Modern approaches to Human Leukocyte Antigen (HLA)-haploidentical (haplo) blood or marrow transplantation (BMT), such as the use of high-dose posttransplantation cyclophosphamide (PTCy), have resulted in low incidence of graft failure, graft-versus-host disease (GVHD), and nonrelapse mortality, making its safety comparable to HLA-matched unrelated donor (MUD) BMT. 1,2 Thus, the potentially curative option of BMT has become a reality to the vast majority of patients. However, posttransplant relapse remains the most common cause of treatment failure. In order to design effective relapse prevention and treatment strategies, we must understand the mechanisms of relapse after the increasingly utilized haplo BMT with PTCy platform3.
HLA complex-mediated presentation of cellular antigens is required for effective T cell-dependent tumor recognition and cytotoxicity. Cancer cells may downregulate HLA expression to evade T-cell surveillance.4 While HLA loss occurs in 70-90% of certain untreated solid tumors,5,6 it is rare in leukemia at presentation.7 However, it has been identified as a mechanism of leukemia immune escape after haplo BMT, occurring in approximately 33% of AML relapses.8,9 Investigators at the San Raffaele Scientific Institute first described acquired uniparental disomy (UPD) of chromosome 6p as a mechanism of genomic HLA loss and evolution in AML patients relapsing after haplo BMT using peripheral blood stem cells (PBSCs) grafts.10 They hypothesized that HLA loss may reflect alloimmune pressure mediated by donor T-cells toward the HLA-mismatches and later identified that active disease at the time of transplant, higher graft T-cell dose, younger patient age, and occurrence of chronic GVHD may represent risk factors.8
However, the conditioning intensity, graft source (PBSCs vs. bone marrow), and type of GVHD prophylaxis (anti-thymocyte globulin vs. PTCy) in the Italian study differ substantially from the PTCy haplo BMT approach.3,11 While single nucleotide polymorphism (SNP) array may detect a copy neutral loss of heterozygosity (CN-LOH) of 6p, this is not routinely performed on relapse samples and its sensitivity is limited in early relapse. Assays to distinguish donor from patient-specific alleles are not clinically available at present. Therefore, the degree to which HLA loss contributes to relapse after the PTCy haplo platform is unknown. Herein, we report the first direct evidence that genomic HLA loss may occur after haplo BMT with PTCy.
Two high-risk AML patients in first complete remission received nonmyeloablative (NMA) conditioning, which consisted of fludarabine 30mg/m2 IV, Days −6 to −2), Cy (14.5 mg/kg IV, Days −6 and −5), and total body irradiation (200 cGy, Day −1). Both patients received a T-cell replete bone marrow (BM) graft with a total nucleated cell (TNC) count of 2.9 × 107 TNC/kg for recipient 1 and 5.92 × 107 TNC/kg for recipient 2 followed by PTCy (50 mg/kg IV on days +3 and +4), mycophenolate mofetil, and tacrolimus (Supplemental Table 1).3 In the setting of 100% donor chimerism, recipient 1 developed isolated testicular relapse eight months posttransplant that was treated with surgery, radiation, and prophylactic intrathecal chemotherapy. At twelve months, BM and PB studies confirmed 100% donor chimerism and remission. Systemic relapse developed at thirteen months with BM revealing 63% myleoblasts and 49% recipient DNA. He underwent reinduction, followed by a second NMA haplo BMT from a different donor. At the time of publication, 1 year after second BMT, he was in remission receiving Vidaza maintenance. For recipient 2, PB and BM studies showed 100% donor chimerism and remission at twelve and eighteen months posttransplant. Systemic relapse developed at twenty-two months when the BM revealed 20% myeloblasts and 36% recipient DNA. At the time of publication, after receiving two salvage regimens, recipient 2 was in remission at Day 90 after second NMA BMT from a mismatched unrelated donor.
