Tight regulation of canonical Wnt/β-catenin signaling is critical for maintaining normal hematopoiesis and its deregulation contributes to leukemia development1. β-catenin, a central molecule in the canonical Wnt/β-catenin signaling pathway, acts as a nuclear transcriptional co-activator with the lymphoid enhancer factor/T cell factor (LEF/TCF) family to stimulate transcription of various target genes2. The intracellular level of β-catenin is controlled by a degradation complex consisting of Apc, Gsk-3β, Axin 1 and Ck-12. We found that loss of Apc leads to bone marrow failure due to exhaustion of hematopoietic stem and progenitor cells in mice3. A marked decrease of Mac-1+Gr-1+ myeloid cells and rapid depletion of granulocyte-macrophage progenitors (GMPs), but not megakaryocyte-erythrocyte progenitors (MEPs) were observed in both Apc-null3 and transgenic mice with expression of stabilized β-catenin4, 5. However, whether this decrease of myeloid cells is due to a blockage of differentiation of GMPs or the depletion of GMPs in vivo or both have not been defined.
PU.1, a member of the Ets family of DNA-binding proteins, is encoded by the Sfpi1 gene and is an important transcriptional regulator that is required for the generation of early myeloid and lymphoid progenitors6. The PUER cells represent an experimentally manipulable cellular model system for analyzing molecular mechanisms that underlie macrophage differentiation in vitro. The PUER cells were originally derived from mouse PU.1−/− myeloid progenitors with expression of a conditionally active PU.1 protein fused to the ligand binding domain of the estrogen receptor (PUER)7. Addition of 4-hydroxy-tamoxifen (4-OHT) leads to activation of PU.1 and differentiation of PUER cells. To test whether activation of Wnt/β-catenin signaling affects macrophage differentiation induced by PU.1 in PUER cells or not, we first expressed stabilized β-catenin mutant (β-catenin-S33Y)8 in PUER cells by retroviral vector Migr1, which co-expresses EGFP (Supplementary Figure 1a). The expression of β-catenin-S33Y was confirmed by Western blot (Supplementary Figure 1b). The PUER cells were derived from the original PUER line that was established in Dr. Singh’s laboratory7. The current cell line underwent monocyte/macrophage differentiation with a low concentration of 4-OHT. Upon 4-OHT addition, PUER protein is induced in PUER cells and the cells undergo differentiation into Mac-1+/F4/80+ macrophages within 3–5 days (Supplementary Figure 2). Of interest, induction of PU.1 expression in PUER cells with expression of the stabilized β-catenin resulted in a significantly low number of Mac1+/F4/80+ cells (Figure. 1a). In the absence of 4-OHT, PUER cells with or without stabilized β-catenin displayed the morphology of myeloid progenitors with a large nucleus (Figure 1b). After treatment of 4-OHT for 5 days, the vector-expressing cells increased in size with an extensive cytoplasm morphologically resembling macrophages. In contrast, the PUER cells with stabilized β-catenin had the same morphology as the cells without 4-OHT treatment (Figure 1b). Taken together, these results suggest that stabilized β-catenin blocks PU.1-induced macrophage differentiation of PUER cells. APC negatively regulates the Wnt/β-catenin pathway by controlling the turnover of β-catenin3. We next explored the consequences of knockdown of the Apc gene on macrophage differentiation in PUER cells. Inhibition of Apc by Apc shRNA 1 or 2 against Apc in PUER cells led to stabilization of β-catenin (Supplementary Figure 3). After induction of PU.1 for 3 days by 4-OHT treatment, PUER cells expressing Apc shRNA1 or 2 had a lower percentage of Mac1+/F4/80+ cells compared with the PUER cells expressing the control vector (Figure 1c), indicating that activation of the Wnt/β-catenin pathway by Apc inhibition also represses PU.1-induced macrophage differentiation of PUER cells. In consistence, we found that stabilized β-catenin also blocked GM-CSF-driven macrophage differentiation of mouse primary bone marrow cells (Figure 1d).
Figure 1. Activated Wnt signaling blocks monocyte/macrophage differentiation in human and mouse progenitor cells.
