Abstract
Unequivocal evidence for pluripotency in which embryonic stem cells contribute to chimeric offspring has yet to be demonstrated in human or nonhuman primates (NHPs). Here, rhesus and baboons ESCs were investigated in interspecific mouse chimera generated by aggregation or blastocyst injection. Aggregation chimera produced mouse blastocysts with GFP-nhpESCs at the inner cell mass (ICM), and embryo transfers (ETs) generated dimly-fluorescencing abnormal fetuses. Direct injection of GFP-nhpESCs into blastocysts produced normal non-GFP-fluorescencing fetuses. Injected chimera showed >70% loss of GFP-nhpESCs after 21 h culture. Outgrowths of all chimeric blastocysts established distinct but separate mouse- and NHP-ESC colonies. Extensive endogenous autofluorescence compromised anti-GFP detection and PCR analysis did not detect nhpESCs in fetuses. NhpESCs localize to the ICM in chimera and generate pregnancies. Because primate ESCs do not engraft post-implantation, and also because endogenous autofluorescence results in misleading positive signals, interspecific chimera assays for pluripotency with primate stem cells is unreliable with the currently available ESCs. Testing primate ESCs reprogrammed into even more naïve states in these inter-specific chimera assays will be an important future endeavor.
Introduction
Pluripotency is now recognized as a spectrum of biological plasticity rather than an ‘on–off’ toggle switch, and criteria for assaying pluripotency range from the most demanding through to less stringent criteria. Certainly, the gold standard assay involves chimera in which pluripotent stem cells, both embryonic stem cells (ESCs); (Lallemand and Brulet, 1990; Nagy et al., 1990; Wood et al., 1993) and more recently PSCs (pluripotent stem cells); (Takahashi et al., 2006; Okita et al., 2007; Wernig et al., 2007) have contributed to both offspring and germ cells after transfer of either normally fertilized embryos or embryos generated using tetraploid complementation (Nagy et al., 1990; Eakin and Behringer, 2003). ESCs are colonies of self-renewing pluripotent cells that demonstrate the ability to differentiate into all three germ layers in the adult body (Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1998). These and other PSCs promise therapeutic applications for human disorders and diseases, and contribute further scientifically as research resources for discovering the fundamental mechanisms of human development and differentiation (reviewed by Riazi et al., 2009). Notwithstanding their potential medical importance, ethical constraints prohibit vital experiments to determine the safety, efficacy and therapeutic potentials of human embryonic stem cells (hESCs); (Daley et al., 2007). Compelling arguments for prohibiting the use of human induced pluripotent stem cells (hiPSCs) in reproductive cloning in chimera have been published (Lo et al., 2010), as have thoughtful considerations of the biological merits and ethical constraints regarding using human: animal chimera for biomedical research (Hyun et al., 2007; Behringer, 2007; Lensch et al., 2007). Consequently, there exist strong rationales for determining the full extent of pluripotentiality, as well as the biological limitations of human- and non-human-primate cells referred to as ‘pluripotent.'
Clinical extrapolations in stem cell medicine rest on the solid scientific foundations of a quarter-century of investigations using mouse embryonic stem cells (mESCs) (reviewed by Evans, 2005). Yet several major concerns remain that cannot be readily answered by studying hESC cell lines in vitro or transplanted into relatively short-lived immunocompromised mice. These questions include whether nhpESCs have full pluripotency as assayed in nonhuman primate (NHP) chimeras, whether differentiated cells remain committed after transplantation and whether ESCs can proliferate or migrate uncontrollably. Recently, important findings have been reported regarding PSC differentiation (Boyd et al., 2008; Trounson, 2006; Vaca et al., 2006; Mizuseki et al., 2003; Elkabetz et al., 2008; Kawasaki et al., 2002; Nakatsuji et al., 2008; Stadtfeld et al., 2008), therapeutic improvements after transplantation (Takahashi, 2006; Takagi et al., 2005); histocompatibility assays (Dighe et al., 2008; Rajesh et al., 2007); and epigenetics (Rugg-Gunn et al., 2005a, 2005b; Zhang et al., 2007; Rugg-Gunn et al., 2007; Fujimoto et al., 2005, 2006; Mitalipov et al., 2007; Mitalipov, 2006). Lastly, the breakthrough discoveries of inducing pluripotency (iPS); (reviewed by Yamanaka, 2008) using human, nonhuman primate (Liu et al., 2008), and mouse cells have dramatically elevated the importance of pluripotency assays for both fundamental developmental biology as well as medicine. Importantly, iPSCs from mice have been demonstrated to result in germ line transmission in both chimeric embryo assays (Okita et al., 2007, 2008; Wernig et al., 2008) as well as in tetraploid complementation experiments (Meissner et al., 2007).
ESC pluripotency has most convincingly been demonstrated in reaggregated embryos where the resultant offspring have ESC contributions to all germ layers and tissues, including the germ line (reviewed by Rossant, 2001). Thus far, only mouse and rat embryonic stem cells (mESCs) aggregated with mouse or rat embryos result in offspring born with demonstrated ESC contribution to all three germ layers and the germ line (Iannaccone and Jacob, 2009). Currently, hESC differentiation is assayed by embryoid bodies (EBs) or teratomas and both contribute to all three germ layers (Conley et al., 2005), but these technologies have limitations—EBs do not mimic 3D axial morphogenesis in vitro accurately and teratomas are a foreign environment that do not produce germ cells. Overwhelming ethical concerns obviously preclude interspecific chimera attempts with hESCs. However, the derivation of nonhuman primate ESCs (nhpESCs) can responsibly bridge gaps in our scientific knowledge between mESCs and hESCs, for example, in the generation of chimeric nonhuman primates with nhpESCs cells, although issues with NHP embryo availability, cost, and complex technical obstacles with chimera production remain (Takada et al., 2002; Schramm and Paprocki, 2004; Scott, 2006; Roberts, 2005). Thus, Mitalipov et al. (Mitalipov et al., 2006) had previously demonstrated that while derived GFP-expressing rhesus pluripotent ESCs injected into 4-to-8-stage fertilized rhesus embryos would incorporate into the trophectoderm and ICM cells of the expanded blastocyst grown in vitro, efforts to produce a chimeric monkey after embryo transfer did not succeed.
Interspecies chimeras have been advanced as an alternative process for exploring early human developmental processes and helping address the basic embryology of hESCs and their potential applications in cell-based therapies (James et al., 2006). The mouse is the best-characterized mammalian model and perhaps a logical choice for studying interspecies chimera: there is an abundance of cheap embryos and recipients; an enormous background literature on mESCs already exists; and mouse–mouse chimeras are well established with regards to genetics, strains and proven techniques. Furthermore, mESCs are unencumbered by the material transfer agreement (MTA) restrictions currently imposed on all NIH Registry hESC lines that explicitly prohibit their use in animal chimera production. Taken together, the answers obtained from testing interspecies mouse–NHP chimeras in utero, after chimeric embryo transfer, and in vitro might provide new and important information on the developmental potentials of embryonic stem cells. In addition, if successful, these intraspecific chimera would open innovative methods for preserving germ lines from endangered species (Songsasen and Wildt, 2007; Pukazhenthi et al., 2006) as well as specialty biomedical research models (Yang et al., 2008; Chan and Yang, 2009).
