Abstract
Invariant natural killer T (iNKT) cells are innate-like T cells that respond to lipid antigens presented by CD1d. These immunoregulatory cells have the capacity for rapid cytokine release after antigen recognition and are essential for the activation of multiple arms of the immune response. HIV-1 infection is associated with iNKT cell depletion in the peripheral blood; however, their role in the gastrointestinal-associated lymphoid tissue (GALT) is less well studied. Our results show that iNKT cells are found at a higher frequency in GALT compared to blood, particularly in HIV-1 elite controllers. The capacity of iNKT cells to produce IL-4 and IL-10 in the GALT was associated with less immune activation and lower markers of microbial translocation, while Treg frequency showed positive associations with immune activation. We hypothesized that the composition of the microbiota would influence iNKT cell frequency and function. We found positive associations between the abundance of several Bacteroides species and iNKT cell frequency and their capacity to produce IL-4 in the GALT but not in the blood. Overall, our results are consistent with the hypothesis that GALT iNKT cells, influenced by certain bacterial species, may play a key role in regulating immune activation in HIV-1 infection.
Introduction
HIV-1 infection leads to the development of chronic inflammation that persists even in antiretroviral (ART)-treated individuals with undetectable viral loads1,2. This inflammation is associated with non-HIV comorbidities, including cardiovascular disease, neurologic disorders, cancers, and an overall increased mortality. It has become apparent that immune activation is a better predictor of HIV-1 disease progression than either peripheral blood CD4+ T-cell count or viral load3, highlighting the importance of chronic immune activation. However, distinct pathways of immune activation (innate vs. adaptive) appear to have differential prognostic capacity, depending on the cohorts4. Importantly, while ART significantly diminishes immune activation (particularly if initiated early after infection5), levels do not normalize to those of uninfected individuals. Invariant natural killer T (iNKT) cells are innate-like T cells that respond to lipid antigens presented on CD1d, an MHC class I-like molecule expressed on antigen presenting cells (APCs)6. iNKT cells are characterized by their expression of the semi-invariant T cell receptor chain Vα24-Jα18 preferentially paired to a Vβ11 chain. Upon stimulation, iNKT cells are capable of rapid production of a vast array of cytokines and chemokines and are instrumental in orchestrating innate and adaptive immune response7. iNKT cells can recruit and modulate other immune cells, including natural killer (NK) cells, dendritic cells (DC), and conventional CD4+ and CD8+ T cells8. Depending on the type of specific interactions between iNKT cells and DCs, the cytokines secreted by activated iNKT cells may either activate or suppress adaptive immune responses.
Mouse studies have shown that the symbiotic microbiota can impact the maturation and function of iNKT cells in the mucosa9,10. A sphingolipid produced by the human commensal Bacteroides fragilis has been shown to bind CD1d and modulate iNKT cells11. When compared to specific pathogen free-mice, germ-free mice have a greater frequency of iNKT cells in intestinal lamina propria and epithelium, but these cells express lower levels of activation and produce less cytokines in response to stimulation12. Therefore, the gut microbiome influences the post-thymic maturation of iNKT cells, and intestinal bacterial reconstitution is a potential strategy for correcting systemic iNKT hypo-responsiveness in individuals with an altered microbial landscape. Dysbiosis of gut microbiota, particularly depletion of Bacteroidia members (including Bacteroides fragilis), has recently been described in the context of untreated HIV-1 infection, and is associated with markers of systemic immune activation and chronic inflammation13.
Mouse studies have revealed a role for iNKT cells in the control of viral infections, but their involvement in viral immunity in humans is less well characterized14,15. Previous studies have shown that iNKT cells in the peripheral blood are selectively and rapidly depleted in early HIV-1 infection16 and in models of SIV-infected non-human primates17. Some studies reported reduced iNKT proliferation and cytokine secretion (IFNγ, TNF, and IL-4in response to αGalCer/IL-2/PMA stimulation in HIV-1 infection, with variable restoration of function on antiretroviral therapy (ART)18-20. The role of iNKT cells in HIV-1 progression, whether defined by viral replication or immune activation is unclear. Furthermore, HIV-1 has evolved to escape direct recognition of infected cells by iNKT cells (Paquin-Proulx et al, unpublished observation). Given their ability to produce IL-10 and activate regulatory T cells (Tregs), iNKT cells have the potential to help control pathologic T cell activation21. In the present study, we investigated the role of peripheral blood and gut iNKT cells in controlling immune activation in HIV-1 infection and the consequences of gut microbial dysbiosis on iNKT frequency and function.
Results
iNKT cells are reduced in the blood but not in the GALT during HIV-1 infection
A total of 23 HIV-1-infected subjects and 10 healthy controls were enrolled in the study and paired blood and gut-associated lymphoid tissue (GALT) samples were obtained (Table 1). Thirteen of the HIV-infected subjects were on ART at the time of sampling, and three of the ten untreated patients met the definition of viremic controllers (viral load below 200 copies/ml). Mononuclear cells were isolated from the samples and flow cytometry was performed. Staining with Vα24, together with PBS57-CD1d tetramer, was used to identify iNKT cells (Figure 1A). iNKT cells were increased in the GALT compared to the blood across all subjects (Supplementary figure 1A). As previously reported, iNKT cells were found at a reduced frequency in the blood of viremic and ART-treated HIV subjects compared to healthy controls (Figure 1B)22-24. There was a trend for increased iNKT frequency in the ART-treated group compared to the viremic group (p=0.07). Surprisingly, no change in iNKT cell frequency was observed in the GALT of viremic and ART-treated HIV-infected individuals compared to healthy controls (Figure 1C). HIV controllers appeared to have preserved iNKT cell frequency in the blood and higher frequency in the GALT compared to all other groups. Next, we investigated the distribution of the CD4+ subset of iNKT cells as this population has been shown to be preferentially depleted in the peripheral blood during HIV infection22,23. No significant differences were observed between HIV-infected individuals and controls (Figure 1D) and between the viremic and ART-treated groups (Supplementary Figure 1B and C) both in the blood and in GALT. However, HIV-infected subjects had a significantly increased proportion of CD4+ iNKT cells in GALT compared to the blood (Figure 1D). Our results confirm the loss of iNKT cell in the blood of HIV-infected individuals that has been reported by several studies before and suggest for the first time, that iNKT cells may be preserved in the GALT in these patients.