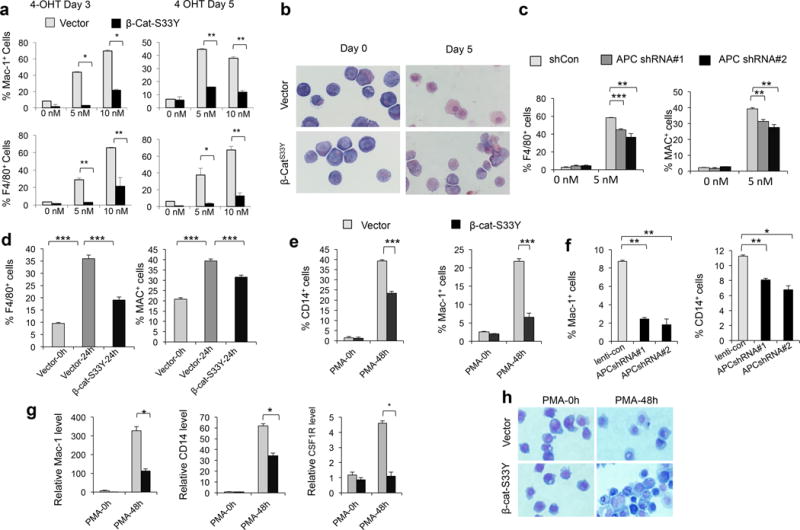
(a) Histograms of flow cytometric analysis of PUER cells expressing Migr1-β-cat-S33Y or Migr1 vector after treatment with 4-OHT for 3 or 5 days. (b) May-Grünwald giemsa staining of PUER cells expressing Migr1-β-cat-S33Y or Migr1 vector after treatment with 5nM 4-OHT. The cells were stained with May-Grünwald giemsa. (c) Summary of flow cytometric analysis of F4/80+ (left panel) and Mac-1+ (right panel) PUER cells expressing shCon (control vector), APC shRNA#1 or APC shRNA#2 after treatment with 5 nM 4-OHT for 3 days. (d) Summary of flow cytometric analysis of percentage of F4/80+ cells and Mac-1+ cells in primary bone marrow (BM) cells expressing Migr1-β-cat-S33Y or Migr1 vector with treatment of 20ng/ml GM-CSF for 24 hours (vector-24h, β-cat-S33Y-24h) or vector-infected BM cells without addition of GM-CSF (vector-0h) in vitro. BM cells, isolated from the C57BL/6 mice, were infected with retrovirus expressing Migr1 or Migr1-β-cat-S33Y. Two days after infection, the cells were received GM-CSF treatment. (e) Summary of flow cytometric analysis of percentage of CD14+ and Mac-1+ in U937 cells expressing Migr1-β-cat-S33Y or Migr1 vector after treatment of 50 ng/ml PMA or vehicle for 48 hours. (f) Summary of flow cytometric analysis of percentage of CD14+ and Mac-1+ in U937 cells expressing ShCon (control), APCshRNA#1 or APC shRNA#2 after treatment of 50 ng/ml PMA or vehicle for 48 hours. (g) qRT-PCR analysis of the expression of Mac-1, CD14 and CSF1R in U937 cells expressing Migr1-β-cat-S33Y or Migr1 vector with or without PMA treatment. (h) May-Grünwald giemsa staining of U937 cells expressing control vector or β-cat-S33Y infected before and after treatment of PMA. All experiments were performed in triplicate. The results are representative of 3 independent experiments. *, p<0.05;**, p<0.01; ***, p< 0.001.
Next, we determined whether activation of Wnt/β-catenin inhibits differentiation of human U973 cells exposed to PMA. We found that either expression of stabilized β-catenin (Supplementary Figure 4a) or knockdown Apc expression (Supplementary Figure 5a,b) significantly reduced the frequency of Mac-1+ and CD14+ cells of U937 cells after exposed to PMA (Supplementary Figure 4b, Figure 1e, f). Furthermore, stabilized β-catenin inhibits mRNA expression of CD14, Mac-1 and Csf1r genes induced by PMA in U937 cells, as determined by qRT-PCR (Figure 1g). As shown in Figure 1h, the morphological features associated with mature monocytes were observed in U937 cells expressing control vector but not β-cat-S33Y after PMA treatment for 48 hours. These regulates suggest that activation of Wnt/β-catenin signaling inhibits PMA-induced monocyte/macrophage differentiation of U937 cells. Together, our studies demonstrate a negative role of β-catenin overexpression in monocyte/macrophage differentiation.