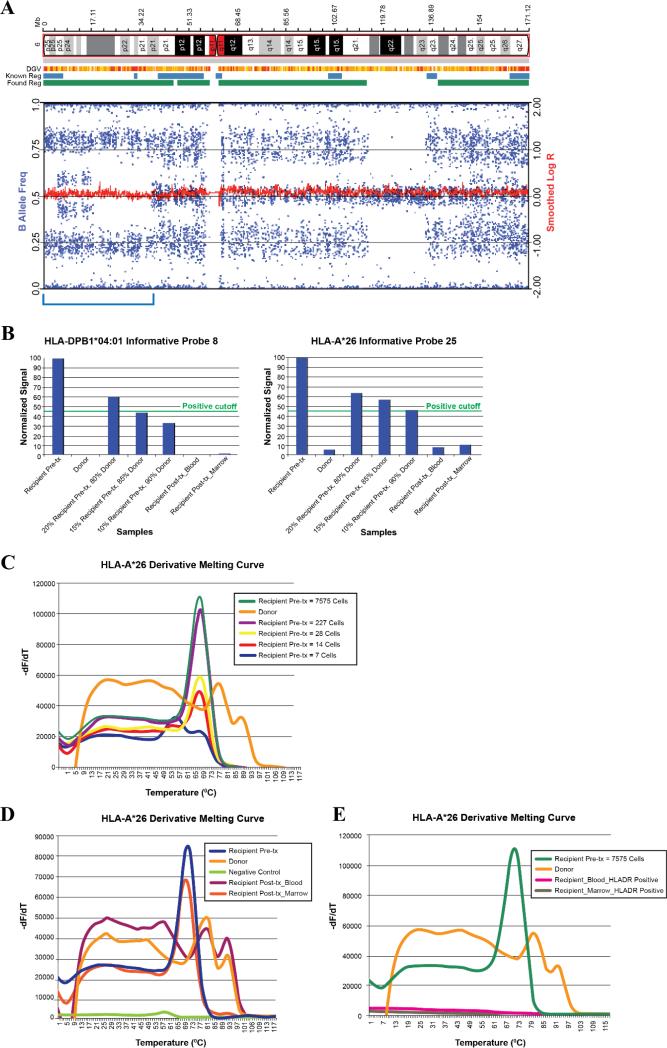
For recipient 1, G-banded karyotype analysis at the time of systemic relapse showed a complex karyotype similar to the findings at diagnosis. Single nucleotide polymorphism (SNP) array was performed on the relapse sample BM and PB and detected the presence of both patient and donor alleles. However, a clonal 38.2 Mb region of CN-LOH was present on the short arm of chromosome 6. Specifically, CN-LOH occurred in two regions of shared recipient and donor haplotype: the terminal end (4.9 Mb from terminal to p25.1) and more proximally (20.8 Mb from p22.3 to p21.2). In addition, CNLOH occurred over 12.5 Mb (from p25.1 to p22.3) in a region where the recipient and donor haplotypes differed (Figure 1A). To confirm putative HLA loss, we performed reverse sequence specific oligonucleotide (rSSO) hybridization and melting curve analysis of the targeted alleles. For rSSO, based on pretransplant HLA typing results (Supplementary Table 1), PB samples from recipient 1 and donor 1 from pretransplantation time points were utilized to determine the informative probes for mismatches between the recipient and donor alleles at HLA-A*26, HLA B*56, HLA-C*01, and DPB1*04:01. DNA mixing experiments were performed to establish a detection threshold (Figure 1B). None of the probes for the patient's mismatched HLA alleles were detected near the threshold level in the PB or BM at relapse. Consistent with the above, melting curve analysis primers designed for the recipient 1-specific HLA-A*26 detected its presence in the recipient's pretransplant PB, but not in the donor PB (Figure 1C). HLA-A*26 was not detected in the recipient's PB collected at the time of relapse, but was detected in the BM relapse sample (Figure 1D). Importantly, the HLA-A*26 patient-specific allele was undetectable in the HLA-DR+ leukemic blasts isolated from the BM and PB (Figure 1E).
Figure 1. Assessment of patient-specific human leukocyte antigen (HLA)-haplotype loss on chromosome 6 using single nucleotide polymorphism array, reverse sequence specific oligonucleotide probe, and melting curve analysis in recipient 1.
