on biological aging as measured by white-blood-cell telomere length. Aging Cell. 2006;5:361–365. doi: 10.1111/j.1474-9726.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M, Lennard A, Brummendorf T, Holyoake T. Telomere shortening correlates with prognostic score at diagnosis and proceeds rapidly during progression of chronic myeloid leukemia. Leuk Lymphoma. 2004;45:1775–1781. doi: 10.1080/10428190410001693542. [DOI] [PubMed] [Google Scholar]
- Effros RB, Allsopp R, Chiu CP, Hausner MA, Hirji K, Wang L, Harley CB, Villeponteau B, West MD, Giorgi JV. Shortened telomeres in the expanded CD28-CD8+ cell subset in HIV disease implicate replicative senescence in HIV pathogenesis. AIDS. 1996;10:F17–22. doi: 10.1097/00002030-199607000-00001. [DOI] [PubMed] [Google Scholar]
- Effros RB, Dagarag M, Valenzuela HF. In vitro senescence of immune cells. Exp Gerontol. 2003;38:1243–1249. doi: 10.1016/j.exger.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Fried J, Perez AG, Clarkson B. Quantitative analysis of cell cycle progression of synchronous cells by flow cytometry. Exp Cell Res. 1980;126:63–74. doi: 10.1016/0014-4827(80)90471-1. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Hori S, Heike Y, Takei M, Maruyama M, Inoue Y, Lee JJ, Kim HJ, Harada Y, Kawai H, Shimosaka A, Kami M, Tanosaki MD, Wakasugi H, Saito S, Takaue Y, Kakizoe T. Freeze-thawing procedures have no influence on the phenotypic and functional development of dendritic cells generated from peripheral blood CD14+ monocytes. J Immunother. 2004;27:27–35. doi: 10.1097/00002371-200401000-00003. [DOI] [PubMed] [Google Scholar]
- Hultdin M, Gronlund E, Norrback K, Eriksson-Lindstrom E, Just T, Roos G. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 1998;26:3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson T, Bakhshi R, Petrova D, Pocock R, Imani M, Seifalian AM. Biological applications of quantum dots. Biomaterials. 2007;28:4717–4732. doi: 10.1016/j.biomaterials.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Kapoor V, Hakim FT, Rehman N, Gress RE, Telford WG. Quantum dots thermal stability improves simultaneous phenotype-specific telomere length measurement by FISH-flow cytometry. J Immunol Methods. 2009;344:6–14. doi: 10.1016/j.jim.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor V, Telford WG. Telomere length measurement by fluorescence in situ hybridization and flow cytometry. Methods Mol Biol. 2004;263:385–398. doi: 10.1385/1-59259-773-4:385. [DOI] [PubMed] [Google Scholar]
- Klapper W, Moosig F, Sotnikova A, Qian W, Schroder JO, Parwaresch R. Telomerase activity in B and T lymphocytes of patients with systemic lupus erythematosus. Ann Rheum Dis. 2004;63:1681–1683. doi: 10.1136/ard.2003.016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka D, Yasuda J, Yoshida K, Yoneda A, Yasuda C, Kingetsu I, Toyokawa Y, Yokoyama T, Saito S, Yamada A. Abnormal telomerase activity and telomere length in T and B cells from patients with systemic lupus erythematosus. J Rheumatol. 2006;33:1102–1107. [PubMed] [Google Scholar]
- Lansdorp PM. Stress, social rank and leukocyte telomere length. Aging Cell. 2006;5:583–584. doi: 10.1111/j.1474-9726.2006.00247.x. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- Lauzon W, Sanchez Dardon J, Cameron DW, Badley AD. Flow cytometric measurement of telomere length. Cytometry. 2000;42:159–164. doi: 10.1002/1097-0320(20000615)42:3<159::aid-cyto1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Law H, Lau Y. Validation and development of quantitative flow cytometry-based fluorescence in situ hybridization for intercenter comparison of telomere length measurement. Cytometry. 2001;43:150–153. [PubMed] [Google Scholar]
- Law HLYL. DNA index and Q Flow FISH measurement of telomere length. Cytometry. 2001;45:80. [Google Scholar]
- Martens UM, Brass V, Sedlacek L, Pantic M, Exner C, Guo Y, Engelhardt M, Lansdorp PM, Waller CF, Lange W. Telomere maintenance in human B lymphocytes. Br J Haematol. 2002;119:810–818. doi: 10.1046/j.1365-2141.2002.03910.x. [DOI] [PubMed] [Google Scholar]
- Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- Monteiro J, Batliwalla F, Ostrer H, Gregersen PK. Shortened telomeres in clonally expanded CD28-CD8+ T cells imply a replicative history that is distinct from their CD28+CD8+ counterparts. J Immunol. 1996;156:3587–3590. [PubMed] [Google Scholar]
- Mosiman VL, Patterson BK, Canterero L, Goolsby CL. Reducing cellular autofluorescence in flow cytometry: an in situ method. Cytometry. 1997;30:151–156. [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q, Baerlocher G, Vulto I, Lansdorp PM. Telomere length in human natural killer cell subsets. Ann N Y Acad Sci. 2007;1106:240–252. doi: 10.1196/annals.1392.001. [DOI] [PubMed] [Google Scholar]