To understand the mechanism by which β-catenin inhibits PU.1-mediated monocyte differentiation, we performed microarray analysis of gene expression profiling of PUER cells with expression of stabilized β-catenin or control vector before and after 24 h induction of PU.1 expression. As shown in Supplementary Figure 6, 505 genes were markedly up-regulated while 390 genes were significantly down-regulated by PU.1 gene expression (≥2 or ≤ 2fold, FDR<0.01) (Supplementary Table 1). Induction of PU.1 expression in PUER cells with expression of stabilized β-catenin for 24 hours resulted in 505 genes up-regulated by PU.1 with 219 genes significantly inhibited by stabilized β-catenin. However, of 390 genes down-regulated by PU.1, 72 genes were up-regulated by stabilized β-catenin (Supplementary Table 2, Figure 2a), indicating that β-catenin significantly impairs PU.1-targeted gene transcription. Notably, more than 32% of PU.1-regulated genes were reversibly regulated by stabilized β-catenin, indicating that β-catenin overexpression antagonizes the function of PU.1 in myeloid progenitors.
Figure 2. Active β-catenin represses transcriptional programs induced by PU.1 in PUER cells and Egr2 partially rescues the blockage induced by stabilized β-catenin.
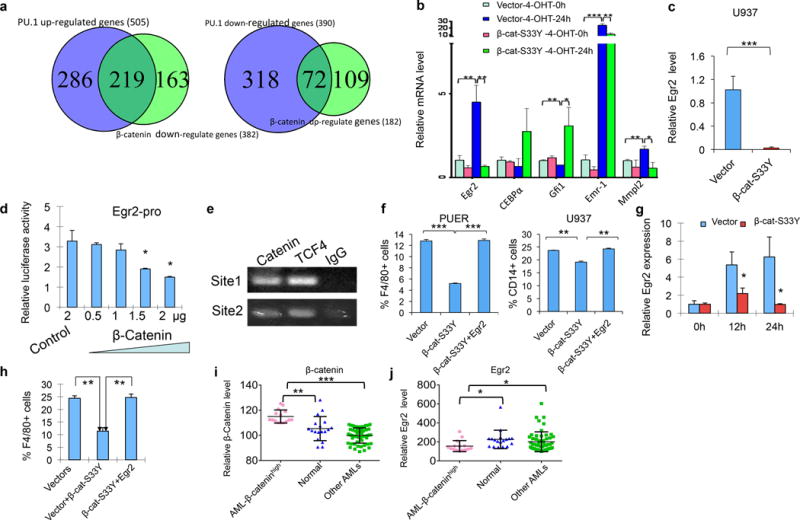
(a) Venn diagrams showing the PU.1-regulated genes that were down-regulated (right panel) or up-regulated (left panel) by stabilized β-catenin in PUER cells. (b) qRT-PCR analysis of expression of the selected genes in PUER cells expressing GFP (Migr1) control vector or Migr1-β-cat-S33Y (right panel) with or without induction of PU.1. Gene expression was normalized initially to Hprt expression. (c) Active β-catenin inhibits Egr2 expression in U937 cells. Egr2 expression was determined by qRT-PCR in U937 cells expressing β-cat-S33Y or control vector. The U937 cells were cultured with PMA for 12 hours before analysis. (d) Egr2 promoter is downregulated by β-catenin. The luciferase reporter plasmid containing Egr2 promoter, control plasmids and plasmids expressing stabilized β-catenin were co-transfected into 293T cells. The amount of β-catenin plasmids were increased gradually with gradually reduced amount of control plasmids to keep constant amount of plasmids added in each sample. Experiments were repeated for three times. (e) β-catenin and TCf4 directly binds the Egr2 promoter. β-catenin, TCF4 antibodies and IgG were used to perform the ChIP assay and PCR products were run on 2% gel. Specific DNA bands were amplified by primers of Site1 and Site2 from DNA immunoprecipitated by β-catenin or TCF4 antibodies but not control IgG. Experiments were repeated for two times. (f) Summary data of flow cytometric analysis of PUER cells and U937 cells. The percentage of F4/80-positive cells was determined in PUER cells expressing control vector, β-catenin or β-catenin and Egr2 after treatment of 5 nM 4-OHT for 24 hours (left panel). The percentage of CD14-positive cells in was determined in U937 cells expressing control vector, β-catenin or β-catenin and Egr2 after treatment of 50 ng/ml PMA for 48 hours (right panel). (g) β-catenin inhibits Egr2 expression in BM cells. Egr2 expression was determined by qRT-PCR in BM cells expressing vector or β-catenin-S33Y before (0h) and after treatment of 20ng GM-CSF for 12 or 24 hours (12h, 24h). BM cells were infected with retrovirus expressing MSCV-neo vector or MSCV-neo-β-catenin-S33Y and cultured in medium with neomycin for 4 days before GM-CSF treatment. (h) Summary of flow cytometric analysis of percentage of F4/80+ cells in BM cells expressing MSCV-neo and Migr1, or Migr1-β-cat-S33Y or MSCV-neo-Egr2 and Migr1-β-cat-S33Y with treatment of 20ng/ml GM-CSF for 24 hours in vitro. (I, J) Egr2 