A) Profile of chromosome 6 in recipient 1. A schematic representation of chromosome 6 is included above the plots of single nucleotide polymorphism (SNP) array. Log R Ratio (red line average) at 0 indicates no copy number alteration and thus copy neutral loss of heterozygosity (CN-LOH) shown in the bracketed region. The wide range of B allele frequencies (blue tracks) represent the presence of both patient and donor alleles; B) Two reverse sequence specific oligonucleotide (rSSO) probes, HLA-A*26 (Probe #25) and HLADPB1 (Probe #8), were shown to be recipient-specific with low-level binding to the donor and decline in recipient-specific binding with serial dilutions of the recipient DNA with donor DNA. Posttransplantation relapse specimens of blood and marrow both show levels of Probe 25 consistent with donor samples, suggesting that the HLA-A was lost from the leukemic blasts. Similar results are shown with HLA-DPB1 Probe 8; C) Melting curve analysis of the dilution series shows its sensitivity in detecting the mismatched HLA-A*26 allele in the recipient diluted sample out to 14 cell equivalents. -dF/dT represents derivative of fluorescence over temperature; D) Melting curve analysis shows high pretransplant levels of HLA-A*26 DNA in the recipient, with no detection of HLA-A*26 in the donor sample. Posttransplant blood samples mirror that of the donor, reflecting loss of genomic HLA in the leukemic blasts at relapse. Bone Marrow specimens, however, showed a level of HLA-A*26 that is lower than the pretransplant recipient levels; E) Melting curve analysis on sorted leukemic blasts (defined as CD45low HLA-DR+) showed no HLA-A*26 DNA, supporting HLA loss.
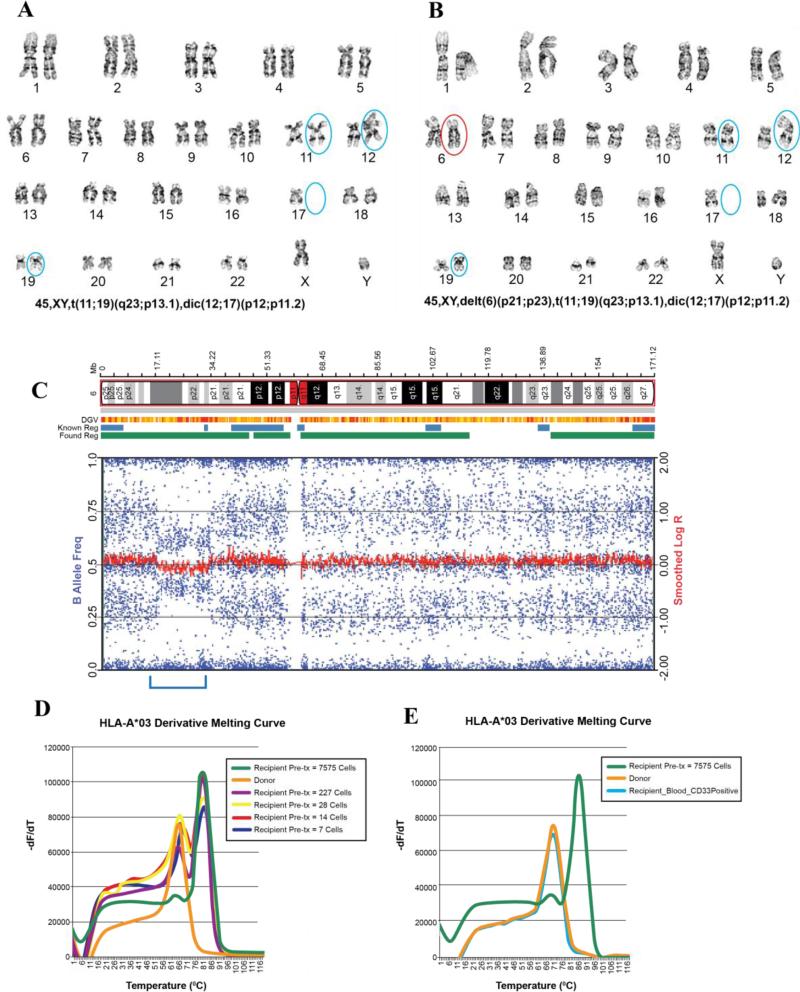
For recipient 2, G-banded karyotype analysis at the time of relapse revealed twelve cells that displayed a normal female karyotype and eight cells (40%) that showed male sex chromosomes. In addition to the translocations t(11;19) and dic(12;17) identified in the diagnostic sample (Figure 2A), a new interstitial deletion on chromosome 6p was detected (Figure 2B). SNP array showed the presence of both donor and recipient allele populations and confirmed a clonal interstitial deletion of 15.9 Mb on chromosome 6p that includes the HLA loci (Figure 2C). Based on pre-transplant HLA typing results, we performed HLA-specific melting curve analysis using primers for the HLA-A*03 allele of recipient 2 (Supplementary Table 1). This allele was present in the recipient pretransplant PB, but not in the donor specimen (Figure 2D). No HLA-A*03 DNA was detected in the CD33+ leukemic blasts at relapse (Figure 2E).