Table 1. Subjects demographics.
| Gender | Age | CD4 count | Viral load | Duration of infection (years) | Time on ART (total years) | |
|---|---|---|---|---|---|---|
| Healthy (n=10) | 9M, 1F | 32.5 (23-59) | 828 (538-1,173) | |||
| HIV (n=23) | ||||||
| viremic (n=7) | 5M, 1M to F, 1F | 46 (31-61) | 458 (257-887) | 13,187 (1,102-305,178) | 6.0 (0-23) | 0 (0-0.97) |
| ART (n=13) | 12M, 1F | 54 (32-66) | 616 (374-1,023) | undectable | 24.5 (1-34) | 7.9 (0.9-19.6) |
| Controllers (n=3) | 2M, 1F | 46 (25-50) | 573 (393-900) | undectable (undectable-141) | 15 (3-27) | 0 (0-0.56) |
Figure 1. Frequency of iNKT cells in the blood and GALT.
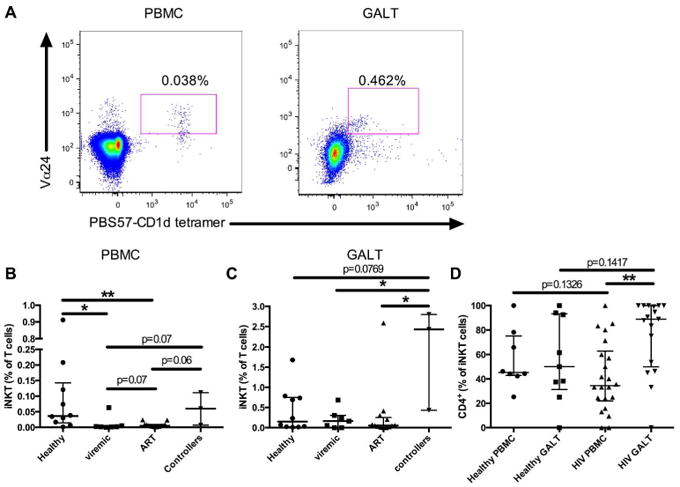
Representative gating strategy, iNKT cells were identified based on Vα24 and PBS57-CD1d tetramer positive stainings (A). Frequency of iNKT cells in the blood (B) and in the GALT (C) for healthy controls (n=10), viremic (n=7), ART-treated (n=13), and viremic controllers HIV-infected subjects (n=3). Frequency of iNKT cells expressing CD4 in the blood and GALT of healthy controls (blood n=8 and GALT n=9) and HIV-infected subjects (blood n=22 and GALT n=17) (D). * indicates p < 0.05 and ** indicates p < 0.01.
iNKT cells in the GALT of HIV-infected individuals have a Th2 cytokine profile
Previous studies demonstrated that cytokine production by iNKT in the blood of HIV-infected subjects is impaired19. However, cytokine production by iNKT cells in the GALT may be more relevant during HIV-1 disease progression. To assess the functional potential of iNKT cells, PBMC and rectal mononuclear cells (RMC) were stimulated with PMA and ionomycin and the production of IFNγ, TNF, IL-4 and IL-10 was evaluated by flow cytometry (Figure 2A). No differences were observed in cytokine production by iNKT cells in the blood and GALT of HIV-individuals compared to healthy controls. The majority of iNKT cells in the blood of healthy controls produced IFNγ and TNF while the frequencies of IL-4 and IL-10 producing iNKT were low (Figure 2B, C, D, and E). However, we observed a trend for a lower frequency of IFNγ+ and TNF+ iNKT cells in the GALT of healthy individuals compared to the blood. This difference was even more marked in the HIV-infected subjects. The percentage of iNKT in the GALT producing IL-4 and IL-10 varied greatly, ranging from undetectable to 100%. There was a trend for higher IL-4+ (p=0.06) and IL-10+ (p=0.06) iNKT cells in the GALT compared to the blood for HIV-infected individuals only. Our results suggest that a greater proportion of GALT iNKT cells in HIV-subjects have a Th2 cytokine profile compared to the blood.
Figure 2. Cytokine production by iNKT cells in the blood and GALT.
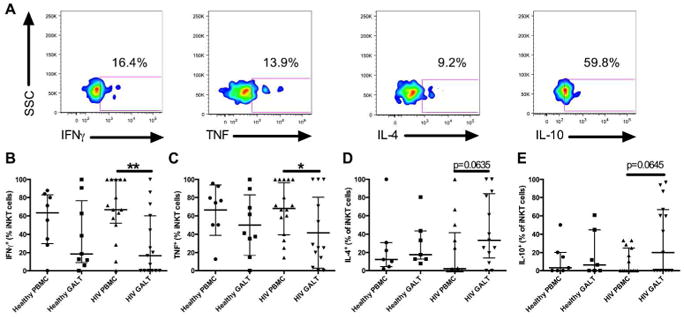
Cells were stimulated with PMA and Ionomycin before intracellular staining for cytokines. The gates were set using unstimulated controls, representative staining for IFNγ, TNF, IL-4 and IL-10 (A). Percentage of iNKT cells producing IFNγ (B, controls: blood n=8, GALT n=9 and HIV-infected subjects: blood n=16, GALT n=15), TNF (C, controls: blood n=8, GALT n=9 and HIV-infected subjects: blood n=16, GALT n=13), IL-4 (D, controls: blood and GALT n= 7 and HIV-infected subjects: blood and GALT n=13) and IL-10 (E, controls: blood and GALT n= 7 and HIV-infected subjects: blood and GALT n= 15). * indicates p < 0.05 and ** indicates p < 0.01.
