Abstract
A combination of the herbicide paraquat (PQ) and fungicide maneb (MB) has been linked to Parkinson's disease. Previous studies show that this involves an additive toxicity with at least two different mechanisms. However, detailed understanding of mixtures is often difficult to elucidate because of the multiple ways by which toxic agents can interact. In the present study, we used a combination of transcriptomics and metabolomics to investigate mechanisms of toxicity of PQ and MB in a neuroblastoma cell line. Conditions were studied with concentrations of PQ and MB that each individually caused 20% cell death and together caused 50% cell death. Transcriptomic and metabolomic samples were collected at time points prior to significant cell death. Statistical and bioinformatic methods were applied to the resulting 30,869 transcripts and 1358 metabolites. Results showed that MB significantly changed more transcripts and metabolites than PQ, and combined PQ + MB impacted more than MB alone. Transcriptome–metabolome-wide association study (TMWAS) showed that significantly changed transcripts and metabolites mapped to two network substructures, one associating with significant effects of MB and the other included features significantly associated with PQ + MB. The latter contained 4 clusters of genes and associated metabolites, with one containing genes for two cation transporters and a cation transporter regulatory protein also recognized as a pro-apoptotic protein. Other clusters included stress response genes and transporters linked to cytoprotective mechanisms. MB also had a significant network structure linked to cell proliferation. Together, the results show that the toxicologic mechanism of the combined neurotoxicity of PQ and MB involves network level interactions and that TMWAS provides an effective approach to investigate such complex mechanisms.
Keywords: Maneb, Metabolomics, Paraquat, Parkinson's disease
1. Introduction
Human disease is complex, and etiologies often involve multiple interacting factors. Loscalzo et al. [1] discussed this in detail for sickle cell disease, in which a single genetic mutation results in different disease phenotypes. Genetic epidemiology studies have consistently provided evidence for multiple factors in disease causation, and current understanding is that only a small fraction of non-infectious disease burden can be directly attributed to a genetic cause. Willett concluded that even when specific etiologic factors remain unclear, non-genetic factors often account for greater than 80% of the total attributable risk [2]. These non-genetic factors include environmental exposures and become particularly evident when there are dramatic changes in disease rates in a country over time [2]. Wild introduced the term “exposome” to include all exposures for an individual from conception to death [3]. Examples of exposures include chemicals arising from the foods we consume, the microbiome, pharmaceuticals and supplements, consumer products, and industrial and agricultural chemicals [4]. To understand the contribution of this complex mixture of exposures to mechanisms of toxicity, approaches are needed to separate and deconvolute the effects of individual factors.
Parkinson's disease (PD) is a multifactorial disease that is characterized by the progressive loss of dopaminergic neurons of the substantia nigra pars compacta (SN). This progressive loss of dopamine input from the SN to the striatum results in degenerative loss of motor function that manifests in bradykinesia, postural instability, tremor and rigidity [5]. This disease remains idiopathic in etiology due to only 5–10% of classical PD cases that result from monogenetic mutations [5]. Epidemiological studies have identified that factors such as living in a rural area, consuming well water, farming, and pesticide exposure may be risk factors for developing PD [7]. These observations have led to the development of an “environmental hypothesis” of PD. This hypothesis states that there are chemicals in the environment that are capable of selectively damaging the dopaminergic neurons of the SN, thus contributing to the development of PD [8].
More recently, Ritz et al. have described an increased risk of PD due to pesticide exposure [9], [10], [11]. Two common pesticides, paraquat (PQ) and maneb (MB), have been demonstrated in vivo to preferentially alter the nigrostriatal dopamine system [12], [13], [14]. These two pesticides alone or in combination have also been shown to increase the risk of developing PD [15]. Our previous mechanistic studies of these two chemicals in neuroblastoma cells [16] showed that PQ and MB act via diverse mechanisms, with increased combined toxicity due to an additive effect of two toxic mechanisms.
Both transcriptomics and metabolomics are useful omic techniques in toxicology studies in the post-genomic era. In recent years, several important omics-based, toxicology-related studies on environmental contaminants in fish models have been published [17], [18], [19], [20]. In the present study, we used information-rich transcriptome and metabolome analyses with advanced computational methods to study the additive toxic mechanisms in combined MB and PQ neurotoxicity in a neuroblastoma cell line. This combined transcriptome–metabolome wide association study (TMWAS) showed multiple network-level interactions. Among the clusters of associated transcripts, two cation transporters and a functionally linked regulatory protein were increased by MB. Each component of this transport network was apparent as a hub for associated metabolites when PQ was also included. In the absence of MB, metabolite clusters were not present with PQ. PQ-related changes in metabolism linked to multiple MB-associated transcript changes showed that toxicity involves network-level interactions. The results further show that TMWAS provides an approach to study complex mechanisms of toxicity, which can, in principle, be extended to study mechanisms in more complex in vivo systems.
2. Materials and methods
2.1. Cell culture
CAD cells, mouse catecholaminergic neuronal cells, were obtained from Health Protection Agency Culture Collections (United Kingdom). Cells were cultured in DMEM/F12 (1:1) with l-glutamine, 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were treated with PQ, MB, or the combination of PQ and MB. The concentrations of pesticide used for each treatment group were as follows: PQ20 (90 μM), PQ50 (270 μM), MB20 (10.5 μM), MB50 (27 μM) or the combination of PQ and MB (90 μM PQ + 10.5 μM MB).
2.2. Gene expression array
CAD cells were cultured in 6-well plates and treated for 4 h with pesticides. At the completion of the treatment period, RNA was isolated using a RNeasy minikit from Qiagen. RNA samples (1 μg total RNA) were delivered to the Cancer Genomics Core facility at Winship Cancer Institute, Emory University where samples were assessed for quality and prepared for gene expression array analysis as per manufacturer protocol. Three biological replicates were used to investigate gene expression using a mouse WG-6 array from Illumina.