Figure 2. Assessment of patient-specific mismatched human leukocyte antigen (HLA) - haplotype loss using single nucleotide polymorphism array and melting curve analysis in recipient 2.
A) Karyotype of the diagnostic sample; B) Karyotype of the relapse sample showing a new finding of deletion in 6p; C) Single nucleotide polymorphism (SNP) array revealed the 15.9 Mb interstitial deletion (bracketed) that includes the HLA loci. The wide range of B allele frequencies (blue tracks) represent the presence of both patient and donor alleles; D) Melting curve analysis shows the ability to detect HLA-A*03 DNA in limited numbers of recipient pretransplant cells, but not in donor cells. -dF/dT represents derivative of fluorescence over temperature; E) Isolated leukemic CD45low CD33+ blasts from the recipient's peripheral blood and bone marrow show no detection of HLA-A*03 DNA, similar to that of the donor, suggesting loss of the recipient's mismatched HLA.
PTCy has improved the safety of haplo BMT, leading to its increased utilization. However, little is known regarding mechanisms for relapse after this transplant approach. While HLA loss has been documented after haplo BMT using PBSCs grafts with methotrexate and anti-thymocyte globulin,8,10 there have been no reports of its occurrence after haplo BMT utilizing BM grafts followed by PTCy. To our knowledge, these represent the first two cases of HLA loss after NMA haplo BMT using PTCy. Both cases were identified at the time of relapse given clinical suspicion and, in recipient 2, suggestion of genomic loss on karyotype analysis. By conducting rSSO hybridization and melting curve analysis of the targeted alleles, we were able to convincingly demonstrate the loss of the mismatched recipient HLA allele on leukemic cells. rSSO, performed in recipient 1, showed evidence of this loss in posttransplant specimens for all mismatched alleles tested. Detecting 7-14 cell equivalents compared to 1212 cell equivalents for the rSSO method, melting curve analysis offers greater specificity and sensitivity. Importantly, melting curve analysis showed absence of the mismatched recipient HLA haplotype on the isolated leukemic blasts in both cases. Interestingly, the cases represent distinct mechanisms of HLA loss. SNP array for recipient 1 demonstrated UPD at 6p, which is the mechanism described by the Italian BMT group.10 However, the karyotype and SNP array for recipient 2 revealed a deletion of 6p that encompassed the mismatched HLA locus. The later represents a different, but similar, genomic mechanism and supports that the leukemic cells may lose the mismatched HLA haplotype through multiple means, all resulting in evasion of the donor immune system.12
Deciphering the incidence of HLA loss after haplo BMT with PTCy requires further study. Currently, no standardized test to detect HLA loss relapse exists. However, our cases clearly demonstrate that loss of the mismatched HLA haplotype operates as a mechanism of AML relapse after haplo BMT irrespective of graft source, conditioning, or posttransplant immune suppression. Given the increasing utilization of the haplo BMT1, there is an urgent need to develop a broadly applicable and sensitive methodology for detecting and monitoring HLA loss. Recognizing HLA loss may allow us to better tailor treatment. For instance, there is growing experience with haplo donor lymphocyte infusion,13,14 which would be ineffective in HLA loss relapse, but retain the risk of GVHD. Therefore avoidance of this potentially toxic therapy is critical in these patients. Further, the selection of a second donor with an alternative haplotype to improve graft-versus-tumor reactivity against the resistant clone may be an effective therapeutic strategy.8,9 In conclusion, HLA loss should be considered in patients with AML relapsing after haplo BMT with PTCy, particularly with late relapses8 and in patients with extramedullary disease.15 These observations support the development of a standardized test to routinely assess for HLA loss relapse after haplo BMT.