Production of IL-4 and IL-10 by iNKT is associated with lower immune activation in the blood of HIV-infected subjects
Next, we evaluated immune activation of CD4+ and CD8+ T cells in the blood and in the GALT by measuring co-expression of CD38 and HLA-DR. In healthy controls, we found a trend towards increased activation of CD4+ T cells in the GALT compared to the blood (Figure 3A, p=0.08). Significantly higher levels of CD38 and HLA-DR co-expression were found on CD4+ and CD8+ T cells in the GALT of HIV-infected individuals when compared to paired blood samples (Figure 3A and B). A non-significant trend for greater cellular activation in the blood was observed when comparing HIV-infected subjects to controls. However, HIV-infected individuals had significantly increased activation of both CD4+ and CD8+ T cells in the GALT compared to healthy controls. Single expression of CD38 was also analyzed (Supplementary Figure 3A and B). iNKT cells are believed to have regulatory functions, which are in part mediated by their capacity to produce cytokines7. Therefore, we investigated if there was any association between cytokine production by iNKT cells and immune activation in HIV-infected subjects. We found no associations between immune activation in the GALT and iNKT cell cytokine production (data not shown). However, IL-10 production by iNKT cells in the blood was associated with lower CD4+ and CD8+ T cell activation in the blood (Figure 3C and D). Furthermore, IL-10 and IL-4 production by GALT iNKT cells were respectively associated with lower CD4+ and CD8+ T cell immune activation in the blood (Figure 3E and F). HIV-infected subjects were then grouped according to the capacity of iNKT cells to produce IL-10 and the levels of immune activation were compared. Subjects with GALT iNKT cells producing IL-10 had significantly lower frequencies of activated peripheral CD4+ T cells (Supplementary Figure 2 A) and subjects with blood iNKT cells producing IL-10 showed a trend for lower levels of activated peripheral CD4+ and CD8+ T cells (Supplementary Figure 2 B and C). In addition, we analyzed CD38 single expression and its associations with iNKT cell cytokine production. CD38 expression was inversely associated with IL-4 and IL-10 production by GALT iNKT cells and IL-10 production by blood iNKT cells (Supplementary Figure 3). These results suggest a role for GALT iNKT cells in dampening the pathological peripheral immune activation in HIV-1 infection.
Figure 3. CD4+ and CD8+ T cell activation in the blood and GALT and associations with iNKT cell cytokine production.
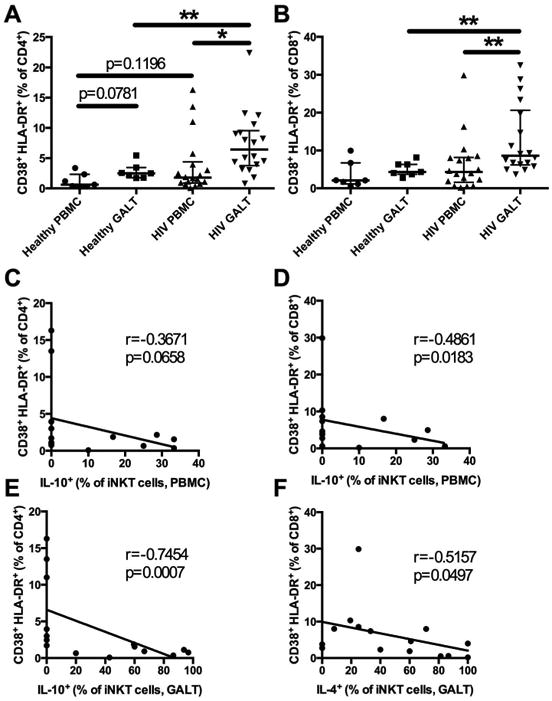
Co-expression of CD38 and HLA-DR on CD4+ T cells (A) and CD8+ T cells (B) in the blood and GALT of controls (n=7) and HIV-infected subjects (n=18). Associations between IL-10+ iNKT cells in the blood and CD38+HLA-DR+ CD4+ (C) and CD8+ (D) T cells of HIV-infected subjects. Associations between IL-10+ and IL-4+ iNKT cells in the GALT of HIV-infected subjects and CD38+HLA-DR+ CD4+ (E) and CD8+ (F) T cells respectively. * indicates p < 0.05 and ** indicates p < 0.01.
In addition to iNKT cells, Tregs are also known to have an important role in modulating immune activation25. Therefore, we analyzed Tregs (defined as CD3+, CD4+, CD25+ and Foxp3+) frequencies in peripheral blood and GALT of HIV-infected individuals. Similar frequencies of Tregs across groups were observed in the blood and in the GALT (Supplementary Figure 4A) and no difference in the frequency of Tregs in the blood compared to the GALT was observed for controls and HIV-infected individuals. We observed a positive association between Tregs frequency in the GALT and CD4+ and CD8+ T cell activation in the GALT of HIV-infected individuals (Supplementary Figure 4B and C). No associations were found between Tregs and iNKT cell frequency or function (data not shown).
Cytokine production by GALT iNKT cells is associated with lower microbial translocation
Damage to the integrity of the gut epithelial barrier by HIV-1 infection has been reported to lead to the presence of microbial products in the circulation, often referred to as microbial translocation (MT). MT has been associated with the pathologic immune activation that is characteristic of HIV-1 disease26. The kynurenine (Kyn) pathway of tryptophan (Trp) catabolism has been demonstrated to be dysregulated in HIV-1 infection, leading to an elevated Kyn/Trp ratio in the blood27. This dysregulation is reported to be associated with changes in the composition of the microbiome of HIV-infected individuals and with established markers of disease progression such as IL-613. We postulated that the immunoregulatory activity of GALT iNKT cells may limit disturbance to the gut barrier and therefore lower immune activation by reducing MT. For this purpose, we measured the concentration of soluble CD14 (sCD14) and LPS-binding protein (LBP) in the plasma as they have been shown to be indirect markers of MT26,28. We also measured intestinal fatty acid-binding protein 2 (I-FABP2, a marker of gut damage29), IL-6 and the Kyn/Trp ratio. As expected, HIV-infected subjects had significantly elevated levels of sCD14, LBP, and I-FABP2 (Figure 4A, B, and C), suggesting gut epithelial barrier dysfunction and MT. The Kyn/Trp ratio was also elevated in HIV-infected individuals (Figure 4D) as well as the levels of IL-6 (Supplementary Figure 5). We then looked for relationships between all of the above parameters and iNKT frequency or function. The capacity of peripheral iNKT cells to produce IL-4 was positively associated with plasma levels of sCD14 and there was a trend for an inverse association between the capacity of intestinal iNKT cells to produce IL-10 and the levels of sCD14 (Figure 4E and F). iNKT cell production of IL-4 and TNF-α in the GALT showed negative associations with the levels of LBP (Figure 4G and H). Finally, IL-4 production by peripheral iNKT cells was associated with elevated Kyn/Trp ratios (Figure 4I). Additionally, HIV-infected subjects were grouped according to capacity of iNKT cells to produce IL-4 or IL-10. We observed that individuals in the group with higher production of IL-4 had lower levels of sCD14 (Supplementary Figure 6). Together, our results show that a higher capacity to produce cytokines by iNKT in the GALT is associated with lower markers of MT, suggesting a role for GALT iNKT cells in modulating this pathological process in HIV-1 infection.