2.3. High performance metabolic profiling
CAD cells were cultured in 6-well plates and treated for 24 h with pesticides (n = 6 per treatment). Cells were washed with PBS and 200 μl of ice cold acetonitrile (ACN) was added to each well to precipitate protein and extract metabolites. Extraction conditions for the routine metabolomics workflow were derived from previous studies comparing ACN extraction and methanol extraction, which showed that these extractions provide similar quality data for the LC-FTMS method as used. The extracts were collected, debris pelleted via centrifugation and supernatants were saved for analysis. Samples were analyzed in triplicate by LC-FTMS (Accela-LTQ Velos Orbitrap; mass-to-charge ratio (m/z) range from 85 to 850) with 10 μl injection volume using a dual chromatography setup (anion exchange and reverse phase (C18)) and a formic acid/ACN gradient as described by Soltow et al. [21]. Electrospray ionization was used in the positive ion mode. Data were extracted using apLCMS [22] as m/z features, where an m/z feature is defined by m/z, RT (retention time) and ion intensity (integrated ion intensity for the peak). Each sample was run in triplicate on both columns. Putative identification of metabolites was made using the Madison Metabolomics Consortium Database (MMCD) [23] and Metlin Mass Spectrometry Database [24]. Metabolites were identified via tandem mass spectrometry (MS/MS) and matching fragmentation patterns to those of known standards.
2.4. Biostatistics and bioinformatics
Statistically significant gene expression and metabolite intensity was determined using the R package Limma [25] using a false discovery rate of 5% [26]. The sparse partial least squares (sPLS) regression method [27], [28] implemented in the R package mixOmics was used to perform integration and visualization of transcriptome × metabolome associations. sPLS is a variable selection and dimensionality reduction method that allows integration of heterogeneous omics data from same set of samples; transcriptome (matrix X) and metabolome (matrix Y) data in this case where matrix X is an n × p matrix that includes n samples and p genes and matrix Y is an n × q matrix that includes n samples and q metabolites. Variable selection for network analysis was performed by using the top 1% of metabolites and top 5% of genes from the first 5 principal components from the sPLS analysis. Venn diagrams were generated using BioVenn [29]. Gene ontology analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [30]. Significant pathway enrichment of genes and metabolites was achieved using MetaCore [31]. All statistical analyses were performed in R.
3. Results
3.1. Transcriptomic analysis
To investigate the transcriptomic signatures of PQ, MB and the combination of PQ and MB, RNA was isolated from CAD neuroblastoma cells after a 4 h incubation period. This time point was selected based upon previous findings that the transcription factor Nrf2 was activated after a 3 h treatment with PQ or MB [16] and the desire to determine gene expression alterations due to exposure rather than repair and recovery. RNA extracts were submitted to the Emory Cancer Genomics gene array facility where the samples were hybridized and analyzed using a mouse WG-6 chip from Illumina. Limma (FDR 0.05) was used to determine which transcripts differed significantly from the control (Supplementary Table S1). Results showed that PQ20 altered the expression of 9 genes while PQ50 altered 310. MB changed the expression of far more genes compared to equivalent toxic doses of PQ (MB20, 573 genes; MB50, 3008 genes). The additive toxic dose of PQ20 plus MB20 altered substantially more genes (1529 genes) than either PQ20 [10] or MB20 (573) alone. Thus, the extensive gene expression changes revealed by the transcriptome data indicate that the cell death endpoint is a relatively poor indicator of the underlying molecular processes involved in the toxic interaction.
Supplementary material related to this article can be found, in the online version, at doi:10.1016/j.toxrep.2014.07.006.
Transcripts found to be significantly altered (FDR 0.05) compared to control.
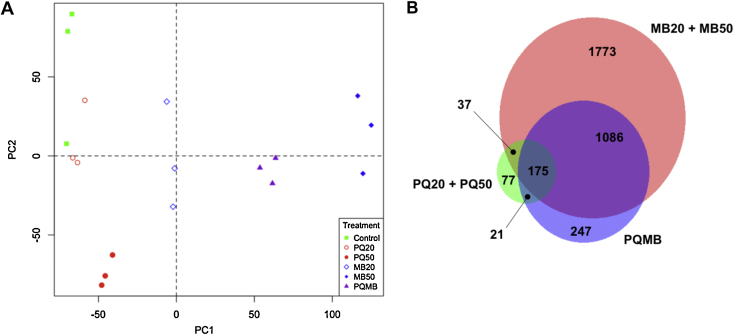
These gene expression data were subjected to principal component analysis (PCA) (Fig. 1A). The scores plot shows that two general clusters were present, with one containing control, PQ20 and PQ50, while the other included all of the MB-treated samples (MB20, MB50 and PQMB). A Venn diagram (Fig. 1B) of the unique genes altered by the treatment conditions showed that the two general clusters observed by PCA shared 233 genes while 77 were unique to the PQ-group and over 3000 were unique to the MB-group. Using traditional concepts of additive and synergistic interactions, one could consider the 233 shared genes to represent ones where additive effects could occur while genes changed by PQ alone or MB alone could represent genes contributing to synergistic effects.
Fig. 1.
Transcriptomic data reveal toxicant-specific data clustering and gene expression. Principal component analysis (A) shows two distinct groupings where control, PQ20, and PQ50 project along principal component 2 (PC2) and MB20, MB50, and PQMB project along principal component 1 (PC1) in a concentration dependent manner. Venn diagram (B) displays the number of genes common between groups as well as the number of genes unique to each toxicant treatment.
Further investigation of the specific function of these unique genes (PQ alone or MB alone) was conducted via gene ontology analysis using DAVID. No significantly enriched functions were found in the PQ-group, but the MB-group included many significantly altered functions. Some of the most significant functions included cell cycle (135 genes, q = 2 × 10−11), cell cycle processes (88 genes, q = 4.2 × 10−7), DNA metabolic processes (91 genes, q = 9 × 10−7), protein catabolism (104 genes, q = 8.4 × 10−6), response to cellular stress (84 genes, q = 8.4 × 10−6), and transcription (270 genes, q = 1.3 × 10−5). Additionally, MetaCore was used to investigate significant pathway enrichment of these genes unique to each grouping. Only one significant pathway, inhibition of ERK, was significantly enriched in the PQ-group. The genes unique to MB were significantly enriched in 139 different pathways and similar to the gene ontology investigation using DAVID; the top 6 significant pathways all involve the cell cycle and cell cycle regulation. These analyses did little to clarify the previous conclusion [16] that combined additive toxicity of PQ and MB involves combination of different toxicological mechanisms.