Supplementary Material
Acknowledgements
This study was supported by National Institutes of Health grants P01 CA015396 and P30 CA006973.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Kanakry CG, Fuchs EJ, Luznik L. Modern approaches to HLA-haploidentical blood or marrow transplantation. Nature reviews Clinical oncology. 2015 doi: 10.1038/nrclinonc.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciurea SO, Zhang MJ, Bacigalupo AA, Bashey A, Appelbaum FR, Aljitawi OS, et al. Haploidentical transplant with posttransplant cyclophosphamide vs matched unrelated donor transplant for acute myeloid leukemia. Blood. 2015;126:1033–40. doi: 10.1182/blood-2015-04-639831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luznik L, O'Donnell P, Symons H, Chen AR, Leffell MS, Zahurak M, et al. HLA-haploidentical bone marrow transplantation for hematologic malignancies using nonmyeloablative conditioning and high-dose, posttransplantation cyclophosphamide. Biology of Blood Marrow Transplant. 2008;14:9. doi: 10.1016/j.bbmt.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Molecular Medicine Today. 1999;5:178–86. doi: 10.1016/s1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera T, Maleno I, Lopez-Nevot M, Redondo M, Fernandez MA, Collado A, et al. High frequency of HLA-B44 allelic losses in human solid tumors. Hum Immunol. 2003;64:941–50. doi: 10.1016/s0198-8859(03)00164-2. [DOI] [PubMed] [Google Scholar]
- 6.Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ. Multiple Genetic Alterations Cause Frequent and Heterogeneous Human Histocompatibility Leukocyte Antigen Class I Loss in Cervical Cancer. The Journal of Experimental Medicine. 2000;191:961–76. doi: 10.1084/jem.191.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda K, Hiraki A, Fujii N, Watanabe T, Tanaka M, Matsue K, et al. Loss or down-regulation of HLA class I expression at the allelic level in freshly isolated leukemic blasts. Cancer Science. 2007;98:7. doi: 10.1111/j.1349-7006.2006.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crucitti L, Crocchiolo R, Toffalori C, Mazzi B, Greco R, Signori A, et al. Incidence, risk factors and clinical outcome of leukemia relapses with loss of the mismatched HLA after partially incompatible hematopoietic stem cell transplantation. Leukemia. 2015;29:1143–52. doi: 10.1038/leu.2014.314. [DOI] [PubMed] [Google Scholar]
- 9.Vago L, Toffalori C, Ciceri F, Fleischhauer K. Genomic loss of mismatched human leukocyte antigen and leukemia immune escape from haploidentical graft-versus-leukemia. Seminars in Oncology. 2012;39:707–15. doi: 10.1053/j.seminoncol.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C, Stanghellini MT, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. New England Journal of Medicine. 2009;361:11. doi: 10.1056/NEJMoa0811036. [DOI] [PubMed] [Google Scholar]
- 11.McCurdy SR, Kanakry JA, Showel MM, Tsai HL, Bolaños-Meade J, Rosner GL, et al. Risk-stratified outcomes of nonmyeloablative HLA-haploidentical BMT with high-dose posttransplantation cyclophosphamide. Blood. 2015;125:3024–31. doi: 10.1182/blood-2015-01-623991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamaki H, Fujioka T, Ikegame K, Yoshihara S, Kaida K, Taniguchi K, et al. Different mechanisms causing loss of mismatched human leukocyte antigens in relapsing t(6;11)(q27;q23) acute myeloid leukemia after haploidentical transplantation. Eur J Haematol. 2012;89:497–500. doi: 10.1111/ejh.12017. [DOI] [PubMed] [Google Scholar]
- 13.Zeidan AM, Forde PM, Symons H, Chen A, Smith BD, Pratz K, et al. HLA-Haploidentical Donor Lymphocyte Infusions for Patients with Relapsed Hematologic Malignancies after Related HLAHaploidentical Bone Marrow Transplantation. Biology of Blood and Marrow Transplantation. 2014;20:314–8. doi: 10.1016/j.bbmt.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiso A, Raiola AM, Gualandi F, Dominietto A, Varaldo R, Van Lint MT, et al. DLI after haploidentical BMT with post-transplant CY. Bone Marrow Transplant. 2015;50:56–61. doi: 10.1038/bmt.2014.217. [DOI] [PubMed] [Google Scholar]
- 15.Stölzel F, Hackmann K, Kuithan F, Mohr B, Füssel M, Oelschlägel U, et al. Clonal Evolution Including Partial Loss of Human Leukocyte Antigen Genes Favoring Extramedullary Acute Myeloid Leukemia Relapse After Matched Related Allogeneic Hematopoietic Stem Cell Transplantation. Transplantation. 2012;93:744–9. doi: 10.1097/TP.0b013e3182481113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.