Here, we explore mouse–nhpESC chimeras produced with classic mouse embryo aggregation or blastocyst injection techniques (Nagy, 2003). Using GFP-expressing rhesus and baboon nhpESCs, we demonstrate that nhpESCs associate with the ICM in expanded mouse blastocysts, but rarely proliferate after outgrowth experiments and do not intermingle with mouse tissues, as determined by in vitro analysis. Furthermore, we show that chimeric mouse–nhpESC blastocysts transferred to pseudopregnant mouse recipients produce fetuses but without detectable contribution from the GFP-expressing nhpESCs, as ascertained by immunohistochemical, PCR and MRI analysis. Collectively, we conclude that interspecies chimera between distant mammals is unfavorable for studying the full pluripotency of primate ESCs, lending intellectual support for intraspecific primate chimeric experimentation.
Results
The rhesus male line nhpESC 2706 was particularly robust following transduction with the EF1α-GFP transgene and could be traced in mouse chimera tissues using monkey-specific primers to the SRY gene. Supplemental Table S1 summarizes the various stem cell lines employed for preparing injection- or aggregation-produced interspecies mouse chimeras. All of the nhpESCs employed were low passage colonies (range: 7–51) of good ESC morphology (Fig. S1A) and maintained their pluripotent characteristics following transduction with various GFP transgenes (Fig. S1B), as determined by ‘stemness’ (Fig. S1C; Table S1) and cell surface marker expression (Table S1), as well as their ability to produce teratomas when injected into NOD-SCID strain mice. Additionally, spontaneous differentiation of GFP-expressing nhpESC colonies in vitro did not silence the transgene, providing confidence that the primate cells would maintain GFP detection within interspecific chimera construction following embryo transfer (Fig S1D–I). Control intraspecific chimeras were produced by a yellow fluorescent variant of R1 mESCs (7ACS/EYFP; ATCC; Manassas, VA) that was germline-competent and stained positive for pluripotency markers (Hadjantonakis et al., 2002).
GFP-expressing Rhesus nhpESC lines (Table 1) used in the aggregation chimera assay produced expanded mouse blastocysts with GFP-expressing nhpESC lines 2706 and 3006 associated with the blastocyst inner cell mass (ICM) cells. We typically combined mouse zona pellucida-free 2-to-8-cell stage embryos with Rhesus or Baboon GFP-expressing nhpESCs (Rhesus: Fig. 1A, arrow) in a depression well and cultured them in vitro until the blastocyst stage (see Figs. S6–S8). After fixation, confocal optical sectioning (Baboon: Fig. 1B: differential interference contrast (DIC); 1C: Hoechst DNA, blue) demonstrated direct fluorescence detection of baboon GFP-expressing nhpESCs within the ICM (Fig. 1D: GFP, green, arrowheads). Here, no GFP-expressing baboon nhp 2706ESCs cells were observed in the outer trophectodermal cells, as demonstrated with a trophectoderm-specific antibody, CDX2 (Fig. 1E: red; arrow, ICM; composite image, Fig. 1F). We observed nearly 27% (129/479) of expanded chimera blastocysts with GFP-expressing Rhesus or Baboon nhpESCs exclusively in the mouse ICM and another 11% (55/479) with GFP-expressing Rhesus or Baboon nhpESCs in both the ICM and trophectoderm. The number of GFP-expressing nhpESCs associated with the mouse ICM was variable, with a majority showing 2–5 NHP cells within the ICM of expanded blastocysts.
Table 1.
Summary of mouse–nhp interspecies chimera fetus production.
| [a] |
[b] |
[c] |
[d] |
[e] |
[f] |
[g] |
[h] |
[i] |
|---|---|---|---|---|---|---|---|---|
| Chimera type |
GFP ESC cell line |
Recipient type |
Total embryo #'s transferred (# of trials) |
Total implantation sites [IP] (% of ET) |
# Normal fetuses recovered (% of total IP) |
# Abnormal fetuses recovered (% of total IP) |
# of reabsorbed/ empty sacs (% of total IP) |
# GFP, YFP or bang particle normal fetuses (% of total IP) |
| Mouse 2N blastocyst- injection |
Rhesus nhp2706 |
ICR | 147 (9) | 56 (38) | 43 (77) | 8 (14) | 5 (9) | 0 |
| Rhesus nhp106 |
ICR | 60 (3) | 25 (42) | 25 (100) | 0 | 0 | 0 | |
| Baboon ESC-4 |
NOD-SCID | 32 (4) | 8 (25) | 5 (63) | 0 | 3 (38) | 0 a | |
| Mouse ESC- YFP |
ICR | 48 (4) | 16 (33) | 12 (75) | 0 | 4 (25) | 5 (31) | |
| Mouse 2N embryo- aggregation |
Rhesus nhp2706 |
ICR | 39 (4) | 37 (95) | 7 (19) | 19 (51) | 11 (30) | 0 b |
| Rhesus nhp 3006 |
ICR | 18 (1) | 18 (100) | 2 (11) | 6 (33) | 10 (56) | 0 b | |
| Mouse ESC- YFP ± bang particles |
ICR | 42 (4) | 14 (33) | 6 (43) | 0 | 8 (57) | 5 (36) | |
| Mouse 4N- injection |
Rhesus nhp2706 |
ICR | 9 (1) | 9 (100) | 0 | 0 | 9 (100) | 0 c |
| Rhesus nhp106 |
ICR | 7 (1) | 0 | 0 | 0 | 0 | 0 | |
| Rhesus nhp3006 |
ICR | 7 (1) | 0 | 0 | 0 | 0 | 0 |
2/4 recipients died prior to fetal analysis on day E12 post transfer. 1 reabsorbing embryo expressed nhpESC-GFP cells.
9/25 abnormal fetal tissues demonstrated surviving nhpESC-GFP cells.
5/9 reabsorbing or empty sacs demonstrated surviving nhpESC-GFP cells.
Figure 1.
Generation of mouse×nhpESC-GFP interspecies aggregation chimera. A: a ‘sandwich’ mouse aggregation chimera prepared by mixing a small clump of GFP-expressing nhp2706 ES cells (green; arrow) with two 2-cell mouse embryos in a depression well (arrowheads). B–F: confocal image of a fixed interspecies aggregation chimera (B: DIC; and C: DNA) produced with GFP-expressing BabESC-4, showing localization of BabESCs (D: green, arrowheads) at the mouse ICM [B,C,E: arrows; E: cdx-2, a trophectoderm specific marker, red] but not in the outer trophectodermal cells (F: merged imaged). Similar aggregation chimera w Bar=20 μm.
Time-lapse video microscopy (TLVM) was used to investigate intraspecific and interspecific chimera formation in vitro (Figs S6–S8). After 24–48 h of aggregation within a depression well, compacted embryos with adhering mouse or Rhesus nhp ESCs were collected and prepared for TLVM recording for development to the expanded blastocyst stage. In intraspecific chimera (mouse×YFP-expressing mouse ESCs), video evidence suggested that the adhering mouse YFP-ESC (Fig. S6A–B) would integrate into the mouse during periods of blastocoel expansion, often as trophectoderm cells were undergoing division. Interestingly, the expanding mouse blastocyst did not collapse during the YFP-ESC incorporation phase, perhaps suggesting that the mouse embryo and mouse ESCs share similar cell surface signaling molecules that could mediate aggregation. This observation was similar to control blastocysts lacking the zona pellucida (not shown), but distinct from observations reported in other rodents (Gonzales et al., 1996).
