Abstract
Reverse genetic analysis can connect a gene and its protein counterpart to a biological function(s) by knockout or knockdown of the specific gene. However, when a protein has multiple biochemical activities, the conventional genetics strategy is incapable of distinguishing which biochemical activity of the protein is critical for the particular biological function(s). Here, we propose a structural reverse genetics strategy to overcome this problem. In a structural reverse genetics study, multiple biochemical activities of a protein are segregated by mapping those activities to a structural element(s) in the atomic resolution tertiary structure. Based on the structural mapping, a mutant lacking one biochemical activity of interest can be produced with the other activities kept intact. Expression of the mutant by knockin or ectopic expression in the knockout strain along with the following analysis can connect the single biochemical activity of interest to a biological function. Using the structural reverse genetics strategy, we have dissected the newly identified GTP-dependent activity of a lipid kinase PI5P4Kβ from its ATP-dependent activity. The GTP-insensitive mutant has demonstrated the existence of the GTP bioenergetic sensor system in mammalian cells and its critical role in tumorigenesis. As structural reverse genetics can identify in vivo significance of individual biochemical activity, it is a powerful approach to reveal hidden biological functions, which could be a novel pharmacological target for therapeutic intervention. Given the recent expansion of choices in structural biological methods and advances in genome editing technologies, the time is ripe for structural reverse genetics strategies.
Keywords: functional network, GTP bioenergetic system, multifunctional proteins, PI5P4Kb, structural reverse genetics
Structural reverse genetics strategy
Decades of accumulated data from large-scale analyses such as genome sequencing, protein–protein interactomes, and genetic interactomes have suggested complex relationships between biochemical activities of a protein and its biological functions. Together with interactome data, it becomes evident that many proteins, especially those in eukaryotes, have more than one biochemical activity (e.g., catalytic activities, and binding or scaffolding activities to other molecules) and are involved in more than one biological function (See Fig. 1A for PI5P4Kβ as an example). Due to this complexity, simple applications of the conventional gene knockout and knockdown experiments cannot distinguish whether the deduced biological function(s) is due to a single biochemical activity of a protein (Fig. 1B). For instance, a ubiquitin-editing enzyme, A20 [tumor necrosis factor alpha-induced protein 3 (TNFAIP3)], has E3 ubiquitin ligase activity to produce Lys-48-linked polyubiquitination as well as deubiquitinase activity for Lys-63-linked polyubiquitin [1]. Knockout of A20 could not reveal the significance of E3 ligase or deubiquitinase activity. Phosphatase and tensin homolog (PTEN) has both lipid and protein phosphatase activities [2,3]. Knockout of PTEN abolishes both activities and thus could not reveal which phosphatase activity is critical for the observed biological functions.