Figure 4. Markers of microbial translocation and associations with iNKT cell cytokine production.
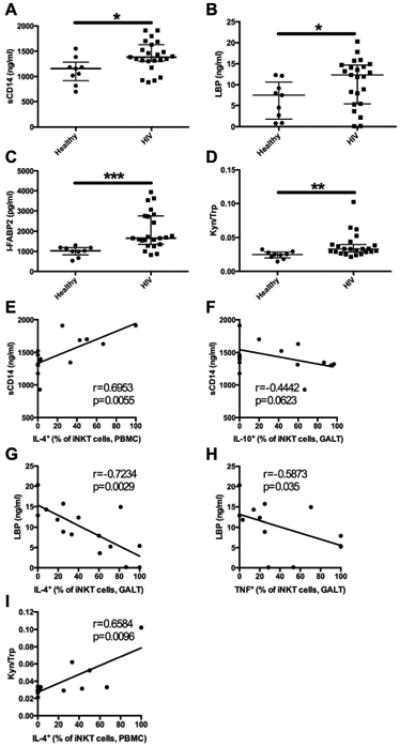
The levels of sCD14 (A), LBP (B), I-FABP2 (C), and Kyn/Trp ratio (D) were determined in the serum of healthy controls (n=9) and HIV-infected subjects (n=23). Associations between the levels of sCD14 in HIV-infected subjects and IL4+ iNKT cells in the blood (E) and IL10+ iNKT cells in the GALT (F). Associations between LBP levels in the serum of HIV-infected subjects and IL4+ (G) and TNF+ (H) iNKT cells in the GALT. Association between Kyn/Trp ratio in HIV-infected subjects and IL4+ iNKT cells in the blood (I). * indicates p < 0.05 and ** indicates p < 0.01.
Bacteroides are associated with iNKT frequency and IL-4 production in the GALT
The composition of the bacterial gut microbiota of untreated HIV-infected subjects is distinct from that of healthy individuals, with ART-treated patients having an intermediate change in the microbiome profile30. One of the genera significantly depleted in HIV-infected individuals is Bacteroides. We confirmed that Bacteroides were reduced in ART-treated HIV-infected subjects in our study (Figure 5A). Next, we performed an unbiased analysis of all gut-resident operational taxonomic units (OTUs) abundances compared to GALT iNKT cell frequency in ART-treated subjects and found that negative correlations existed between GALT iNKT cell frequency and OTUs in the Prevotella genus (Benjamini-Hochberg Q value < 0.15, Supplementary file), a genus that has been shown to be increased in abundance in HIV-infected subject gut microbiomes and associated with elevated mucosal immune activation31. Given that Bacteroides fragilis expresses a glycolipid that can activate human iNKT cells11, we investigated whether the abundance of (OTUs) within the Bacteroides genus was associated with frequencies of iNKT cells in both peripheral blood and GALT within ART-treated HIV-infected subjects, all study subjects combined, and uninfected subjects only (Figure 5B and C, Supplementary file). Fewer OTUs reached P<0.10 for comparisons to peripheral blood as opposed to GALT iNKT frequencies, and no trends were consistent across all subject groupings for comparisons to peripheral blood. However, when comparing OTU abundances to GALT iNKT frequencies, consistent positive associations were found between several Bacteroides OTUs and iNKT frequencies across subject groups. Finally, we looked for associations between Bacteroides OTUs and cytokine production by iNKT cells. We found a positive association between specific Bacteroides OTUs and the capacity of GALT iNKT cells to produce IL-4 (Figure 5D, Supplementary file), though these observations exhibited Benjamini-Hochberg false discovery rate Q values > 0.70. These results suggest that loss of the Bacteroides genus in HIV-infected individuals could influence both the frequency and function of iNKT cells in the gut.
Figure 5. Change in microbiota in HIV-infected individuals and associations with iNKT frequency and function.
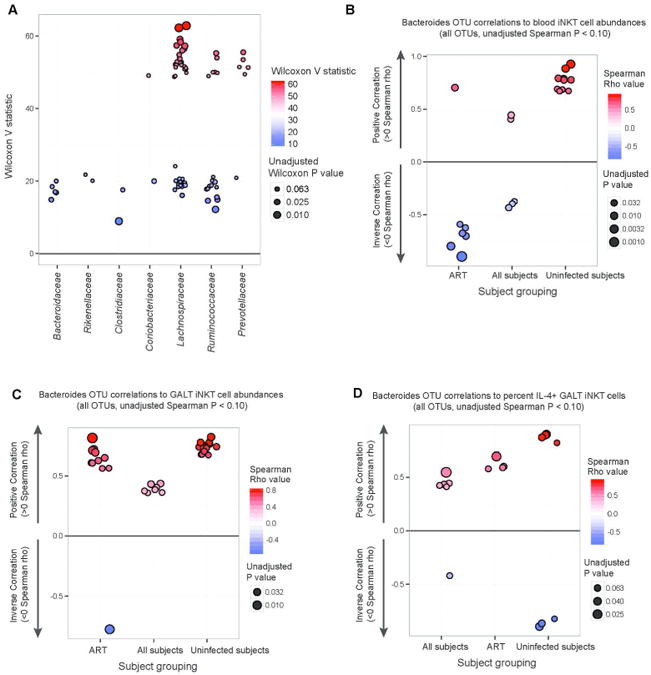
Wilcoxon rank-sum tests were performed comparing gut mucosal OTU abundances between HIV-infected subjects undergoing ART and uninfected subjects. OTUs with p < 0.15 are shown including taxonomic families to which each OTU belongs. Wilcoxon V statistics (y-axis) provide non-parametric enrichment/depletion information, and unadjusted P values are depicted by point sizes (A). Abundance of all detected OTUs within the Bacteroides genus were compared to blood iNKT cell percent abundances for three subject groups (ART HIV-infected, uninfected, and all subjects combined). Spearman rho values depict directionality of correlation, and all OTUs with p < 0.10 shown (B). Abundance of all Bacteroides OTUs were compared to GALT iNKT cell percent abundances for the same subject groups (C, p < 0.10 shown). Spearman correlations were performed comparing all Bacteroides OTU relative abundances and proportions of IL-4+ GALT iNKT cells following PMA and ionomycin stimulation (D, p < 0.10 shown).