3.2. Metabolomic analysis
To assess alterations in the metabolome due to PQ, MB and PQ + MB treatments, cells were treated for 24 h to ensure that gene transcription, protein translation and enzymatic activity could occur and a metabolic signature could be evaluated. Metabolites were extracted using acetonitrile and these extracts were analyzed using a high performance metabolic profiling workflow as previously described [21]. Metabolites were separated using C18 chromatography and ions were detected using a LTQ-Velos Orbitrap mass spectrometer. After data extraction using apLCMS [22], the data were filtered to include only the ions (m/z) with a coefficient of variation (%CV) of less than 10%. Limma (FDR 0.05) was used with the resulting 1358 ions to select metabolites that were significantly different from control samples. Analysis of PQ-treated samples showed that PQ20 and PQ50 altered 21 and 464 metabolites, respectively. Similar to the transcriptome results, MB altered the intensity of many more metabolites compared to control (MB20, 352; MB50, 590). Importantly, the combination of PQ20 and MB20 altered 454 metabolites, greater than PQ20 [22] or MB20 (352) alone.
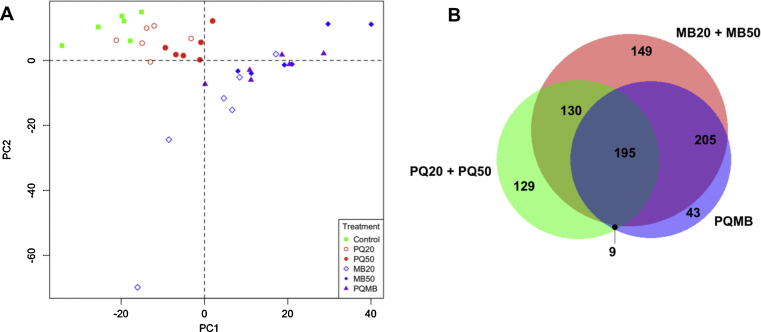
The metabolomics data were also evaluated using PCA (Fig. 2A) and provided results similar to the transcriptome data. Two distinct clusters were present that included control, PQ20 and PQ50 in one cluster and the MB treated samples (MB20, MB50, PQMB) in the other. A Venn diagram of the metabolomics data (Fig. 2B) showed that 129 m/z were unique to the PQ-group, 397 m/z unique to the MB-group and 334 m/z shared by both. The lists of m/z unique to the two groups were matched to KEGG identifiers (ID) by searching the Metlin database for the following adducts: M+H, M+Na, M+K, M+H–H2O, M+H–2H2O, M+ACN+H. These KEGG ID were then used with MetaCore for pathway enrichment analysis. Using the KEGG ID unique to the PQ group, significant enrichment was found in various pathways such cholesterol and sphingolipid transport (−Log P = 13.096), pathways of His, Glu, and Gln metabolism (−Log P = 3.962), and tyrosine metabolism (−Log P = 2.213). Pathways significantly enriched in the MB-group included estrogen biosynthesis (−Log P = 5.219) and pathways of branched chain amino acid (Leu, Ile, Val) metabolism (−Log P = 2.978). Thus, the metabolomics data were consistent with the interpretation that additive MB and PQ toxicity is due to different mechanisms of toxicity.
Fig. 2.
Metabolomic data demonstrates toxicant-specific clustering and metabolite patterns. Principal component analysis (A) shows two distinct groupings where control, PQ20, and PQ50 project along principal component 1 (PC1) and MB20, MB50, and PQMB project along both PC1 and PC2 in a concentration dependent manner. Venn diagram (B) displays the number of genes common between groups as well as the number of genes unique to each toxicant treatment.
3.3. Integration of transcriptome and metabolome
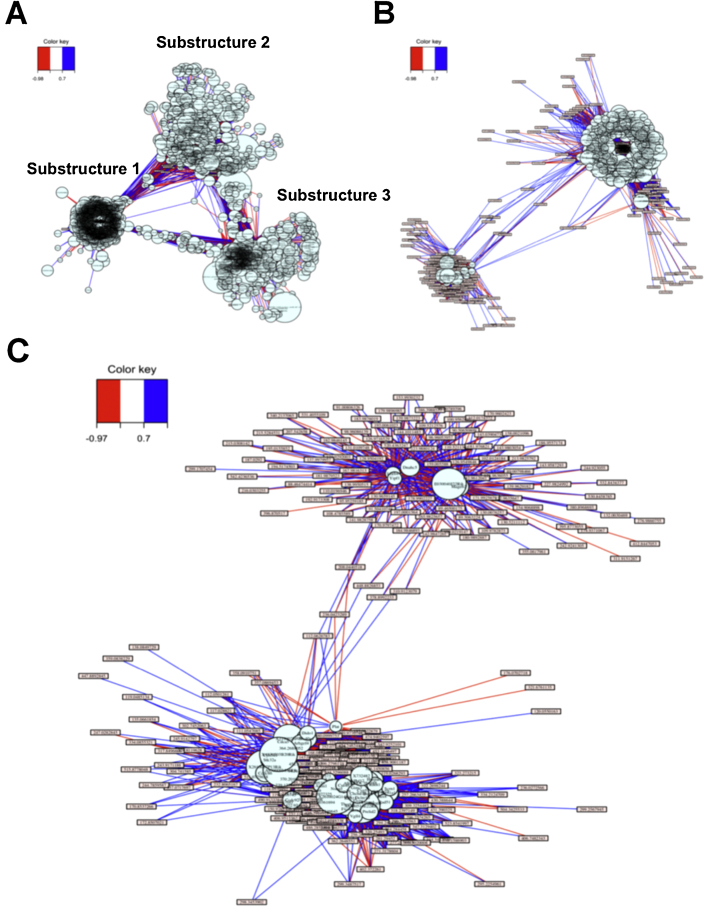
To investigate the gene and metabolite interactions occurring due to MB and paraquat interaction, we examined interactions for the PQ20, MB20 and PQMB transcriptome and metabolome data using the sPLS method implemented in the mixOmics R package (details in Section 2). An association network comprising of 340 metabolites and 7625 genes was generated at a threshold of 0.5 for association score using sPLS. The overall interaction network showed three major substructures (Fig. 3). One substructure [1] was very dense suggesting an overall homogeneity of most interactions. This substructure had distal associations with the other more diffuse and less dense substructures [2], [3]. Substructure 2 had few intermediate elements connecting with Substructure 1, while Substructures 2–3 and 1–3 had many intermediate elements. This relationship suggests that Substructure 2 may represent a second-order interaction with Substructure 1 linked principally through Substructure 3.