is downregulated in BM cells from AML patients with β-catenin overexpression as compared to BM cells from healthy individuals. The signal intensities of Egr2 (probe ID: ILMN_1743199) and β-catenin (probe ID: ILMN_1808436) (probe ID: ILMN_2392043) from published microarray dataset (Series GSE34823) of BM mononuclear cells from 15 AML samples with top-level expression of β-catenin, 54 other AML samples and 18 control samples from healthy individuals were analyzed. The experiments were performed in triplicate. Data are representative of 2–3 independent experiments. *, p<0.05;**, p<0.01; ***, p< 0.001.
Consistent with our microarray data, qRT-PCR analysis revealed that expression of a conditional version of PU.1 induced Egr2, Mmpl2, Emr1 and inhibited Gfi1 in PUER cells (Figure 2b). Notably, stabilized β-catenin inhibited PU.1-induced Egr2, Mmpl2 and Emr1 and significantly increased Gfi1 expression. C/EBPα, which antagonizes PU.1-induced macrophage differentiation9, was also markedly up-regulated by stabilized β-catenin in PUER cells after induction of PU.1. It is likely that β-catenin overexpression antagonizes PU.1-contolled gene transcription, partially through up-regulating CEBPα and GFI1 expression.
Egr2 is a transcriptional master regulator that is regulated by PU.1 and cooperates with PU.1 in promoting monocyte-macrophage differentiation. It activates macrophage specific genes including Emr-1 and Mmpl2 and represses genes that promote neutrophil lineage differentiation, including Gfi17. To determine whether stabilized β-catenin inhibited Egr2 expression in a human cell line during monocyte/macrophage differentiation, we expressed β-catenin-S33Y in U937 cells. EGR2 expression is determined by qRT-PCR before and after induction of monocyte/macrophage differentiation of U937 cells by PMA. Similarly, β-catenin-S33Y markedly suppressed PMA-induced EGR2 expression during U937 cell differentiation (Figure 2c). Stabilized β-catenin significantly inhibited Egr2 expression in the absence of PU.1 gene expression, raising the possibility that β-catenin also directly regulates Egr2 expression. There are several predicted TCF4 binding sites identified on the promoter region of Erg2 (Supplementary Figure 7). Luciferase reporter showed that Egr2 promoter activity was repressed significantly with an increased amount of plasmids expressing stabilized β-catenin (Figure 2d). Next, we performed chromatin immunoprecipitation (ChIP) assay followed by PCR. Our results revealed that both TCF4 and β-catenin binds directly to the predicted TCF4 binding sites in Egr2 promoter (Figure 2e). To further determine whether down-regulation of Egr2 plays a critical role in mediating the inhibitory function of β-catenin in monocyte differentiation, we co-expressed Egr2 and β-catenin-S33Y in PUER and U937 cells. As shown in Figure 2f, forced-expression of Egr2 reversed β-catenin-S33Y-induced inhibition of monocyte/macrophage differentiation of both PUER and U937 cells. In mouse primary BM cells, Egr2 expression was markedly increased at 12 hours and 24 hours after GM-CSF treatment. Notably, β-catenin-S33Y significantly blocked GM-CSF-induced Egr2 expression (Figure 2g). Moreover, forced-expression of Egr2 reversed the inhibitory effect of stabilized β-catenin on monocyte/macrophage differentiation of primary BM cells (Figure 2h). Together, stabilized β-catenin inhibits Egr2 expression in mouse primary BM cells, PUER and human U937 cells while forced expression of Egr2 released the blockage of monocyte-macrophage differentiation induced by stabilized β-catenin in these cells, suggesting an important role of Egr2 in mediating the negative effect of β-catenin overexpression in monocyte-macrophage differentiation.