Discussion
Several studies have examined the frequency and function of peripheral blood iNKT cells in HIV-1 infection but limited information is available for the GALT, an important target in HIV-1 pathogenesis. We found that iNKT cells were depleted in the blood but not in the GALT of HIV-infected subjects and that GALT iNKT cells consisted of an increased proportion of the CD4+ subset. This is in contrast to a previous study that reported that CD4+ iNKT cells are lost in the gut of HIV-1-infected individuals32. The discrepancy between our results and those of Ibarrondo et al. could be explained by important differences in the HIV-1 cohorts and the methods used to identify iNKT cells. The majority of our HIV-1-infected individuals had an undetectable viral load (ART-suppressed or controllers) and we used a CD1d tetramer to identify iNKT cells while the cohort of Ibarrondo et al. consisted exclusively of untreated subjects and iNKT cells were identified using antibodies against the invariant TCR. Altogether, this would suggest that viral suppression prevents the depletion of the CD4+ subset of iNKT cells in the GALT. Although based on a small number of subjects, our results suggest that HIV-1 elite controllers maintain normal iNKT cell frequency in the blood and have high levels of iNKT cells in the GALT. Further studies enrolling more controllers will be required to confirm the validity of these results.
CD4+ iNKT are known to produce more Th2 cytokines than the CD4- subsets33-35. Therefore, given our finding of higher percentages of CD4+ iNKT in the gut as compared to blood in HIV-infected subjects, it is not surprising that we observed more production of IL-4 and IL-10, two Th2 cytokines, and less IFNγ and TNFα by iNKT cells in the GALT compared to the blood. Our results suggest that iNKT cells can regulate the pathologic chronic immune activation in HIV-1 infection by their production of IL-4 and IL-10 in the GALT as well as IL-10 in the blood. We could not detect iNKT cell production of IL-10 by a significant proportion of HIV-infected individuals, suggesting that they could be separated in two groups based on the capacity of iNKT cells to produce IL-10. However, this could be due to the detection limit of the assay. Further studies with more subjects in each group are required to resolve this matter. Our findings are consistent with the model proposed by Rout et al who have shown that sooty mangabeys, natural hosts of SIV that do not progress to AIDS, exhibit a preservation of iNKT cell frequency and function after SIV infection. They have thus suggested that iNKT cell dysfunction has a role in AIDS pathogenesis36. MT has been proposed as a major component driving immune activation in HIV-infected individuals. We found that production of IL-4, IL-10 and TNF by iNKT cells in the GALT, but not in the blood, were associated with lower MT as measured by sCD14 and LBP, suggesting that iNKT cell production of IL-10 in the blood could have a localized effect on immune activation while production of the same cytokine in the GALT could have a more generalized effect by reducing MT. In contrast, the frequency of Tregs in the GALT was positively associated with CD4+ and CD8+ T cell activation in the GALT. We saw no association between Tregs and markers of MT, suggesting a specific role for iNKT cells. How production of IL-4, IL-10 and TNF by GALT iNKT cells contributes to reduced MT remains to be determined. It is a likely possibility that the ability of iNKT cells to interact with APCs to shape the adaptive immune response may be involved. Moreover, mice lacking IL-10 have been shown to have increased gut permeability caused by an excessive Th1 response against enteric bacteria37. On the other hand, the positive associations between IL-4 production by peripheral iNKT and Kyn/Trp ratio and sCD14 might represent in part their response to MT.
To our knowledge, this is the first study looking at the influence of the microbiome on iNKT cell frequency and function in humans. A study performed in mice showed that a sphingolipid derived from Bacteroides fragilis can inhibit iNKT cell activation and that Bacteroides fragilis colonization reduces the frequency of iNKT cells in the colon but not in other organs or in the blood10. A different group reported that Bacteroides fragilis produced a sphingolipid that can activate iNKT cells11. In our hands, multiple Bacteroides OTUs exhibited positive associations with GALT iNKT cells and only one OTU presented a negative association in HIV-infected subjects (with the later association not being present in uninfected subjects). This would suggest that Bacteroides produce an antigen that can activate and expand iNKT cells in the GALT. The reported greater abundance of Bacteroides in HIV elite controllers compared to viremic patients38 could therefore contribute to the higher frequency of iNKT in the GALT of elite controllers in our study. Mouse studies have shown that animals kept in germ-free conditions have lower IFNγ, TNF and IL-4 production by iNKT cells following stimulation with α-Gal-Cer12, but the bacteria responsible for the functional maturation of iNKT cells were not identified. In this study, we have found a positive association between Bacteroides and IL-4 production by GALT iNKT cells. While fetal human iNKT cells have been reported to mature and acquire function in the small intestine before colonization by the normal microbiota39, our results are consistent with the hypothesis that in adults the normal microbiota may provide signals that support GALT iNKT cell frequency and functionality. These findings would be strengthened by further study in larger human cohorts. Vα24- cells specifically binding to α-Gal-Cer loaded CD1d tetramer have been detected in PBMC following in vitro expansion40. However, the limited amount of GALT material obtained in our study did not allow us to study this population of NKT cells.
Based on our results, we propose a model where iNKT cells in the GALT have an important role in limiting MT and chronic pathologic immune activation in HIV-1 infection. This role of GALT iNKT is influenced by the composition of the gut microbiota, with loss of the Bacteroides genus in HIV-infected individuals possibly affecting both iNKT frequency and function. This suggests that strategies boosting GALT iNKT cells could reduce the MT and persistent immune activation that are important factors in the morbidity caused by HIV-1.
Materials and Methods
Study subjects
PBMCs and GALT samples were obtained from participants in the San Francisco-based HIV-1-infected SCOPE cohort. Samples from HIV-1-seronegative controls were obtained from healthy volunteers. The study was approved by the local Institutional Review Board (University of California San Francisco Committee on Human Research), and individuals gave written informed consent. Samples were obtained from the following numbers and categories of HIV-1-infected individuals: 3 untreated virologic “controllers” (viral load, <200 HIV-1 copies/ml), 13 HAART-suppressed patients (viral load, <50 copies/ml), and 7 untreated “virologic noncontrollers” (viral load, >1,000 copies/ml). All had CD4+ T cell counts of >250 cells/mm3. See Table 1 for baseline subject characteristics.