Fig. 3.
Toxicological response networks generated via integration of transcriptomic and metabolomic data. These networks represent (A) all genes and metabolites, (B) top 500 hub genes (0.7 threshold), and (C) top 50 hub genes (threshold 0.7).
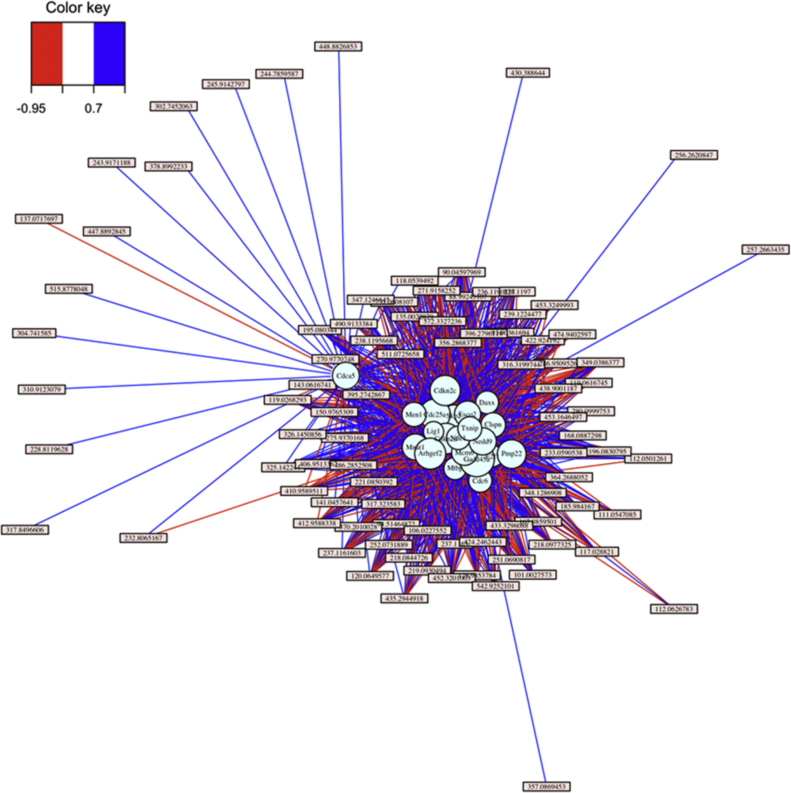
Substructure detail was further investigated by sequentially imposing filters for the number of hub genes and strength of gene–metabolite associations using association scores from sPLS. With 500 hub genes and 0.5 as a threshold filter for association score, the large dense cluster containing most of the genes and metabolites was lost and only 2 subclusters remained (not shown). With higher stringency (500 hub genes and a threshold of 0.7), this substructure was preserved (Fig. 3B) and only a small number of metabolites connected the hub genes. Restriction to the top 50 hub genes (Fig. 3C) simplified these relationships, and manual searching of the genes against Supplementary Table S1 showed that both modules in Fig. 3C contained genes significantly associated with MB treatment as visualized in Fig. 1. Importantly, the smaller module in Fig. 3C (Top cluster) contained metabolites associated with PQMB and not MB alone or PQ alone. Thus, the interaction of the transcriptome and metabolome data has a network organizational structure linked to MB treatment with additional distinguishing transcript–metabolite modules specifically linked to PQ in MB-treated cells.
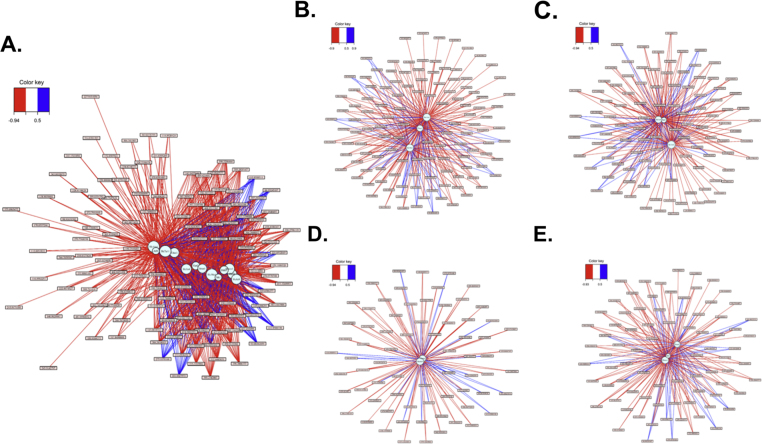
Examination of the PQMB module showed that 4 clusters (Cluster 1: Chac1, Slc11a1, Slc30a1; Cluster 2: Gpt2, Slc1a4, Slc7a11; Cluster 3: Pparg, Slc6a9; Cluster 4: Hmox1, Mt1, Srxn1) were present among 13 genes (Fig. 4). Cluster 1 contained two cation transporters and Chac1, a homolog of E. coli cation-transporter regulator-1. Chac1 is a proapoptotic protein of the unfolded protein response pathway [32] that has glutamylcyclotransferase activity specific for glutathione [33]. Slc11a1 is a divalent cation transporter with activity for iron and manganese, and Slc30a1 is a zinc transporter. Thus, Cluster 1 is a cation-related gene-metabolite response cluster with known activities, which could contribute to combined PQMB toxicity. The metabolites positively and negatively associated with Cluster 1 were largely the same as those for Clusters 2 and 3 so these are discussed together below.