β-catenin overexpression is frequently detected in AML samples and it is associated with an adverse prognosis10, 11. By analysis of a published set of microarray data from BM cells from 69 AML patients and 18 control healthy individuals12, we found that a group of AML patients (15 out of 69) had a markedly increased β-catenin expression as compared to healthy individuals (P=0.001) and the other AML patients (P=5.65E-10). Of interest, this group of AML patients with a high β-catenin expression displayed a reduced Egr2 expression in BM cells as compared to healthy individuals (P=0.0156) and the other AML patients (P=0.034) (Figure 2i, j). These data suggest that an increased expression of β-catenin likely inhibits Egr2 expression in BM cells in AML patients.
In summary, we have shown that β-catenin overexpression induced blockage of monocyte-macrophage differentiation by inhibiting PU.1-targeted gene transcription including Egr2 expression in myeloid progenitor cells. A recent study showed that minimal PU.1 reduction induces myeloid-biased preleukemic stem cells and promotes subsequent transformation to AML in the context of Msh2-deficiency13. Therefore, compromised PU.1-targeted gene transcription induced by β-catenin overexpression, at least partially, may mediate a pathogenic role of β-catenin in myeloid leukemia.
Supplementary Material
Acknowledgments
We would like to acknowledge the funding support from by the National Institute of Health grants RO1 CA140979 (to Z.Q.) and the China Scholarship Council (to W.J.).
Footnotes
Authors’ contributions
ZQ, YS, WJ: Study design; ZQ, YS, WJ, YH, XQ, JL: acquisition of data, analysis and interpretation of data; ZQ, HO, YS, WJ, YH, manuscript preparation
Online Supplementary Material
Supplementary Material and Methods and Data can be found with this article online.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Lento W, Congdon K, Voermans C, Kritzik M, Reya T. Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol. 2013 Feb;5(2) doi: 10.1101/cshperspect.a008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009 Jul;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian Z, Chen L, Fernald AA, Williams BO, Le Beau MM. A critical role for Apc in hematopoietic stem and progenitor cell survival. J Exp Med. 2008 Sep 1;205(9):2163–2175. doi: 10.1084/jem.20080578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006 Oct;7(10):1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 5.Baba Y, Yokota T, Spits H, Garrett KP, Hayashi S, Kincade PW. Constitutively active beta-catenin promotes expansion of multipotent hematopoietic progenitors in culture. J Immunol. 2006 Aug 15;177(4):2294–2303. doi: 10.4049/jimmunol.177.4.2294. [DOI] [PubMed] [Google Scholar]
- 6.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007 Oct 15;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 7.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006 Aug 25;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 8.Ming M, Wang S, Wu W, Senyuk V, Le Beau MM, Nucifora G, et al. Activation of Wnt/beta-catenin protein signaling induces mitochondria-mediated apoptosis in hematopoietic progenitor cells. J Biol Chem. 2012 Jun 29;287(27):22683–22690. doi: 10.1074/jbc.M112.342089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003 Oct;4(10):1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 10.Chung EJ, Hwang SG, Nguyen P, Lee S, Kim JS, Kim JW, et al. Regulation of leukemic cell adhesion, proliferation, and survival by beta-catenin. Blood. 2002 Aug 1;100(3):982–990. doi: 10.1182/blood.v100.3.982. [DOI] [PubMed] [Google Scholar]
- 11.Gandillet A, Park S, Lassailly F, Griessinger E, Vargaftig J, Filby A, et al. Heterogeneous sensitivity of human acute myeloid leukemia to beta-catenin down-modulation. Leukemia. 2011 May;25(5):770–780. doi: 10.1038/leu.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Bletiere DR, Blanchet O, Cornillet-Lefebvre P, Coutolleau A, Baranger L, Genevieve F, et al. Routine use of microarray-based gene expression profiling to identify patients with low cytogenetic risk acute myeloid leukemia: accurate results can be obtained even with suboptimal samples. BMC Med Genomics. 2012;5:6. doi: 10.1186/1755-8794-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, da Silva Ferreira M, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015 Oct;21(10):1172–1181. doi: 10.1038/nm.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