Immunocytochemistry analysis of fixed aggregation chimera at the end of the TLVM showed evidence of successful YFP-mouse ESC incorporation near the site of the mouse ICM (Supplemental Fig. S6C–F). We next explored interspecific aggregation chimera formation produced by combining mouse embryos with unlabeled Rhesus nhp 2706 ESC line. In aggregation chimera that failed to incorporate the nhpESCs, the mouse morula stage rapidly expands into a fully expanded blastocyst. No evidence of breaching the trophectoderm or incorporation of nhp2706 ESCs into the embryo proper was observed (Fig. S7). However, incorporation of nhp 2706 ESCs into the expanding mouse blastocyst appeared to occur just after a rapid blastocyst collapse (Fig. S8D–E and 8I–J, arrowheads). These cells appeared to move rapidly towards the animal pole, or ICM region. Blastocyst collapse is perhaps caused by the breeching and interjection of nhpESCs through the tight junction of the adhering outer mouse trophoblast cells.
We transferred mouse 2N embryo aggregation chimera produced with GFP-expressing nhp 2706 or 3006 ES cell lines into ICR strain recipients to investigate fetal contributions in vivo (Table 1). Of 57 total aggregation chimera transferred to 5 recipients, we observed 55 (96%; Table 1, column e) implantation sites but only 9 (16%; Table 1, column f) normal fetuses. The vast majority of the tissues recovered were either abnormal (25 total; 45%; Table 1, column g) or being reabsorbed (21 total; 38%; Table 1, column h). None of the fetuses recovered demonstrated GFP-expressing nhpESCs (Table 1, column i). Microscopic analysis of the fetal tissues recovered from embryos produced with rhesus GFP-expressing nhp2706 cell line showed many instances of axial abnormalities (head–trunk: Fig. 2A) and delayed fetal development (Fig. 2C) but no detectable GFP-expressing cells (Fig. 2B–D). In several instances, surviving GFP-expressing 2706 ES cells were observed in reabsorbing implantation tissues (Fig. 2E, brightfield; 2F, GFP, green, arrowheads). Thus, aggregation chimeras with GFP-expressing nhpESCs produced mosaic blastocysts with varying number of GFP cells associated with the mouse ICM and high numbers of abnormal fetuses following embryo transfer to pseudopregnant recipients.
Figure 2.
Developmental abnormalities in interspecies aggregation chimera at E10.5 prepared with GFP-expressing nhp 2706 ESCs. A–B: head–trunk axial deformity (A: BF; arrowheads, A: anterior head region; P: posterior trunk region) in an aggregation chimera. No GFP expression was observed (B: green). C–D: severely delayed embryonic development (C: BF), but no discernible GFP expression (D: green). The tail region was slightly damaged during dissection from the decidual sac. E–F: an implantation site without a definable embryo (E: BF) but with a few GFP expressing cells (F: green, arrowheads). BF: brightfield optics; A: anterior head region; P: posterior trunk region. Bars=500 μm.
Next, we explored interspecies chimera after GFP-expressing nhp 2706 or 106 ESCs were injected into the expanded mouse blastocysts. We first determined the survival of injected nhp2706 ESCs within the mouse blastocoel niche. Chimeras were produced by microinjecting a known number of GFP-expressing nhp2706 ESCs into expanded mouse blastocysts (Fig. S2A), placing the GFP-expressing cells adjacent to the mouse ICM (Fig. S2B). As shown, within 4–6 h, the re-expanded mouse blastocysts (Fig. S2C) demonstrated GFP-expressing nhp2706 ESCs in the blastocoel, some localized at the mouse ICM (Fig. S2D: GFP, green). However, only 43% [10/23] of injected blastocysts retained any GFP-positive nhp2706 ESCs after 21 h of in vitro culture and fluorescent analysis of surviving GFP-nhp2706 ESCs revealed >70% loss of the total number of cells. We then performed embryo transfers of injected interspecies chimeric blastocysts to pseudopregnant ICR or NOD-SCID mice after using either rhesus or baboon GFP-expressing ESCs. We observed several implantation sites (ICR: 81/207 [39%]; NOD-SCID: 8/32 [25%]; Table 1, columns d–e) and a high percentage of normal E10.5 fetuses at recovery (ICR: 68/81 [84%]; NOD-SCID: 5/8 [63%]; also Fig. S3). However, none of the normal fetuses expressed GFP (Table 1; column i; Fig. S3). Conversely, from 48 embryo transfers using control intraspecific chimeric blastocysts produced with YFP-expressing mESCs, 16 implantation sites (33%; Table 1, column e) and 12 normal fetuses (75%; Table 1, column f) were recovered, with 5 fetuses expressing YFP (31%; Table 1, column i; see also, Fig. S3). Microscopic analysis of injection chimeric embryos produced with GFP-expressing Rhesus nhp 2706 ESCs (Fig. 3) or GFP-BabESC4 cell lines (Fig. S4) in either ICR or NOD-SCID recipients demonstrated no GFP expression in the tissues of recovered normal fetuses (Rhesus 2706: Fig. 3A–B; BabESC-4: Fig. S4A–B). Various abnormal embryos were largely negative for GFP detection also (Rhesus 2706: Fig. 3A–B), although occasional GFP ‘dots’ were observed in some recovered abnormal tissues (Rhesus 2706: Fig. 3E, brightfield; 3F, GFP, green, arrowheads; Fig. S4E, brightfield; Fig. S4F, GFP, green, arrowhead; BabESC-4: Fig. S4C, brightfield; S4D: GFP, green, arrowheads). Mouse intraspecific embryos produced with YFP-expressing mESCs and transferred to pseudopregnant recipients showed extensive fluorescence throughout the E10.5 day fetus (Fig. 3G–H; YFP, yellow).
Figure 3.
Chimeric blastocyst injection embryos at E10.5 day post embryo transfer. A–B: Normal embryo produced from a mouse×GFP nhp2706-ESC injection chimera (A: BF), but without GFP expression (B: green). C–D: An abnormal mouse embryo (C: BF) derived from the transfer of a mouse×GFP-expressing nhp2706-ESCs injection chimeric. No GFP expressing cells are seen in the disorganized tissue (D: green, GFP). E–F: a reabsorbing mouse embryo (E: BF) derived from a mouse×GFP-expressing nhp2706-ESCs injection chimeric. A few GFP-expressing cells are observed in the fetus (F: green, arrowheads). G–H: Control chimeric embryo derived from a mouse×YFP-mouse ESCs blastocyst transfer into an ICR recipient. The normal embryo (I: BF) expresses YFP in many tissues (J: YFP). BF: bright field; GFP: green fluorescent protein. All embryo transfer were performed in ICR recipients. All chimera were produced with GFP-expressing rhesus 2706 male ESC line. Bar=500 μm.
Mouse–nhpESC chimeras were also prepared using the mouse tetraploid complementation assay (Nagy, 2003). We electrofused mouse 2-cell embryos to produce 4N embryos and permitted these to develop to the expanded blastocyst stage before injecting them with GFP-expressing nhpESCs and performing embryo transfers to ICR pseudopregnant recipients. Harvest of fetal material around E12.5 showed high implantation sites with chimera prepared with nhpESC 2706 ESCs (Table 1, column e), but no normal fetal development and mostly necrotic or reabsorbing implantation sites upon sacrifice (Table 1, column h).