Fig. 4.
Transcriptome–metabolome associations present the PQMB module. 13 genes present in the PQMB module are displayed (A). Cluster 1 (B), Cluster 2 (C), Cluster 3 (D), and Cluster 4 (E) are also displayed.
Cluster 2 included genes encoding a module associated with amino acid transport and metabolism: Gpt2 (glutamate–pyruvate transaminase), an enzyme catalyzing reversible transamination of glutamate and pyruvate in amino acid metabolism and gluconeogenesis; Slc1a4 (glutamate/neutral amino acid transporter) and Slc7a11 (light chain, xc− system). The xc− system supports supply of precursors Glu and cystine for GSH synthesis, so this may represent a response to oxidative stress.
Cluster 3 included peroxisome proliferator-activated receptor gamma (PPAR-γ; Pparg) and a glycine transporter, Slc6a9. PPAR-γ is a nuclear receptor that forms heterodimers with retinoid X receptors (RXRs) and regulates transcription of various genes, especially involved in fatty acid storage and glucose metabolism. Glycine increases PPAR-γ expression [34], suggesting that this could also be part of an adaptive or protective response rather than a toxic response.
Finally, Cluster 4 contained stress response genes, heme oxygenase-1 (Hmox1), metallothionein-1 (Mt1), and sulfiredoxin-1 (Srxn1), which respond to a range of cell stresses including oxidative stress and metal toxicities. Together, the genes and associated metabolites represent a network response for PQMB toxicity rather than a single molecular target of toxicity for either PQ or MB.
Database matches to the metabolites positively and negatively correlating with all four clusters were largely the same and are summarized in Table 1; PPAR-γ had the largest number of associations >|0.3|. Co-enzyme metabolites associated with mitochondrial function and energy metabolism included methylnicotinamide, riboflavin, thiamine and vitamin D, which were positively associated, and pyridoxamine, pantetheine, 2-(alpha-hydroxyethyl)thiamine diphosphate and dihydropteroate, which were negatively associated. Methylnicotinamide is a methylated product derived from NAD and is also derived from polyADP-ribose polymerase activation, involved in regulation of chromatin structure and activation of cell death. Dihydropteroate is an intermediate in 1-carbon metabolism, and other 1-carbon metabolism intermediates also had negative association, including methionine (Met) and S-adenosylhomocysteine. One-carbon metabolism is important in nucleotide metabolism, and both pyrimidine (cytosine, dCDP, dihydrouracil, dihydroorotate)- and purine (hypoxanthine)-related metabolites varied in association with the 13 transcripts (Table 1).
Table 1.
Metabolites associated with four transcript clusters (Fig. 4) changed by paraquat in maneb-treated cells. Metabolite matches were obtained by searching the Metlin database with high resolution m/z within 10 ppm. Associations were similar for all genes and clusters and are summarized together for those with positive and negative associations. PPAR-γ in cluster 3 was most representative and associated metabolites are noted with an asterisk (*).
| Pathways | Positive | Adduct | m/z | Negative | Adduct | m/z |
|---|---|---|---|---|---|---|
| Coenzyme metabolism | *Methylnicotinamide | M+H | 137.0718 | *Pyridoxamine | M | 168.0887 |
| Riboflavin | M+H | 377.1441 | Pantetheine | M+HCOONa | 347.1247 | |
| Thiamine | M(C13)+H | 267.1219 | *2-(alpha-Hydroxyethyl)thiamine diphosphate | M(C13)+2H | 236.1191 | |
| Vitamin D3 | M+NH4 | 402.3723 | *Dihydropteroate | M+Na | 337.1007 | |
| Nucleoside metabolism | *Cytosine | M+H | 112.0501 | dCDP | M+2H | 195.0156 |
| *Hypoxanthine | M(C13)+H | 138.0482 | *Dihydrouracil | M+2Na–H | 173.0298 | |
| *Dihydroorotate | M(C13)+H | 160.0420 | ||||
| Amino acid metabolism | *Methylpyrrolinium | M+HCOONa | 153.0765 | *Methionine | M+NH4 | 167.0860 |
| *Acetylornithine | M+2H | 88.0574 | *Threonine | M+H | 120.0650 | |
| *Dicarboxypropyllysine | M+2H | 139.0733 | *N-Acetyl-l-glutamate 5-semialdehyde | M(S34)+H | 176.0703 | |
| *Hydroxyindoleacetaldehyde | M(C13)+H | 89.0413 | Oxoadipate | M+H | 160.0372 | |
| *Phenylpyruvate | M+H | 165.0541 | *Urocanate | M(S34)+H | 141.0458 | |
| *Phenylacetate | M+H–H2O | 119.0485 | *Indoleacetic acid | M+H | 236.0273 | |
| *Methyl oxopentanoic acid | M+HCOOK | 215.5285 | ||||
| *Imidazole-4-acetaldehyde | M+H | 111.0547 | ||||
| *S-Adenosylhomocysteine | M+HCOONa | 453.1135 | ||||
| Antioxidant systems | *α-Tocopherol | M+H | 431.3903 | |||
| *γ-Glutamylcysteine | M+H–H2O | 233.0591 | ||||
| Energy metabolism | Oxaloacetate | M+2Na | 88.9925 | Phosphoenolpyruvate | M | 167.9816 |
| *Lactate | M+2Na–H | 135.0023 | ||||
| *Creatinine | M+ACN+Na | 177.0731 | ||||
| Lipid metabolism | 7-Dehydrocholesterol | M+NH4 | 402.3723 | Glycerol | M+K | 131.0110 |
| Glycerol 1-P | M+ACN+Na | 236.0273 | ||||
| *Oxooctanoyl-CoA | M(C13)+2H | 455.1100 | ||||
| *Sphingosine or dehydrosphinganine | M+Na | 322.2698 | ||||
| *Sphingosine 1-P | M+H–H2O | 334.2125 | ||||
In addition to the amino acid metabolites indicated above, multiple cationic amino acid (Lys, His) metabolites were associated, both positively and negatively, with the transcripts. Notably, phenylalanine (Phe) metabolites were positively associated with the transcripts; phenylalanine has been previously associated with changes in PPAR-γ expression. Along with Met, Phe, Lys and His above, methylpyrrolinium is metabolically linked to acetylornithine, which is a component of the urea cycle and used by mitochondria as a precursor for arginine (Arg) and citrulline. Other negative associations included threonine and metabolites of tryptophan (indoleacetate) and branched chain amino acids (methyl-2-oxopentanoic acid). Thus, the data show a generalized association of essential amino acids and related metabolites to changes in PPAR-γ and other associated transcripts.