Blood and GALT samples
5 ml of blood was collected in BD Vacutainer EDTA coated tubes for PBMC and plasma isolation purposes. After centrifuging at 400g for 10 min without braking, the plasma layer was removed and frozen at -80°C for ELISA quantification. The cellular fraction from the first spin was used to isolate PBMC by centrifugation over a ficoll-paque (GE Healthcare, Uppsala, Sweden) layer at 800g for 25 min without braking. The PBMC layer was then removed and washed twice in RPMI with L-glutamine, Penicillin/Streptomycin, Hepes, and 10% Fetal bovine serum (referred now on as R-10) at 400g for 10 min with braking. GALT from rectosigmoid biopsy specimens were placed on a shaking incubator at 37°C with a digestion mix of RPMI with +L-glutamine, Hepes, Penicillin/Streptomycin, and Collagenase Type II (0.25mg/ml) (Sigma-Aldrich, St. Louis, MO, USA). After one digestion of 30 minutes, sample was strained through a 70 uM cell strainer and washed through with cold R-10. Undigested biopsies were transferred into the collagenase digestion mix for repeat digestion of 30 min. Strained and digested biopsies were washed in R-10 and spun down at 700g for 6 minutes at 4°C to isolate the RMC. PBMCs and RMCs were then re-suspended and counted using Guava Viacount (Millipore, Brillerica, MA, USA) on the Accuri C6 (BD Biosciences, San Jose, CA, USA).
Flow cytometry and mAbs
For surface staining, cells were stained with surface markers for 30 min on ice and washed twice with FACS buffer (PBS with 2% FBS and 2 mM EDTA buffer). After, cells were fixed and permeabilized with Fix/Perm buffer (BD Biosciences) for 20 min on ice, washed twice with BD Perm/Wash, and stained with the intracellular antibodies for 60 min on ice. For PoxP3 staining, cells were fixed and permeabilized with Fix/Perm Buffer (eBiosciences, San Diego, CA, USA) for 60 min on ice, washed twice with Perm/Wash buffer (eBiosciences) and stained with intracellular antibodies for 60 min on ice. Subsequently, the cells were washed twice with the respective Perm/Wash buffer and kept in 2% paraformaldehyde. Antibodies used: PBS57-CD1d tet APC (kindly donated by NIH tetramer resource facility), Vα24 FITC, and CD3 ECD were from Beckman Coulter (Fullerton, CA), CD4 Qdot655, CD8 Qdot 605, and the viability marker AmCyan were from Life Technologies (Carlsbad CA, USA), CD25 APC, CD38 PE, HLA-DR PerCP, IFNγ V450, TNF Alexa700, IL-10 PE, and IL-4 PE-Cy7 were all from BD bioscience. Data were acquired on a BD LSRFortessa instrument (BD Biosciences) and analyzed using FlowJo Version 9.8.5 software (TreeStar, Ashland, OR, USA).
ELISA
Il-6, sCD14, LBP, and FABP-2 levels were detected in plasma isolated from peripheral blood with a commercially available enzyme-linked immunosorbant assay (all from R&D Systems, Minneapolis, MN, USA) and performed according to the standard protocol. For sCD14 and IL-6, commercially available quality controls (R&D Systems) were also performed to ensure accurate detection of the kits.
Microbiome sample processing and analysis
DNA was extracted from gut biopsy samples using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Each DNA sample was PCR amplified in triplicate using primer pairs that targeted the V4 hypervariable region of the 16S rRNA gene, contained a unique barcode sequence to enable demultiplexing of pooled samples, and contained an adapter sequence that enables the amplicon to bind to the MiSeq flow cell. Successful amplicons were pooled in approximately equal molar concentrations and sequenced on the Illumina MiSeq platform. Paired sequencing reads were quality filtered and demultiplexed using the QIIME software package. Briefly, assembled sequencing read pairs were binned into OTUs (operational taxonomic units using a 97% similarity to the Greengenes database) and reads that either did not cluster to the Greengenes database or that were chimeric were removed from subsequent analyses. OTUs that had a cumulative read count across all samples of less than 0.001% of the total reads were removed from downstream analysis. Sample read numbers were rarefied to the read number of the lowest sample after processing (94,780) resulting in a rarefied OTU table. OTU abundances were compared between uninfected and ART-treated HIV-infected subjects using a custom R script in conjunction with the “exactRankTests” package. Spearman correlations were performed using the “Hmisc” R package using a custom script. Data visualizations were performed using the R package “ggplot2”.
Tryptophan metabolism
Liquid chromatography–tandem mass spectrometry was used to assess kynurenine and tryptophan levels as previously described41.
Statistical analysis
All statistical analysis was performed using Graph Pad Prism version 6.0f for Mac OSX (GraphPad Software, La Jolla, CA). Groups where compared using the Mann-Whitney test, paired blood and GALT samples were compared using Wilcoxon marched-pairs signed rank test. Associations between groups were determined by Spearman's rank correlation. P values < 0.05 were considered statistically significant.
Supplementary Material
Supplementary Figure 1. Frequency of CD4+ iNKT cells in the blood and GALT of HIV-infected individuals. Percentage of iNKT cells in the blood and the GALT of all subjects (A, n=33). Percentage of the CD4+ subset of iNKT cells in the blood (B, controls n=8, HIV-untreated n=6, HIV-ART treated n=13, and HIV-controllers n=3) and GALT (C, controls n=9, HIV-untreated n=5, HIV-ART treated n=9, and HIV-controllers n=3). *** indicates p < 0.001.
Supplementary Figure 2. CD4 T cell immune activation in HIV-infected individuals with or without IL-10 production by GALT iNKT cells. Comparison between the levels of CD38+HLA-DR+ CD4+ T cells in the blood of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by GALT iNKT cells (A). Comparison between the levels of CD38+HLA-DR+ CD4+ (B) and CD8+ (C) T cells in the blood of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by blood iNKT cells. *** indicates p < 0.001.
Supplementary Figure 3. IL-4 and IL-10 production by iNKT cell are associated with lower CD38 levels. Expression of CD38 on CD4+ T cells (A) and CD8+ T cells (B) in the blood and GALT of controls (n= 7) and HIV-infected subjects (n=18). Associations between IL-4+ iNKT cells in the GALT and CD38+ expression on CD4+ T cells in the GALT (C) and CD8+ T cells in the blood (D) and GALT (E) of HIV-infected subjects. Associations between IL-10+ iNKT cells in the GALT and CD38 expression by CD4+ T cells in the blood (F) of HIV-infected subjects. Associations between IL-10+ iNKT cells in the blood and CD38 expression by CD4+ T cells in the GALT (G), CD38 expression by CD8+ T cells in the blood (H) and GALT (I) of HIV-infected subjects. Comparison between the expression of CD38 by CD4+ T cells in the blood of HIV-infected subjects with (n=6) or without (n=7) IL-10 production by GALT iNKT cells (J). Comparison between the expression of CD38 by CD4+ T cells in the GALT (K), CD8+ T cells in the blood (L) and GALT (M) of HIV-infected subjects with (n=6) or without (n=7) IL-10 production by blood iNKT cells. * indicates p < 0.05 and *** indicates p < 0.001.