To analyze mouse–nhpESC chimeras at the cellular level, a few normal E12.5 fetuses were selected for immunohistochemical analysis (Fig. S5). We counterstained 10 μm sections with anti-GFP antibody to compare with any detected GFP expression. Preliminary analysis of ectoderm (spinal cord tissue: Fig. S5A1), mesoderm (pericardial tissue: Fig. S5B1) and endoderm (urogenital tissue; Fig. S5C1) layers in an interspecies fetus suggested extensive survival of GFP-expressing cells that co-localized precisely with anti-GFP staining (Fig. S5A2, S5B2, and S5C2). However, control tissue sections from a fertilized E12.5 day mouse embryo suggested extensive autofluorescence following fluorescein and rhodamine excitation in a variety of tissues, rendering fluorescent analysis unreliable (fluorescein excitation: Fig. S5A3, S5B3, and S5C3; rhodamine excitation: Figs. S5A4, S5B4, and S5C4). Efforts to control for endogenous fluorescence by using various blocking agents prior to application of primary and secondary antibodies were not successful (our unpublished data).
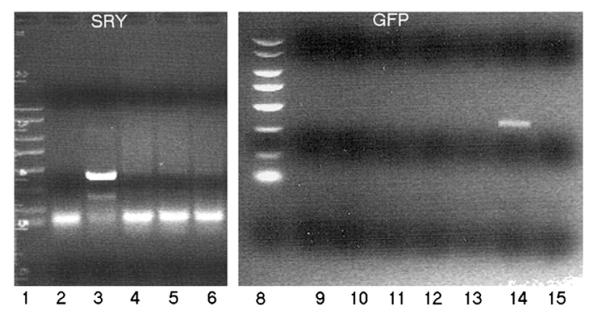
On selected interspecies embryos produced with GFP-expressing rhesus nhp2706 ESCs (a male line), we explored if SRY and GFP DNA could be detected by PCR analysis (Fig. 4). For a positive control, we used DNA isolated from a transgenic male monkey carrying the GFP transgene (ANDi) (Chan et al., 2000), demonstrating the detection of SRY DNA (Fig. 4, lane 3) and GFP DNA (Fig. 4, lane 14). However, no DNA from the embryonic tissues of these interspecies chimeras produced positive bands with primers from either SRY (Fig. 4, lanes 4–6) or GFP (Fig. 4, lanes 9–13), suggesting that no nhpESCs had survived in the developing mouse fetuses. Analysis of an interspecies chimeric blastocyst outgrowth produced with GFP-expressing nhp2706 ESCs also did not detect SRY DNA after 1 month in culture (Fig. 4, lane 2). This particular colony did not demonstrate GFP expression in surviving cells after a few days of culturing in vitro.
Figure 4.
PCR analysis of SRY (Left) and GFP (Right) DNA in embryos derived from mouse blastocyst injected with GFP nhp ESC 2706 male line. Lane 1: DNA marker; lane 2: chimeric outgrowth without GFP positive cell expression; lane 3: positive DNA control (transgenic monkey cells from ANDi); lanes 4–6: embryos derived from an injection chimera attempt (mouse×GFP nhpESC 2706 cell line); lane 8: DNA marker; lanes 9–13: embryos derived from an injection chimera (mouse×GFP nhpESC 2706 cell line); lane 14: positive DNA control (ANDi cells); lane 15: blank.
Finally, we investigated GFP-expressing nhpESC survival, proliferation and integration with mouse cells in vitro following blastocyst outgrowth on sterile coverslips (Fig. 5). Chimeric injection blastocysts outgrown for 3 days showed that GFP-expressing cells remained largely clustered together without significant intermixing with mouse cells (Fig. 5A–C). Likewise, aggregation chimeric blastocyst outgrown for 17 days in vitro (Fig. 5D–F) demonstrated that while the GFP-expressing nhpESCs proliferated over the 2 weeks in culture, the nhpESCs did not integrate into the mouse ICM cellular area (Fig. 5D–F, * indicates mouse differentiated cells derived from the mouse ICM). Regardless, the survival of pluripotent, GFP expressing nhpESCs in mouse chimeric blastocyst outgrowths were low (~2.5%) overall.
Figure 5.
Interspecies chimeric outgrowth. A–C: Confocal imaging of a mouse blastocyst injected with male GFP-expressing nhp2706 ESCs at day 3 post-outgrowth. The GFP-nhpESCs remain clustered together (A: green) without intermixing with mouse ICM cells. B: Hoechst DNA; C: merged image. D–F: Sequential fluorescent and Hoffman Modulation Contrast (HMC) images of a mouse aggregation chimeric blastocyst produced with male GFP nhp2706 ESCs. At day 2 post outgrowth, the chimeric blastocyst attaches onto the MEF feeders with extensive GFP nhpESCs at the mouse ICM (D: green). On day 8 post-outgrowth (E), the expanding mouse ESCs (E: *) are distinctly separate from the expanding GFP-nhpESCs (E: green; inset, details of GFP-nhpESCs). On day 17 days post-outgrowth, rapid proliferation of the GFP-expressing nhpESCs along a distinct border of the largely differentiated mouse ESC colony (*) is observed, with little intermixing of the mouse: monkey ESCs. Mag=100×; Bar=20 μm.
Discussion
The contribution of ESCs and other PSCs to chimeric offspring resulting in germ-line transmission is the most stringent assay for demonstrating biological pluripotency (reviewed by Behringer, 2007). This chimera assay has resulted in significant insights into the various categories of PSCs, even with mice, since embryonal carcinoma, embryonic germ, ESCs and PSCs generated by induced pluripotency all pass this test, whereas stem cells from epiblasts do not (Tesar et al., 2007; Brons et al., 2007). Epiblast SCs, in which Lif signaling was introduced transgenically (Bao et al., 2009), were shown to have regained the ability to participate in chimeric development and transmit to the germ-line, demonstrating that the loss of this signaling cascade during post-implantation development results, in part, with this diminishment of pluripotency.
Chimera have been generated in the lab exclusively as interspecific chimera between mouse species (Rossant and Frels, 1980) and recently rats (review by Iannaccone and Jacob, 2009). Even within the rodent family, intergeneric chimera between mice and voles did not succeed (Mystkowska, 1975a, 1975b). In domestic species, intergeneric chimera were first generated between sheep and goat embryos more than sixty years ago (Warwick and Berry, 1949) while interspecific chimera between European and indigenous Asian cattle have also been generated (Williams et al., 1990). Pregnancies generated by the ovine–caprine intergeneric chimera succeed to term but only at low frequencies (Gustafson et al., 1993; Jaszczak et al., 1999) demonstrating the loss of chimera proportions as these animals age post-natally.