Associations with energy intermediates were also present (Table 1). Several metabolites related to lipid metabolism were also negatively associated. Consistent with earlier studies of PQ-induced toxicity, the antioxidant α-tocopherol and the GSH precursor, γ-glutamylcysteine were negatively associated with the transcripts.
The larger module from Fig. 3C with hub genes and metabolites principally associated with MB was enriched in genes functioning in cell cycle regulation (Fig. 5). Metabolites associated with this hub overlapped with the metabolites associated with the 13 transcripts but did not have clearly discernible pathway associations. Surprisingly, there were no apparent associations with pyrimidine bases, dCDP or hypoxanthine. Similarly, there was no apparent association with 1-carbon metabolism energy metabolism or other amino acid degradation products. There were weak associations of some of the genes with the polyamine, spermine.
Fig. 5.
Network structure and gene/metabolite associations associated with genes involved in cell cycle regulation.
4. Discussion
The etiology of PD is complex and the overwhelming majority of cases are idiopathic in nature. Exposures to environmental chemicals, like pesticides, have been linked to an increased risk of PD [6], [7]. In regards to human exposure to PQ and MB, Costello et al. have reported that residential exposure to PQ or MB from agricultural applications resulted in an odds ratio of 2.27, while those who reported exposure to both PQ and MB had an odds ratio of 4.17 [15]. Additionally, ambient exposure to a combination of ziram, MB and PQ resulted in an odds ratio of 3.09 [10]. In vivo and in vitro studies have also demonstrated the neurotoxic properties of PQ and MB. In mice, repeated exposure to PQ and MB results in the loss of tyrosine hydroxylase positive neurons, decreased striatal dopamine levels, and behavioral deficits [12], [14]. In vitro studies have also demonstrated that MB can enhance the toxicokinetics of PQ [35], which may contribute to the enhanced neurotoxicity observed in vivo when both pesticides are administered in combination. Together, these data demonstrated that pesticides, in particular the combination of PQ and MB, increase the risk of developing PD in humans, and in vitro and in vivo studies have brought to light prospective mechanisms of toxicity.
In a previously published study [16], we characterized the individual mechanisms involved in PQ, MB, and PQMB toxicity in vitro. Using SH-SY5Y neuroblastoma cells, we observed that PQ and MB act via distinct redox mechanisms. For example, PQ caused increased ROS production, oxidation of mitochondrial thioredoxin 2 and peroxiredoxin 3, lesser oxidation of cytoplasmic thioredoxin 1 and peroxiredoxin 1, and no oxidation of cellular GSH/GSSG. In contrast, MB alone at a similar toxic dose did not cause ROS production or oxidation of thioredoxin or peroxiredoxin isoforms as observed with PQ. MB treatment did cause an increase in cellular GSH, which was attributed to the ability of MB to cause nuclear localization of Nrf2 and transcriptional activation. The results of these studies showed that MB potentiation of PQ-mediated neurotoxicity does not occur via enhancement of oxidative stress, but suggests that increased toxicity can be attributed to a combination of divergent mechanisms that perhaps involve PQ-mediated oxidation and thiol alkylation by MB [36]. The present studies clarify and extend these data by showing that multiple network responses are involved in the combined neurotoxic response. As discussed below, a toxic cation-response hub (Cluster 1) co-exists with an amino acid/antioxidant response hub (Cluster 2), an adaptive PPAR-γ response hub (Cluster 3) and an early Hmox-1 stress-response hub (Cluster 4). Note that this clustering is defined by the interaction network of the transcriptome and metabolome and therefore cannot be assumed to reflect transcription factors functioning independently.
Transcriptomic analyses of CAD cell response to PQ, MB, or PQMB showed that expression of substantially more genes were altered in response to MB treatment compared to control or PQ treated cells. PCA analysis of this data shown in Fig. 1A revealed two groupings: one that included all MB treated cells (MB20, MB50, PQMB) and the other that included both PQ treatments (PQ20, PQ50) and the controls. This result shows that (1) few significant effects are observed with PQ alone due to the clustering with control samples and (2) supports our previously published research describing two divergent mechanisms in which MB increased GSH while PQ caused GSH oxidation [16]. Also consistent with these previous studies, some of the most significantly altered genes in the MB treated groups were genes regulated by the Nrf2-Keap-1 system. These genes included heme oxygenase-1, sulfiredoxin-1, and Slc7a11. All of the Nrf2-regulated genes were increased in a concentration dependent manner and the expression in the PQMB group was slightly higher than that of the MB20 group. It should also be noted that these Nrf2-regulated genes were not significantly changed in the groups treated with only PQ. Metabolomics data showed very similar patterns to transcriptomic data. For example, MB treatments significantly changed many more metabolites compared to PQ treatments and controls. PCA analysis was also similar to that found in the transcriptomic data (Fig. 2A). Two groupings were observed and the pattern was identical to the gene expression data, where MB treated samples (MB20, MB50, PQMB) formed one group and the second group consisted of the PQ-only treated samples and the controls. Together, the transcriptomic and metabolomics data are in agreement and further suggest that distinct mechanisms are involved with each toxicant.