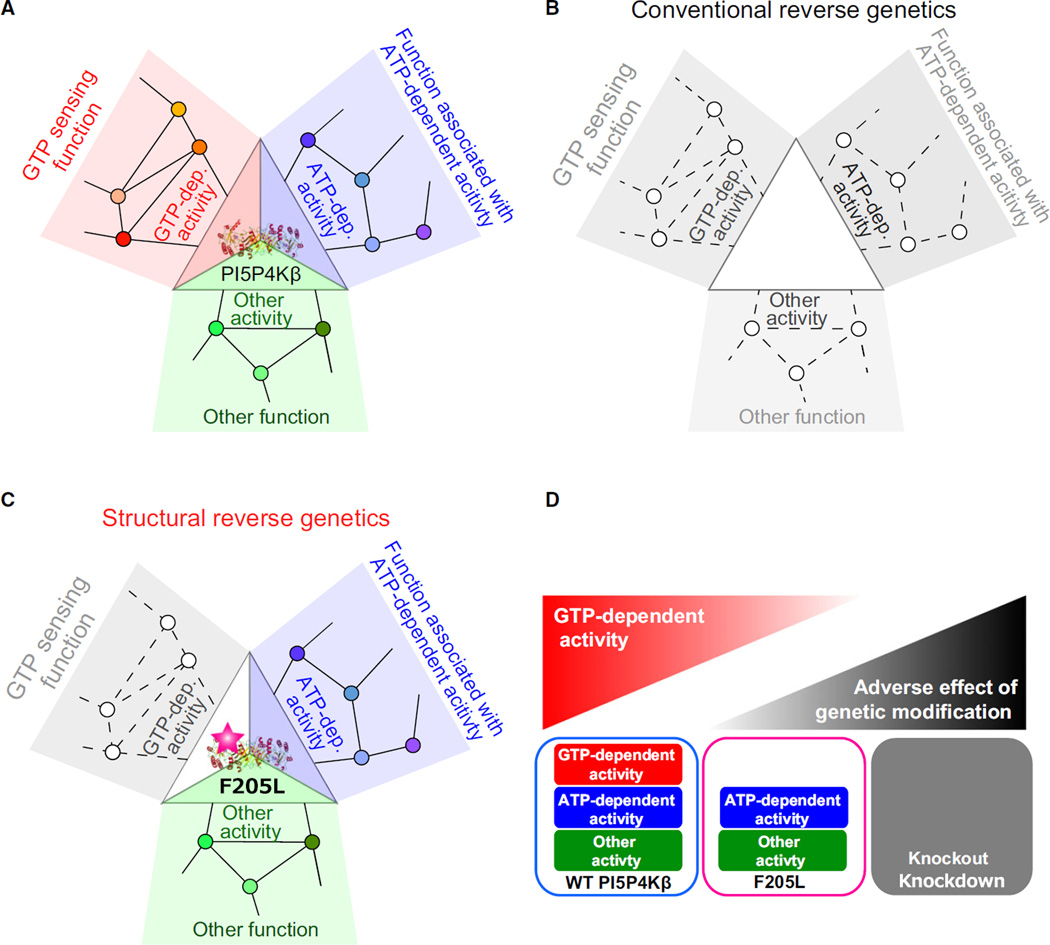
Fig. 1.
Strength of structural reverse genetics strategy. The significance of the structural reverse genetics strategy is exemplified by revealing the role of the GTP-dependent activity of PI5P4Kβ in the GTP-sensing function. (A) Schematic representation of links between three biochemical activities of PI5P4Kβ and associated biological functions. The signaling networks associated with each PI5P4Kβ activity are also indicated. Small circles indicate other proteins or biomolecules and lines connecting small circles indicate the functional interactions between them (not necessarily be the direct molecular interaction). (B) The conventional reverse genetics strategy affects whole biochemical activities of PI5P4Kβ, and all functions associated with PI5P4Kβ along with the cellular signaling networks would be affected. (C) The structural reverse genetics strategy removes only the GTP-dependent activity of PI5P4Kβ without affecting others, therefore revealing the in vivo significance of the associating GTP-sensing function. (D) Comparison of the structural reverse genetics strategy with the conventional reverse genetic strategy. Unlike the structural reverse genetics strategy that specifically disrupts the biochemical activity of interest, the conventional reverse genetic strategy might have adverse effect from the genetic modification if the protein has multiple biochemical activities.
The structural reverse genetics strategy aims to solve the problem of multifunctionality of proteins; it can analyze the relationship between biological functions of cells and a biochemical activity of a protein, via modulating the single biochemical activity of interest in a protein without affecting others (Fig. 1C). This strategy is more straightforward and definitive than the conventional reverse genetic strategies in revealing the in vivo significance of the biochemical activity of a protein of interest, especially in the context of the highly complex cellular signaling networks. However, such a modulation generally cannot be introduced without having atomic resolution structures of the target protein, which is utilized to map the biochemical activities to the structural elements. In case of PI5P4Kβ, the structural reverse genetics strategy was key to segregating the guanosine triphosphate (GTP)-dependent activity of PI5P4Kβ from that of adenosine triphosphate (ATP). This demonstrates the in vivo significance of the GTP-sensing activity of PI5P4Kβ and the presence of the GTP bioenergetic system in mammalian cells.