Among the several rationales for this study, four are most prominent. First, it is important to understand the developmental biology of embryonic stem cells, as well as other lines now classified within the constellation of pluripotent stem cells. While the fundamental science of this field is on firm foundations with the decades of confirmed reports using mouse ESCs, results in other species, including humans and other primates, rest on less sure footings. Related to this point, the enormous expansion of the PSC field as well as understandable regulatory constraints on using hESC chimera assays has resulted in the proliferation of numerous alternative pluripotency assays with various degrees of leniency. Indeed, if pluripotency is viewed as a scale in which assays are rated from greatest stringency to most permissive, then germ-line transmission in tetraploid embryo complementation would be considered at the most reliable. Perhaps less stringent would be fertilized embryo chimera. Owing to the interest in human ESCs, in which only one group has reported chimeric assays (James et al., 2006), teratoma assays serve as the most stringent test for pluripotency in which tissues from the three germ layers are examined. Notwithstanding the practicality of these teratoma assays, organogenesis and patterning are chaotic and the extent of germ layer contributions is rarely quantified. Embryoid bodies and in vitro differentiation, either spontaneous or directed, are perhaps mid-scale on this pluripotency assay ruler. The detection of pluripotency markers by fluorescence (i.e., Oct-4, NANOG, SSEAs, and Tra-1-antigens) is problematic due to problems of autofluorescence, cross-reactivity as well as non-specific expression. RT-PCR is extraordinarily sensitive which forces questions about whether minute numbers of contaminating cells might generate misleading results. Notwithstanding the power of transcriptional analysis and its potential contributions for system biology, the reliability of these in silica approaches for unequivocal demonstrations of biological pluripotency remains to be confirmed. Consequently, the prime rationale for this investigation was to determine in a relevant biological assay the post-implantation potential of nhpESCs in murine chimera.
Secondly, the field of pluripotency is rapidly influencing the design of future medical approaches. With mice, few concerns are raised as to whether a transgenic insertion of GFP might influence the outcome of experiments, thus the importance of more reliably understanding various increases in perturbations as fundamental studies move towards clinical applications. Against this background, James et al. (2006) conducted a complicated set of experiments in which they first established a unique hESC line which was free from the MTA (material transfer agreements) of the traditional stem cell supplies, since those MTAs prohibit the introduction of hESCs into the reproductive systems of mammals or the combination of hESCs with embryos for reproductive purposes. Also, they were able to conduct their investigations without federal funding restrictions that preclude these types of experiments. This study suggested that human ESCs introduced into mouse blastocysts by either aggregation or blastocyst injection survived within the mouse ICM niche and proliferated into differentiated human derivatives. Furthermore, the human ESCs were described as integrated into early embryonic mouse tissues following embryo transfer to pseudopregnant females. Notwithstanding heroic efforts in performing these investigations, questions remain regarding whether the introduced hESCs proliferated and participated in post-implantation development. Perhaps they were ‘bystanders’ surviving on the sidelines and swept up in the morphogenetic migrations. Questions have been raised as to whether the foci detected by fluorescence might even have been adventitious. Perhaps the hESC line generated from anonymously-donated clinically-discarded specimens might have been subprime owing to its origins. Consequently, we undertook these studies using embryos generated by fertile pedigreed primates for the express purpose of generating top-quality ESC lines with the best chances for full biological pluripotency.
Third, beyond fundamental and preclinical significance, the importance of chimeric assays using nonhuman primates extends into the realm of invaluable research resources. Investigations using nonhuman primates are expensive and cumbersome, yet important to bridge the gap from fundamental discoveries in mouse models to clinical investigations. Were PSCs from NHPs to turn out useful in generating chimeric offspring, a significant number of investigations could be performed in vitro with only the last confirmatory studies conducted on specialty primates, as is the case with mice. The opportunity to modify primate research resource requirements using chimera would be rather significant, especially as innovative research models (reviewed by Schatten and Mitalipov, 2009) are emerging, including transgenic primates (Yang et al., 2008; Chan et al., 2000), NHPs with discordant mitochondrial and nuclear genetics (Tachibana et al., 2009), and perhaps reproductive clones soon.
Fourth, bioethical considerations regarding chimera between human cells and animal embryos, related concerns involving transfer of human nuclei into enucleated animal oocytes [cybrids], as well as actual hybrids are topics of active debate (Chapman and Hiskes, 2008; St John and Lovell-Badge, 2007). To help ground these bioethical conversations on a firmer foundation, this investigation, using nonhuman primate stem cells chimerized into mouse embryos, was designed to address the biological feasibilities of this assay. It is important to note that in contrast to the human ESCs available, these primate lines were all generated from fertile pedigreed primates where the best quality embryos were selected from ESC derivations. Should chimeric fetuses or animals be generated using pluripotent stem cells and either interspecific or intraspecific animal embryos, then the biological foundation for this experimental manner of reproduction would underscore the recent calls to prohibit hiPSCs for reproductive cloning (Lo et al., 2010).
Here, we demonstrate that chimeric embryos generated by combining mouse embryos with nhpESCs from either rhesus or baboons are not detected after implantation, even when the primate cells localize to the ICM in both aggregated and injected embryos. Aggregation chimera display numerous nhpESCs at the mouse ICM but fetal development after embryo transfers is significantly impaired (Fig. 2; Table 1), while chimera generated by blastocyst injection have fewer nhpESC within ICM but the surviving fetuses are more developmentally normal (Fig. 3; Table 1). Consequently, we suggest that these interspecific chimeric embryos may have limited pre-clinical utility for analyzing the pluripotent status and developmental capabilities of primate ESC.
Autofluorescence in mouse tissues raises significant concerns of ‘false positive’ interpretations. In Fig. 6, we present an immunohistochemistry analysis of fetal tissues both before and after introduction of the anti-GFP antibody. The danger of premature enthusiasm from extensive mouse fetal autofluorescence is quite high (Fig. 4). However, the survival and/or participation of monkey ESCs in fetal mice were either below detection sensitivity or perhaps precluded by biological incompatibilities. PCR analysis using specific primers to the monkey SRY gene or GFP did not detect the presence of male GFP-expressing nhpESCs in the tissues of mid-stage embryos (Fig. 5). Additionally, we explored tracing ESC participation in developing mouse chimera using high magnetic field MRI microscopy. Mouse YFP-expressing ESCs pre-labeled with Bangs™ beads, superparamagnetic microparticles detectable by MRI (Shapiro et al., 2004), were used to prepare mouse chimeric blastocysts following aggregation or injection into mouse blastocysts. However, despite 38% mouse chimeric production following transfer to pseudopregnant females (Table 1), as assayed by GFP expression, we could not detect the Bangs beads by MRI microscopy as have been previously reported (Shapiro et al., 2004) imaging, perhaps owing to particle levels below MRI detectable thresholds within the tissues. Newer methods coupling transgene reporters with ferritin may be more suitable for investigating individual cell contribution to chimeric tissue and organs (Ahrens et al., 2006; Mills and Ahrens, 2009; Genove et al., 2005).
It is tempting to speculate that differences in the cellular adhesiveness between rodents and primates preclude their migration during gastrulation and beyond. It appears that ESCs prefer to adhere with cells of their own species and perhaps this specific–specific differential adhesion accounts for the results here. When outgrowths of these interspecific nhpESC–mouse chimeras are established, the murine cells appear to grow separately from the growing nhpESC ones, i.e. the surviving colonies do not intermix, but self-select to ‘like’ cells (Fig. 6). Perhaps the adhesive requirements for cells to remain attached during the morphogenetic movements at gastrulation block nhpESC contribution to the developing fetus because their association with the murine cells is too weak.