Supplementary Figure 4. Frequency of Tregs in the blood and GALT of HIV-infected individuals. Tregs were defined as CD4+CD25+Foxp3+ T cells and their frequency was measured in the blood and GALT of healthy controls (n=8) and HIV-infected subjects (n=22) (A). Association between CD38+HLA-DR+ CD4+ T cells and Tregs frequency in the GALT of HIV-infected subjects (B). Association between CD38+ CD8+ T cells and Tregs frequency in the GALT of HIV-infected subjects (C).
Supplementary Figure 5. IL-6 levels in HIV-infected individuals. IL-6 was measured by ELISA in the plasma of healthy controls (n=9) and HIV-infected subjects (n=22). * indicates p < 0.05.
Supplementary Figure 6. Markers of microbial translocation in HIV-infected individuals with or without production of IL-4 or IL-10 by iNKT cells. Comparison between the levels of sCD14 in the plasma of HIV-infected subjects with iNKT cells producing IL-4 >10% (n=6) or <10% (n=9) (A). Comparison between the levels of sCD14 in the plasma of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by GALT iNKT cells (B). Comparison between the Kyn/Trp ratio in the plasma of HIV-infected subjects with iNKT cells producing IL-4 >10% (n=6) or <10% (n=9) (C). ** indicates p < 0.01.
Acknowledgments
This work was supported by NIAID (R01 AI52731), the Delaney AIDS Research Enterprise (DARE; AI096109), NIAID (K24 AI069994), the National Institutes of Health, University of California, San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research, P30-AI027763, the UCSF Clinical and Translational Research Institute Clinical Research Center (UL1 RR024131), the Center for AIDS Prevention Studies (P30 MH62246), the DC CFAR (P30AI117970 and P30AI087714), the Fundação de Amparo a Pesquisa do Estado de São Paulo (04/15856-9 and 2010/05845-0), CNPq/CAPES 056/2012, and the Peter and Shelagh Godsoe Family Foundation through the AIDS Research Institute at UCSF, and the CFAR Network of Integrated Systems (R24 AI067039). I.V.C. was supported by CHRP D13-SF-388 and NSF 1144247. M.S. was supported by NIH NCI K23 CA157929. D.S. was supported by NIH NIAID K08 A120071. L.L. was supported by NH&MRC CJ Martin Fellowship. We acknowledge the NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of CD1d tetramers. We thank Montha Pao, Monica Deswal, Rebecca Hoh and Melissa Krone for SCOPE study coordination.
Footnotes
Author contributions: DPP analyzed data and wrote the manuscript; CC performed experiments and helped write the manuscript; IVC analyzed microbiome data, helped write the manuscript and helped obtain grant funding; DF performed microbiome profiling; LL helped with flow cytometry panels; YH performed kynurenine/tryptophan assays; MS performed GALT biopsies; SVL oversaw microbiome analyses; PWH and DFN gave input on overall study design, analysis, and helped write the manuscript; DS designed and obtained grant funding for the study, performed experiments and helped write the manuscript.
Disclosure: The authors declare that no competing interests exist.
References
- 1.Miedema F, et al. Immune activation and collateral damage in AIDS pathogenesis. Front Immunol. 2013;4:298. doi: 10.3389/fimmu.2013.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatano H. Immune activation and HIV persistence: considerations for novel therapeutic interventions. Curr Opin HIV AIDS. 2013;8:211–216. doi: 10.1097/COH.0b013e32835f9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, et al. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 4.Utay NS, Hunt PW. Role of immune activation in progression to AIDS. Curr Opin HIV AIDS. 2016 doi: 10.1097/COH.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain V, et al. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J Infect Dis. 2013;208:1202–1211. doi: 10.1093/infdis/jit311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godfrey DI, et al. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin Immunol. 2010;22:61–67. doi: 10.1016/j.smim.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Kaer L, Parekh VV, Wu L. Invariant natural killer T cells: bridging innate and adaptive immunity. Cell Tissue Res. 2011;343:43–55. doi: 10.1007/s00441-010-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingender G, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diana J, Lehuen A. NKT cells: friend or foe during viral infections? Eur J Immunol. 2009;39:3283–3291. doi: 10.1002/eji.200939800. [DOI] [PubMed] [Google Scholar]
- 15.Juno JA, Keynan Y, Fowke KR. Invariant NKT cells: regulation and function during viral infection. PLoS Pathog. 2012;8:e1002838. doi: 10.1371/journal.ppat.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Vliet HJ, et al. Selective decrease in circulating V alpha 24+V beta 11+ NKT cells during HIV type 1 infection. J Immunol. 2002;168:1490–1495. doi: 10.4049/jimmunol.168.3.1490. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez CS, et al. Peripheral NKT cells in simian immunodeficiency virus-infected macaques. J Virol. 2009;83:1617–1624. doi: 10.1128/JVI.02138-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Vliet HJ, et al. Cutting edge: Rapid recovery of NKT cells upon institution of highly active antiretroviral therapy for HIV-1 infection. J Immunol. 2006;177:5775–5778. doi: 10.4049/jimmunol.177.9.5775. [DOI] [PubMed] [Google Scholar]
- 19.Snyder-Cappione JE, et al. Lower cytokine secretion ex vivo by natural killer T cells in HIV-infected individuals is associated with higher CD161 expression. AIDS. 2009;23:1965–1970. doi: 10.1097/QAD.0b013e32832b5134. [DOI] [PubMed] [Google Scholar]
- 20.Moll M, et al. Severe functional impairment and elevated PD-1 expression in CD1d-restricted NKT cells retained during chronic HIV-1 infection. Eur J Immunol. 2009;39:902–911. doi: 10.1002/eji.200838780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roelofs-Haarhuis K, Wu X, Gleichmann E. Oral tolerance to nickel requires CD4+ invariant NKT cells for the infectious spread of tolerance and the induction of specific regulatory T cells. J Immunol. 2004;173:1043–1050. doi: 10.4049/jimmunol.173.2.1043. [DOI] [PubMed] [Google Scholar]