Metabolomic data, in combination with the transcriptome data, show that the toxicologic mechanism for the combined toxicity of PQ and MB occur at a network level. Without PQ, MB caused extensive changes in gene expression associated with cell cycle, but without readily discernible pathway effects. Without MB, PQ had little discernable character to effects on either the transcriptome or the metabolome. However, in cells treated with MB, a new network structure was apparent with PQ that included 13 transcripts with multiple associated groups of metabolites. These groups included metabolites associated with oxidative stress and mitochondrial energy metabolism, consistent with prior findings concerning GSH concentration and oxidative stress effects on GSH and thioredoxin redox systems. Additionally, the associations included extensive effects on essential amino acid metabolism and 1-carbon metabolism.
Together with previous evidence, the combined transcriptomic and metabolomic data clarify mechanisms underlying the combined toxicities of MB and PQ. Specifically, the data show that MB has broad effects on transcription, especially affecting cell cycle genes and causing a new set point for nuclear functions. Among the transcripts changed, three related to cation transport appear to be especially important to enhance uptake of paraquat and oxidative metals. Mitochondrial dysfunction and oxidative stress are apparent in the metabolome. Network effects are apparent from the changes in amino acid metabolism, energy metabolism and lipid metabolism. Importantly, these include metabolites of phenylalanine previously associated with PPAR-γ activation. Schumacher et al. showed that high Phe activated peroxisome proliferator-activated receptor-γ (PPAR-γ) and changed gene expression in a manner comparable to the PPAR-γ agonist, rosiglitazone [37]. Associated inhibition of cell proliferation led to the conclusion that neurodevelopmental effects of Phe in PKU may occur due to interruption of PPAR-γ signaling. The extrapolation of results from our in vitro studies to in vivo effects must be made with caution, however, because of the single time points used for analysis. The selection of time points was made in an attempt to optimize findings based upon previous studies, and more complete time course data will be needed to fully understand the temporal nature of the network responses.
The present study emphasizes the need to approach environmental contributions to human disease within a complex systems approach to human pathobiology. The simplistic paradigm of additive and synergistic mechanisms of toxicity, which we previously used to interpret the interactions of MB and PQ toxicity, are inadequate to address the multiple network level interactions revealed by the combined TMWAS. With the availability of powerful omic tools, investigation of such network level interactions can be expected to provide new means to detect adverse human exposures, improve understanding of critical underlying mechanisms, and facilitate remediation and therapeutic strategies to minimize human toxicity.
In summary, the present study used a combination of transcriptomics and metabolomics to investigate mechanisms of toxicity of PQ and MB in a neuroblastoma cell line. The results show that the toxicologic mechanism of the combined neurotoxicity of PQ and MB involves network level interactions with effects on proliferation, transport systems and fundamental metabolic pathways of intermediary metabolism. Importantly, the combination of transcriptomics and metabolomics (TMWAS) provides an effective approach to investigate complex toxicologic mechanisms.
Author contribution
Experimental design (JRR, DPJ); Data acquisition (JRR, YP, VT); Data analysis (KU, JRR, DPJ); Manuscript preparation (JRR, KU, DPJ); Review of manuscript (JRR, KU, YP, DPJ).
Conflict of interest
None declared.
Transparency document
Acknowledgements
The research in this manuscript was supported by grant funds from the National Institutes of Health ES019821 and ES022266 (JR); ES009047, ES019776, ES023485, HL113451 and AG038756 (DJ).
Footnotes
Available online 24 July 2014
References
- 1.Loscalzo J., Kohane I., Barabasi A.L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol. Sys. Biol. 2007;3:1–11. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willett W.C. Balancing life-style and genomics research for disease prevention. Science. 2002;296:695–698. doi: 10.1126/science.1071055. [DOI] [PubMed] [Google Scholar]
- 3.Wild C.P. The exposome: from concept to utility. Int. J. Epidemiol. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 4.Jones D.P., Park Y., Ziegler T.R. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foltynie T., Brayne C., Barker R.A. The heterogeneity of idiopathic Parkinson's disease. J. Neurol. 2002;249:138–145. doi: 10.1007/pl00007856. [DOI] [PubMed] [Google Scholar]
- 6.Hatcher J.M., Pennell K.D., Miller G.W. Parkinson's disease and pesticides: a toxicological perspective. Trends Pharmacol. Sci. 2008;29:322–329. doi: 10.1016/j.tips.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priyadarshi A., Khuder S.A., Schaub E.A., Priyadarshi S.S. Environmental risk factors and Parkinson's disease: a metaanalysis. Environ. Res. 2001;86:122–127. doi: 10.1006/enrs.2001.4264. [DOI] [PubMed] [Google Scholar]
- 8.Drechsel D.A., Patel M. Role of reactive oxygen species in the neurotoxicity of environmental agents implicated in Parkinson's disease. Free Radic. Biol. Med. 2008;44:1873–1886. doi: 10.1016/j.freeradbiomed.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritz B., Yu F. Parkinson's disease mortality and pesticide exposure in California 1984–1994. Int. J. Epidemiol. 2000;29:323–329. doi: 10.1093/ije/29.2.323. [DOI] [PubMed] [Google Scholar]