Perhaps cell cycle differences between primate and rodent pluripotent stem cells preclude primate ESCs from participation in development after implantation. The time course of development also differs significantly between rodents and primates. Blastocysts develop in mice within 3.5–4 days, whereas human blastocysts require 5–6 days and nhp primates a week or more. Perhaps differences in cell cycle influence the relative proliferation of nhpESCs within the differently timed mouse embryo. It is also worth noting that whereas mouse gestation is around three weeks, rhesus and baboons require over a half-year.
Recent evidence has shown that human ESCs have similar characteristics to mouse epiblast stem cells (EpiSCs) rather than mouse ESCs. Like hESCs, mouse EpiSCs demonstrate similar dependence on bFGF/Activin signaling, grow in flatter colony morphologies with slower growth patterns compared to mESCs, and show similarities in X-chromosome inactivation (Tesar et al., 2007; Brons et al., 2007). Also, since mouse EpiSCs do not typically produce chimera in intraspecific chimera assays, it may be that nonhuman primate ESCs will also be poor candidates for chimera production in monkey: monkey chimera attempts. However, methods are now being discovered that permit intraconvertibility of human ESCs into more murine ESC-like states (Nichols and Smith, 2009; Buecker et al., 2010; Kerr and Cheng, 2010; Xu et al., 2010). Hanna et al. (Hanna et al., 2010) recently demonstrated that hESCs can be reprogrammed into a more mouse ESC like states with regards to gene expression profiles, X chromosome inactivation in female lines, and Lif/Stat3 signaling. While human PSCs cannot be used for intraspecific chimera attempts, reprogramming nonhuman primate ESCs into this naïve state can be tested to determine if it improves interspecific chimera results for the demonstration of full pluripotentiality as well as for enhanced biomedical utility of these preclinical research resources.
Conclusions
ESCs from both baboons and rhesus integrate into the ICM of mouse blastocysts in both aggregation and injection chimera, but they are lost after implantation. This suggests that interspecies chimera may have limited pre-clinical diagnostic utility for determining the developmental potentials of cells from primates. Regardless, the likelihood that primate ESCs may participate in chimeric development in intraspecific embryos remains both with currently available nhpESCs and perhaps more successfully with naïve nhpESCs.
Material and methods
Mice
Female F1 B6D2F1 mice (Harlan Sprague Dawley, Indianapolis, IN) were hormonally superstimulated, bred to fertile males, and collected as described previously (Simerly and Schatten, 1993). ICR or NOD-SCID strain females mated to vasectomized ICR males (Harlan) produced pseudopregnant female recipients for embryo transfers (ETs) (Nagy, 2003).
ESC lines
Mouse YFP-expressing embryonic stem cells (mESCs) were obtained commercially (ATCC; Manassas, VA) and cultured as described (Hadjantonakis et al., 2002). Nonhuman primate embryonic stem cell lines were derived and maintained on mitomycin-C inactivated primary mouse embryonic fibroblasts (MEFs) on 0.1% gelatin-coated dishes as previously described (Navara et al., 2007a; Simerly et al., 2009). Supplemental Table S1 list the pluripotency marker characteristics and teratoma data outcomes for 3 transduced Rhesus lines (nhp 2706; nhp 3006; and nhp 106) and 1 baboon line (BabESC-4) employed for this study. All NhpESCs colonies were mechanically passaged weekly with culture medium changed every 48 hours. GFP transduction of nhpESC lines is described in the Supplemental data.
Interspecies chimera production
Colonies of YFP-mESCs or GFP-nhpESCs were briefly treated with 0.05% trypsin-EDTA (45 s) at 37 °C, mechanically scraped, and washed once. Aggregation chimeras were prepared by combining zona pellucida-free mouse embryos (4-to-8 cell stage) with small clumps (~10–50 cells) of transduced mouse or monkey ESCs in depression wells (Nagy, 2003) using the appropriate stem cell media. Aggregations were grown until the expanded blastocyst stage and analyzed for incorporation at the mouse blastocyst ICM using attenuated fluorescent exposure (<5 s) on a Nikon TE-300 inverted microscope.
For injection chimeras, small clumps of transduced mouse or monkey ESCs were microinjected into expanded mouse blastocysts using beveled pipettes (17 μm; Humagen, Charlottesville, VA). Injected mouse blastocysts were recovered 6 h before ETs into pseudopregnant recipients. Methods for producing tetraploid mouse chimeric embryos are provided in the Supplemental Data.
Outgrowths
The zona pellucida of aggregation- or injection-produced expanded chimeric mouse blastocysts were removed with acid Tyrode's (Specialty Media, Millipore Corporation, Bedford, Mass), recovered for 30 min, and then plated onto MEFs in rhesus stem cell media (Navara et al., 2007b). Media changes were performed every 48 h and images taken by inverted HMC and fluorescence optics.
Embryo transfers and fetal recoveries
Chimeric blastocysts produced by aggregation or injection methods were transferred into the uterine horns of day 2.5 pseudopregnant ICR or NOD-SCID females using aseptic techniques (Nagy, 2003). Analysis of fetal development in recipients was performed between E12–17.5 days. Brightfield and fluorescent photographs were taken of excised fetuses using a Nikon Digital Sight DS-5MC CCD on a Nikon SMZU dissecting scope (Nikon USA, Melville, NY). Digital images were archived using MediaView software (Molecular Devices, Sunnyvale, CA). Abnormal embryos were given GD (growth disorganization) scores as described Poland et al. (Poland et al., 1981).
Immunocytochemistry, Immunohistochemistry, PCR, and Time-lapse Video Microscopy (TLVM) details are described in the Supplemental data.
Supplementary Material
Acknowledgments
We are grateful to Tony Battelli (MWRI&F) for animal care support and thank Angela Palermo Lauff for the editorial assistance. We also thank Drs. R. Pedersen (Cambridge University) and P. Donovan (University of California—Irvine) for the advice. The support of this research to GS by grants from the National Institutes of Health (HD047675; and RR013632) is gratefully acknowledged.
Abbreviations
- PSC
pluripotent stem cells
- ESC
embryonic stem cells
- iPSC
induced pluripotent stem cells
- nhpESC
nonhuman primate embryonic stem cells
- ICM
inner cell mass cells
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10.1016/j.scr.2011.03.002.