- 22.Sandberg JK, et al. Selective loss of innate CD4(+) V alpha 24 natural killer T cells in human immunodeficiency virus infection. J Virol. 2002;76:7528–7534. doi: 10.1128/JVI.76.15.7528-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motsinger A, et al. CD1d-restricted human natural killer T cells are highly susceptible to human immunodeficiency virus 1 infection. J Exp Med. 2002;195:869–879. doi: 10.1084/jem.20011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez CS, Kelleher AD, Finlayson R, Godfrey DI, Kent SJ. NKT cell depletion in humans during early HIV infection. Immunol Cell Biol. 2014;92:578–590. doi: 10.1038/icb.2014.25. [DOI] [PubMed] [Google Scholar]
- 25.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 27.Hunt PW, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014;210:1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nockher WA, Bergmann L, Scherberich JE. Increased soluble CD14 serum levels and altered CD14 expression of peripheral blood monocytes in HIV-infected patients. Clin Exp Immunol. 1994;98:369–374. doi: 10.1111/j.1365-2249.1994.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelsers MM, et al. Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue distribution and clinical utility. Clin Biochem. 2003;36:529–535. doi: 10.1016/s0009-9120(03)00096-1. [DOI] [PubMed] [Google Scholar]
- 30.Lozupone CA, et al. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 31.Dillon SM, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibarrondo FJ, et al. Preferential depletion of gut CD4-expressing iNKT cells contributes to systemic immune activation in HIV-1 infection. Mucosal Immunol. 2013;6:591–600. doi: 10.1038/mi.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan AC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol. 2013;172:129–137. doi: 10.1111/cei.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med. 2002;195:637–641. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rout N, et al. Loss of effector and anti-inflammatory natural killer T lymphocyte function in pathogenic simian immunodeficiency virus infection. PLoS Pathog. 2012;8:e1002928. doi: 10.1371/journal.ppat.1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madsen KL, et al. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. 1999;5:262–270. doi: 10.1097/00054725-199911000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Nowak P, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015 doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 39.Loh L, Ivarsson MA, Michaelsson J, Sandberg JK, Nixon DF. Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal Immunol. 2014;7:1233–1243. doi: 10.1038/mi.2014.13. [DOI] [PubMed] [Google Scholar]
- 40.Gadola SD, Dulphy N, Salio M, Cerundolo V. Valpha24-JalphaQ-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4(+) and CD8alphabeta(+) T lymphocytes. J Immunol. 2002;168:5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, et al. A simple LC-MS/MS method for determination of kynurenine and tryptophan concentrations in human plasma from HIV-infected patients. Bioanalysis. 2013;5:1397–1407. doi: 10.4155/bio.13.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Frequency of CD4+ iNKT cells in the blood and GALT of HIV-infected individuals. Percentage of iNKT cells in the blood and the GALT of all subjects (A, n=33). Percentage of the CD4+ subset of iNKT cells in the blood (B, controls n=8, HIV-untreated n=6, HIV-ART treated n=13, and HIV-controllers n=3) and GALT (C, controls n=9, HIV-untreated n=5, HIV-ART treated n=9, and HIV-controllers n=3). *** indicates p < 0.001.
Supplementary Figure 2. CD4 T cell immune activation in HIV-infected individuals with or without IL-10 production by GALT iNKT cells. Comparison between the levels of CD38+HLA-DR+ CD4+ T cells in the blood of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by GALT iNKT cells (A). Comparison between the levels of CD38+HLA-DR+ CD4+ (B) and CD8+ (C) T cells in the blood of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by blood iNKT cells. *** indicates p < 0.001.
Supplementary Figure 3. IL-4 and IL-10 production by iNKT cell are associated with lower CD38 levels. Expression of CD38 on CD4+ T cells (A) and CD8+ T cells (B) in the blood and GALT of controls (n= 7) and HIV-infected subjects (n=18). Associations between IL-4+ iNKT cells in the GALT and CD38+ expression on CD4+ T cells in the GALT (C) and CD8+ T cells in the blood (D) and GALT (E) of HIV-infected subjects. Associations between IL-10+ iNKT cells in the GALT and CD38 expression by CD4+ T cells in the blood (F) of HIV-infected subjects. Associations between IL-10+ iNKT cells in the blood and CD38 expression by CD4+ T cells in the GALT (G), CD38 expression by CD8+ T cells in the blood (H) and GALT (I) of HIV-infected subjects. Comparison between the expression of CD38 by CD4+ T cells in the blood of HIV-infected subjects with (n=6) or without (n=7) IL-10 production by GALT iNKT cells (J). Comparison between the expression of CD38 by CD4+ T cells in the GALT (K), CD8+ T cells in the blood (L) and GALT (M) of HIV-infected subjects with (n=6) or without (n=7) IL-10 production by blood iNKT cells. * indicates p < 0.05 and *** indicates p < 0.001.
Supplementary Figure 4. Frequency of Tregs in the blood and GALT of HIV-infected individuals. Tregs were defined as CD4+CD25+Foxp3+ T cells and their frequency was measured in the blood and GALT of healthy controls (n=8) and HIV-infected subjects (n=22) (A). Association between CD38+HLA-DR+ CD4+ T cells and Tregs frequency in the GALT of HIV-infected subjects (B). Association between CD38+ CD8+ T cells and Tregs frequency in the GALT of HIV-infected subjects (C).
Supplementary Figure 5. IL-6 levels in HIV-infected individuals. IL-6 was measured by ELISA in the plasma of healthy controls (n=9) and HIV-infected subjects (n=22). * indicates p < 0.05.
Supplementary Figure 6. Markers of microbial translocation in HIV-infected individuals with or without production of IL-4 or IL-10 by iNKT cells. Comparison between the levels of sCD14 in the plasma of HIV-infected subjects with iNKT cells producing IL-4 >10% (n=6) or <10% (n=9) (A). Comparison between the levels of sCD14 in the plasma of HIV-infected subjects with (n=8) or without (n=7) IL-10 production by GALT iNKT cells (B). Comparison between the Kyn/Trp ratio in the plasma of HIV-infected subjects with iNKT cells producing IL-4 >10% (n=6) or <10% (n=9) (C). ** indicates p < 0.01.