- 10.Wang A., Costello S., Cockburn M., Zhang X., Bronstein J., Ritz B. Parkinson's disease risk from ambient exposure to pesticides. Eur. J. Epidemiol. 2011;26:547–555. doi: 10.1007/s10654-011-9574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock D.B., Martin E.R., Mayhew G.M., Stajich J.M., Jewett R., Stacy M.A., Scott B.L., Vance J.M., Scott W.K. Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol. 2008;8(6):1471–2377. doi: 10.1186/1471-2377-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiruchelvam M., Richfield E.K., Baggs R.B., Tank A.W., Cory-Slechta D.A. The nigrostriatal dopaminergic system as a preferential target of repeated exposures to combined paraquat and maneb: implications for Parkinson's disease. J. Neurosci.: Off. J. Soc. Neurosci. 2000;20:9207–9214. doi: 10.1523/JNEUROSCI.20-24-09207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiruchelvam M., McCormack A., Richfield E.K., Baggs R.B., Tank A.W., Di Monte D.A., Cory-Slechta D.A. Age-related irreversible progressive nigrostriatal dopaminergic neurotoxicity in the paraquat and maneb model of the Parkinson's disease phenotype. Eur. J. Neurosci. 2003;18:589–600. doi: 10.1046/j.1460-9568.2003.02781.x. [DOI] [PubMed] [Google Scholar]
- 14.Thiruchelvam M., Brockel B.J., Richfield E.K., Baggs R.B., Cory-Slechta D.A. Potentiated and preferential effects of combined paraquat and maneb on nigrostriatal dopamine systems: environmental risk factors for Parkinson's disease? Brain Res. 2000;873:225–234. doi: 10.1016/s0006-8993(00)02496-3. [DOI] [PubMed] [Google Scholar]
- 15.Costello S., Cockburn M., Bronstein J., Zhang X., Ritz B. Parkinson's disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 2009;169:919–926. doi: 10.1093/aje/kwp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roede J.R., Hansen J.M., Go Y.M., Jones D.P. Maneb and paraquat-mediated neurotoxicity: involvement of peroxiredoxin/thioredoxin system. Toxicol. Sci.: Off. J. Soc. Toxicol. 2011;121:368–375. doi: 10.1093/toxsci/kfr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams T.D., Turan N., Diab A.M., Wu H., Mackenzie C., Bartie K.L., Hrydziuszko O., Lyons B.P., Stentiford G.D., Herbert J.M., Abraham J.K., Katsiadaki I., Leaver M.J., Taggart J.B., George S.G., Viant M.R., Chipman K.J., Falciani F. Towards a system level understanding of non-model organisms sampled from the environment: a network biology approach. PLoS Comput. Biol. 2011;7:e1002126. doi: 10.1371/journal.pcbi.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams T.D., Wu H., Santos E.M., Ball J., Katsiadaki I., Brown M.M., Baker P., Ortega F., Falciani F., Craft J.A., Tyler C.R., Chipman J.K., Viant M.R. Hepatic transcriptomic and metabolomic responses in the stickleback (Gasterosteus aculeatus) exposed to environmentally relevant concentrations of dibenzanthracene. Environ. Sci. Technol. 2009;43:6341–6348. doi: 10.1021/es9008689. [DOI] [PubMed] [Google Scholar]
- 19.Santos E.M., Ball J.S., Williams T.D., Wu H., Ortega F., van Aerle R., Katsiadaki I., Falciani F., Viant M.R., Chipman J.K., Tyler C.R. Identifying health impacts of exposure to copper using transcriptomics and metabolomics in a fish model. Environ. Sci. Technol. 2010;44:820–826. doi: 10.1021/es902558k. [DOI] [PubMed] [Google Scholar]
- 20.Katsiadaki I., Williams T.D., Ball J.S., Bean T.P., Sanders M.B., Wu H., Santos E.M., Brown M.M., Baker P., Ortega F., Falciani F., Craft J.A., Tyler C.R., Viant M.R., Chipman J.K. Hepatic transcriptomic and metabolomic responses in the Stickleback (Gasterosteus aculeatus) exposed to ethinyl-estradiol. Aquat. Toxicol. 2010;97:174–187. doi: 10.1016/j.aquatox.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Soltow Q.A., Strobel F.H., Mansfield K.G., Wachtman L., Park Y., Jones D.P. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu T., Park Y., Johnson J.M., Jones D.P. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markley J.L., Anderson M.E., Cui Q., Eghbalnia H.R., Lewis I.A., Hegeman A.D., Li J., Schulte C.F., Sussman M.R., Westler W.M., Ulrich E.L., Zolnai Z. New bioinformatics resources for metabolomics. Pac. Symp. Biocomput. 2007:157–168. [PubMed] [Google Scholar]
- 24.Smith C.A., O’Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 25.Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;(20):3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y., Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
- 27.Le Cao K.A., Rossouw D., Robert-Granie C., Besse P. A sparse PLS for variable selection when integrating omics data. Stat. Appl. Genet. Mol. Biol. 2008;7(35) doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 28.Le Cao K.A., Gonzalez I., Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulsen T., de Vlieg J., Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Ekins S., Nikolsky Y., Bugrim A., Kirillov E., Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 32.Mungrue I.N., Pagnon J., Kohannim O., Gargalovic P.S., Lusis A.J. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response. downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A., Tikoo S., Maity S., Sengupta S., Sengupta S., Kaur A., Bachhawat A.K. Mammalian proapoptotic factor ChaC1 and its homologues function as gamma-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13:1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alarcon-Aguilar F.J., Almanza-Perez J., Blancas G., Angeles S., Garcia-Macedo R., Roman R., Cruz M. Glycine regulates the production of pro-inflammatory cytokines in lean and monosodium glutamate-obese mice. Eur. J. Pharmacol. 2008;599:152–158. doi: 10.1016/j.ejphar.2008.09.047. [DOI] [PubMed] [Google Scholar]
- 35.Barlow B.K., Thiruchelvam M.J., Bennice L., Cory-Slechta D.A., Ballatori N., Richfield E.K. Increased synaptosomal dopamine content and brain concentration of paraquat produced by selective dithiocarbamates. J. Neurochem. 2003;85:1075–1086. doi: 10.1046/j.1471-4159.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 36.Roede J.R., Jones D.P. Thiol-reactivity of the fungicide maneb. Redox Biol. 2014;2:651–655. doi: 10.1016/j.redox.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher U., Lukacs Z., Kaltschmidt C., Freudlsperger C., Schulz D., Kompisch K., Muller R., Rudolph T., Santer R., Lorke D.E., Ullrich K. High concentrations of phenylalanine stimulate peroxisome proliferator-activated receptor gamma: implications for the pathophysiology of phenylketonuria. Neurobiol. Dis. 2008;32:385–390. doi: 10.1016/j.nbd.2008.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcripts found to be significantly altered (FDR 0.05) compared to control.