References
- Ahrens ET, Srinivas M, Capuano S, Simhan HN, Schatten GP. Magnetic resonance imaging of embryonic and fetal development in model systems. Methods Mol. Med. 2006;124:87–101. doi: 10.1385/1-59745-010-3:87. [DOI] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer RR. Human–animal chimeras in biomedical research. Cell Stem Cell. 2007;1:259–262. doi: 10.1016/j.stem.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Boyd AS, Wu DC, Higashi Y, Wood KJ. A comparison of protocols used to generate insulin-producing cell clusters from mouse embryonic stem cells. Stem Cells. 2008;26:1128–1137. doi: 10.1634/stemcells.2007-0762. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, de Sousa Lopes S.M. Chuva, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K, Geijsen N. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Yang SH. Generation of transgenic monkeys with human inherited genetic disease. Methods. 2009;49:78–84. doi: 10.1016/j.ymeth.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Luetjens CM, Schatten GP. Sperm-mediated gene transfer. Curr. Top. Dev. Biol. 2000;50:89–102. doi: 10.1016/s0070-2153(00)50005-2. [DOI] [PubMed] [Google Scholar]
- Chapman A, Hiskes AL. Unscrambling the eggs: cybrid research through an Embryonic Stem Cell Research Oversight Committee (ESCRO) lens. Am. J. Bioeth. 2008;8:44–46. doi: 10.1080/15265160802559245. [DOI] [PubMed] [Google Scholar]
- Conley BJ, Denham M, Gulluyan L, Olsson F, Cole TJ, Mollard R. Mouse embryonic stem cell derivation, and mouse and human embryonic stem cell culture and differentiation as embryoid bodies. Curr. Protoc. Cell Biol. 2005;23 doi: 10.1002/0471143030.cb2302s28. Chapter, Unit 23 2. [DOI] [PubMed] [Google Scholar]
- Daley GQ, Ahrlund Richter L, Auerbach JM, Benvenisty N, Charo RA, Chen G, Deng HK, Goldstein LS, Hudson KL, Hyun I, Junn SC, Love J, Lee EH, McLaren A, Mummery CL, Nakatsuji N, Racowsky C, Rooke H, Rossant J, Scholer HR, Solbakk JH, Taylor P, Trounson AO, Weissman IL, Wilmut I, Yu J, Zoloth L. Ethics. The ISSCR guidelines for human embryonic stem cell research. Science. 2007;315:603–604. doi: 10.1126/science.1139337. [DOI] [PubMed] [Google Scholar]
- Dighe V, Clepper L, Pedersen D, Byrne J, Ferguson B, Gokhale S, Penedo MC, Wolf D, Mitalipov S. Heterozygous embryonic stem cell lines derived from nonhuman primate parthenotes. Stem Cells. 2008;26:756–766. doi: 10.1634/stemcells.2007-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin GS, Behringer RR. Tetraploid development in the mouse. Dev. Dyn. 2003;228:751–766. doi: 10.1002/dvdy.10363. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. Embryonic stem cells: a perspective. Novartis Found. Symp. 2005;265:98–103. discussion 103-6, 122-8. [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fujimoto A, Mitalipov SM, Clepper LL, Wolf DP. Development of a monkey model for the study of primate genomic imprinting. Mol. Hum. Reprod. 2005;11:413–422. doi: 10.1093/molehr/gah180. [DOI] [PubMed] [Google Scholar]
- Fujimoto A, Mitalipov SM, Kuo HC, Wolf DP. Aberrant genomic imprinting in rhesus monkey embryonic stem cells. Stem Cells. 2006;24:595–603. doi: 10.1634/stemcells.2005-0301. [DOI] [PubMed] [Google Scholar]
- Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat. Med. 2005;11:450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- Gonzales DS, Boatman DE, Bavister BD. Kinematics of trophectoderm projections and locomotion in the peri-implantation hamster blastocyst. Dev. Dyn. 1996;205:435–444. doi: 10.1002/(SICI)1097-0177(199604)205:4<435::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gustafson RA, Anderson GB, BonDurant RH, Sasser GR. Failure of sheep–goat hybrid conceptuses to develop to term in sheep–goat chimaeras. J. Reprod. Fertil. 1993;99:267–273. doi: 10.1530/jrf.0.0990267. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple noninvasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun I, Taylor P, Testa G, Dickens B, Jung KW, McNab A, Robertson J, Skene L, Zoloth L. Ethical standards for human-to-animal chimera experiments in stem cell research. Cell Stem Cell. 2007;1:159–163. doi: 10.1016/j.stem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Iannaccone PM, Jacob HJ. Rats! Dis. Model. Mech. 2009;2:206–210. doi: 10.1242/dmm.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blasto-cysts. Dev. Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Jaszczak K, Parada R, Guszkiewicz A. Cytogenetic study of some tissues and age-related changes in cell proportions in a goat–sheep chimera. Cytogenet. Cell Genet. 1999;84:55–57. doi: 10.1159/000015214. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Suemori H, Mizuseki K, Watanabe K, Urano F, Ichinose H, Haruta M, Takahashi M, Yoshikawa K, Nishikawa S, Nakatsuji N, Sasai Y. Generation of dopaminergic neurons and pigmented epithelia from primate ES cells by stromal cell-derived inducing activity. Proc. Natl. Acad. Sci. U S A. 2002;99:1580–1585. doi: 10.1073/pnas.032662199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CL, Cheng L. Multiple, interconvertible states of human pluripotent stem cells. Cell Stem Cell. 2010;6:497–499. doi: 10.1016/j.stem.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Brulet P. An in situ assessment of the routes and extents of colonisation of the mouse embryo by embryonic stem cells and their descendants. Development. 1990;110:1241–1248. doi: 10.1242/dev.110.4.1241. [DOI] [PubMed] [Google Scholar]
- Lensch MW, Schlaeger TM, Zon LI, Daley GQ. Teratoma formation assays with human embryonic stem cells: a rationale for one type of human–animal chimera. Cell Stem Cell. 2007;1:253–258. doi: 10.1016/j.stem.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Lo B, Parham L, Alvarez-Buylla A, Cedars M, Conklin B, Fisher S, Gates E, Giudice L, Halme DG, Hershon W, Kriegstein A, Kwok PY, Wagner R. Cloning mice and men: prohibiting the use of iPS cells for human reproductive cloning. Cell Stem Cell. 2010;6:16–20. doi: 10.1016/j.stem.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U S A. 1981;78:7634–1638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mills PH, Ahrens ET. Enhanced positive-contrast visualization of paramagnetic contrast agents using phase images. Magn. Reson. Med. 2009;62:1349–1355. doi: 10.1002/mrm.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov SM. Genomic imprinting in primate embryos and embryonic stem cells. Reprod. Fertil. Dev. 2006;18:817–821. doi: 10.1071/rd06112. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Clepper L, Sritanaudomchai H, Fujimoto A, Wolf D. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. Stem Cells. 2007;25:581–588. doi: 10.1634/stemcells.2006-0120. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Sakamoto T, Watanabe K, Muguruma K, Ikeya M, Nishiyama A, Arakawa A, Suemori H, Nakatsuji N, Kawasaki H, Murakami F, Sasai Y. Generation of neural crest-derived peripheral neurons and floor plate cells from mouse and primate embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2003;100:5828–2833. doi: 10.1073/pnas.1037282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mystkowska ET. Preimplantation development in vivo and in vitro in bank voles, Clethrionomys glareolus, treated with PMSG and HCG. J. Reprod. Fertil. 1975a;42:287–292. doi: 10.1530/jrf.0.0420287. [DOI] [PubMed] [Google Scholar]
- Mystkowska ET. Development of mouse-bank vole inter-specific chimaeric embryos. J. Embryol. Exp. Morphol. 1975b;33:731–744. [PubMed] [Google Scholar]