- Pinto LA, Trivett MT, Wallace D, Higgins J, Baseler M, Terabe M, Belyakov IM, Berzofsky JA, Hildesheim A. Fixation and cryopreservation of whole blood and isolated mononuclear cells: Influence of different procedures on lymphocyte subset analysis by flow cytometry. Cytometry B Clin Cytom. 2005;63:47–55. doi: 10.1002/cyto.b.20038. [DOI] [PubMed] [Google Scholar]
- Plunkett FJ, Soares MV, Annels N, Hislop A, Ivory K, Lowdell M, Salmon M, Rickinson A, Akbar AN. The flow cytometric analysis of telomere length in antigen-specific CD8+ T cells during acute Epstein-Barr virus infection. Blood. 2001;97:700–707. doi: 10.1182/blood.v97.3.700. [DOI] [PubMed] [Google Scholar]
- Potter AJ, Wener MH. Flow cytometric analysis of fluorescence in situ hybridization with dye dilution and DNA staining (flow-FISH-DDD) to determine telomere length dynamics in proliferating cells. Cytometry A. 2005;68:53–58. doi: 10.1002/cyto.a.20181. [DOI] [PubMed] [Google Scholar]
- Roos G, Hultdin M. Flow cytometric determination of telomere length. Cytometry. 2001;45:79–80. doi: 10.1002/1097-0320(20010901)45:1<79::aid-cyto1147>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Rufer N, Brummendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L, Schulzer M, Lansdorp PM. Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. J Exp Med. 1999;190:157–167. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- Rufer N, Reichenbach P, Romero P. Methods for the ex vivo characterization of human CD8+ T subsets based on gene expression and replicative history analysis. Methods Mol Med. 2005;109:265–284. doi: 10.1385/1-59259-862-5:265. [DOI] [PubMed] [Google Scholar]
- Schmid I, Dagarag MD, Hausner MA, Matud JL, Just T, Effros RB, Jamieson BD. Simultaneous flow cytometric analysis of two cell surface markers, telomere length, and DNA content. Cytometry. 2002;49(3):96–105. doi: 10.1002/cyto.10163. Describes the application of the procedure presented in the Basic Protocol to the simultaneous measurement of two cell surface markers and telomere length. [DOI] [PubMed] [Google Scholar]
- Schmid I, Schmid P, Giorgi JV. Conversion of logarithmic channel numbers into relative linear fluorescence intensity. Cytometry. 1988;9:533–538. doi: 10.1002/cyto.990090605. [DOI] [PubMed] [Google Scholar]
- Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- Son NH, Murray S, Yanovski J, Hodes RJ, Weng N. Lineage-specific telomere shortening and unaltered capacity for telomerase expression in human T and B lymphocytes with age. J Immunol. 2000;165:1191–1196. doi: 10.4049/jimmunol.165.3.1191. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, Aviv A, Spector TD. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela HFE, RB . Telomere measurement and replicative senescence. In: Barnett YA, Barnett CR, editors. Aging: Methods and Protocols. otowa, NJ: Humana Press; 2000. pp. 23–52. [Google Scholar]
- Van Ziffle JA, Baerlocher GM, Lansdorp PM. Telomere length in subpopulations of human hematopoietic cells. Stem Cells. 2003;21:654–660. doi: 10.1634/stemcells.21-6-654. [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- Wang SY, Hsu ML, Tzeng CH, Hsu HC, Ho CK. The influence of cryopreservation on cytokine production by human T lymphocytes. Cryobiology. 1998;37:22–29. doi: 10.1006/cryo.1998.2094. [DOI] [PubMed] [Google Scholar]
- Weng NP, Granger L, Hodes RJ. Telomere lengthening and telomerase activation during human B cell differentiation. Proc Natl Acad Sci U S A. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser M, Stadler G, Bohm E, Borth N, Katinger H, Grillari J, Voglauer R. Nuclear flow FISH: isolation of cell nuclei improves the determination of telomere lengths. Exp Gerontol. 2006;41:230–235. doi: 10.1016/j.exger.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Wiley L, Ashok D, Martin-Ruiz C, Talbot DC, Collerton J, Kingston A, Davies K, Chinnery PF, Catt M, Jagger C, Kirkwood TB, von Zglinicki T. Reactive oxygen species production and mitochondrial dysfunction in white blood cells are not valid biomarkers of ageing in the very old. PLoS One. 2014;9:e91005. doi: 10.1371/journal.pone.0091005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Hsieh SC, Li KJ, Lu MC, Yu CL. Premature telomere shortening in polymorphonuclear neutrophils from patients with systemic lupus erythematosus is related to the lupus disease activity. Lupus. 2007;16:265–272. doi: 10.1177/0961203307077155. [DOI] [PubMed] [Google Scholar]