- Nagy ZP. Micromanipulation of the human oocyte. Reprod. Biomed. Online. 2003;7:634–640. doi: 10.1016/s1472-6483(10)62085-8. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nakatsuji N, Nakajima F, Tokunaga K. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 2008;26:739–740. doi: 10.1038/nbt0708-739. [DOI] [PubMed] [Google Scholar]
- Navara CS, Redinger C, Mich-Basso J, Oliver S, Ben-Yehudah A, Castro C, Simerly C. Derivation and characterization of nonhuman primate embryonic stem cells. Curr. Protoc. Stem Cell Biol. 2007a;1 doi: 10.1002/9780470151808.sc01a01s1. Chapter, Unit 1A 1. [DOI] [PubMed] [Google Scholar]
- Navara CS, Mich-Basso JD, Redinger CJ, Ben-Yehudah A, Jacoby E, Kovkarova-Naumovski E, Sukhwani M, Orwig K, Kaminski N, Castro CA, Simerly CR, Schatten G. Pedigreed primate embryonic stem cells express homogeneous familial gene profiles. Stem Cells. 2007b;25:2695–2704. doi: 10.1634/stemcells.2007-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Poland BJ, Miller JR, Harris M, Livingston J. Spontaneous abortion. A study of 1,961 women and their conceptuses. Acta. Obstet. Gynecol. 1981:5–32. [PubMed] [Google Scholar]
- Pukazhenthi B, Comizzoli P, Travis AJ, Wildt DE. Applications of emerging technologies to the study and conservation of threatened and endangered species. Reprod. Fertil. Dev. 2006;18:77–90. doi: 10.1071/rd05117. [DOI] [PubMed] [Google Scholar]
- Rajesh D, Chinnasamy N, Mitalipov SM, Wolf DP, Slukvin I, Thomson JA, Shaaban AF. Differential requirements for hematopoietic commitment between human and rhesus embryonic stem cells. Stem Cells. 2007;25:490–499. doi: 10.1634/stemcells.2006-0277. [DOI] [PubMed] [Google Scholar]
- Riazi AM, Kwon SY, Stanford WL. Stem cell sources for regenerative medicine. Methods Mol. Biol. 2009;482:55–90. doi: 10.1007/978-1-59745-060-7_5. [DOI] [PubMed] [Google Scholar]
- Roberts RM. Embryo culture conditions: what embryos like best. Endocrinology. 2005;146:2140–2141. doi: 10.1210/en.2005-0221. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells in the mammalian blastocyst. Harvey Lect. 2001;97:17–40. [PubMed] [Google Scholar]
- Rossant J, Frels WI. Interspecific chimeras in mammals: successful production of live chimeras between Mus musculus and Mus caroli. Science. 1980;208:419–421. doi: 10.1126/science.7367871. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Human embryonic stem cells as a model for studying epigenetic regulation during early development. Cell Cycle. 2005a;4:1323–1326. doi: 10.4161/cc.4.10.2076. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat. Genet. 2005b;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 2007;16(Spec No. 2):R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- Schatten G, Mitalipov S. Developmental biology: transgenic primate offspring. Nature. 2009;459:515–516. doi: 10.1038/459515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm RD, Paprocki AM. Strategies for the production of genetically identical monkeys by embryo splitting. Reprod. Biol. Endocrinol. 2004;2:38. doi: 10.1186/1477-7827-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CT. Chimeras in the crosshairs. Nat. Biotechnol. 2006;24:487–490. doi: 10.1038/nbt0506-487. [DOI] [PubMed] [Google Scholar]
- Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proc. Natl. Acad. Sci. U S A. 2004;101:10901–10906. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly C, Schatten G. Techniques for localization of specific molecules in oocytes and embryos. Methods Enzymol. 1993;225:516–553. doi: 10.1016/0076-6879(93)25035-z. [DOI] [PubMed] [Google Scholar]
- Simerly CR, Navara CS, Castro CA, Turpin JC, Redinger CJ, Mich-Basso JD, Jacoby ES, Grund KJ, McFarland DA, Oliver SL, Ben-Yehudah A, Carlisle DL, Frost P, Penedo C, Hewitson L, Schatten G. Establishment and characterization of baboon embryonic stem cell lines: an Old World Primate model for regeneration and transplantation research. Stem Cell Res. 2009;2:178–187. doi: 10.1016/j.scr.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songsasen N, Wildt DE. Oocyte biology and challenges in developing in vitro maturation systems in the domestic dog. Anim. Reprod. Sci. 2007;98:2–22. doi: 10.1016/j.anireprosci.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John J, Lovell-Badge R. Human–animal cytoplasmic hybrid embryos, mitochondria, and an energetic debate. Nat. Cell Biol. 2007;9:988–992. doi: 10.1038/ncb436. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic Beta cells into induced pluripotent stem cells. Curr. Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada T, Suzuki Y, Kondo Y, Kadota N, Kobayashi K, Nito S, Kimura H, Torii R. Monkey embryonic stem cell lines expressing green fluorescent protein. Cell Transplant. 2002;11:631–635. doi: 10.3727/000000002783985350. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Takahashi J, Saiki H, Morizane A, Hayashi T, Kishi Y, Fukuda H, Okamoto Y, Koyanagi M, Ideguchi M, Hayashi H, Imazato T, Kawasaki H, Suemori H, Omachi S, Iida H, Itoh N, Nakatsuji N, Sasai Y, Hashimoto N. Dopaminergic neurons generated from monkey embryonic stem cells function in a Parkinson primate model. J. Clin. Invest. 2005;115:102–109. doi: 10.1172/JCI21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J. Stem cell therapy for Parkinson's disease. Ernst Schering Res. Found. Workshop; 2006. pp. 229–244. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Ichisaka T, Yamanaka S. Identification of genes involved in tumor-like properties of embryonic stem cells. Methods Mol. Biol. 2006;329:449–458. doi: 10.1385/1-59745-037-5:449. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Trounson A. The production and directed differentiation of human embryonic stem cells. Endocr. Rev. 2006;27:208–219. doi: 10.1210/er.2005-0016. [DOI] [PubMed] [Google Scholar]
- Vaca P, Martin F, Vegara-Meseguer JM, Rovira JM, Berna G, Soria B. Induction of differentiation of embryonic stem cells into insulin-secreting cells by fetal soluble factors. Stem Cells. 2006;24:258–265. doi: 10.1634/stemcells.2005-0058. [DOI] [PubMed] [Google Scholar]
- Warwick BL, Berry RO. Inter-generic and intra-specific embryo transfers in sheep and goats. J. Hered. 1949;40:297–303. doi: 10.1093/oxfordjournals.jhered.a105963. illust. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Williams TJ, Munro RK, Shelton JN. Production of interspecies chimeric calves by aggregation of Bos indicus and Bos taurus demi-embryos. Reprod. Fertil. Dev. 1990;2:385–394. doi: 10.1071/rd9900385. [DOI] [PubMed] [Google Scholar]
- Wood SA, Allen ND, Rossant J, Auerbach A, Nagy A. Non-injection methods for the production of embryonic stem cell-embryo chimaeras. Nature. 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. U S A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41(Suppl 1):51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Zhao JG, Li H, Liu HF, Huang Y, Huang SZ, Zeng F, Zeng YT. Effect of individual heifer oocyte donors on cloned embryo development in vitro. Anim. Reprod. Sci. 2008;104:28–37. doi: 10.1016/j.anireprosci.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li J, Villemoes K, Pedersen AM, Purup S, Vajta G. An epigenetic modifier results in improved in vitro blastocyst production after somatic cell nuclear transfer. Cloning Stem Cells. 2007;9:357–363. doi: 10.1089/clo.2006.0090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.