Structural reverse genetics identifies a cellular GTP sensor system
Cellular concentrations of energy molecules vary substantially in response to cellular status and pathological states. Changes in the concentrations of energy molecules evoke complex but concerted responses in cell metabolism to cope with the energy crisis. For example, in the GTP energy responses, the salvage and the de novo GTP biosynthetic pathways intercommunicate with one another. Mutation of a key enzyme in the GTP salvage pathway resulted in up-regulation of the de novo purine synthesis pathway [4]. Although the mechanism of intercommunication remains unclear, it is reasonable to predict that the cells have an energy sensor that receives the GTP concentration as a biological cue and converts it into cellular signaling and/or gene expression, for metabolic adaptations. Such energy sensors have been found for ATP and other metabolites [5–8]. However, whether mammalian cells have an energy sensor(s) for GTP remained unknown until the year of 2016.
PI5P4K, or Type II PIPK, is a class of phosphoinositide kinases, which phosphorylates phosphatidylinositol 5-phosphate (PI(5)P) to generate phosphatidylinositol 4,5-diphosphate (PI(4,5)P2) [9]. Mammalian genomes contain three distinct genes encoding PI5P4Kα, β, and γ isoforms. PI5P4Kα knockout mice do not exhibit any significant phenotypes under normal housing conditions and PI5P4Kγ knockout mice have not been reported. In contrast, PI5P4Kβ knockout mice displayed reduced body weight, and the knockdown of PI5P4Kβ in breast cancer cell lines suppressed cell growth and tumor formation [10–12]. Thus, PI5P4Kβ, which controls the levels of a lipid second messenger, PI(5)P, is an emerging target for cancer therapy [11–14]. Recently, we have discovered that PI5P4Kβ is the yet unknown energy sensor for cellular GTP concentration [15]. Our analyses reveal that PI5P4Kβ has (a) an ability to bind GTP; (b) the appropriate Km value (88 µm for GTP and 256 µm for ATP) so that its activity is regulated by physiological changes of the cellular GTP concentration; and (c) an ability to evoke a signal to control cell metabolism and proliferation via its substrate PI(5)P [15]. The (a) and (b) biochemical properties of PI5P4Kβ are a good match to the requirements for a GTP sensor. Thus, it is predicted that PI5P4Kβ should detect the changes in cellular GTP concentration and convert them into lipid second messenger signaling. However, testing the (c) property – the in vivo significance of the GTP-sensing function – was the real challenge. We could not take a conventional knockout or knockdown approach because these approaches would disrupt both the ATP- and GTP-dependent kinase activity of PI5P4Kβ (Fig. 1D). To overcome this problem, we utilized structural reverse genetics to prove the in vivo significance of the GTP-sensing activity of PI5P4Kβ.
Crystallization and structure determination of the PI5P4Kb–nucleotide complexes
Among the 19 phosphoinositide kinases, the crystal structure of PI5P4Kβ is the first to be solved at 3.0 Å resolution in 1998 [16]. However, structure of nucleotide-bound PI5P4Kβ has not been solved for nearly 20 years. Therefore, we decided to determine the crystal structures of the PI5P4Kβ–nucleotide complexes with a resolution sufficient to distinguish the GTP- and ATP-binding modes. These structural data must be useful to rationally design a mutant lacking GTP-sensing activity. While PI5P4Kβ was crystallized easily by the earlier reported method [16], crystal quality was insufficient to obtain atomic resolution structures of the nucleotide complexes.
To improve the resolution of the PI5P4Kβ crystals, we applied a multistep soaking method [17]. Initially, cryoprotectants for the PI5P4Kβ crystal were screened. Among 14 cryoprotectants, ethylene glycol (EG, 25% v/v) and polyvinylpyrrolidone K15 (PVP, 25% v/v) occasionally showed diffractions better than 3 Å in the apo form crystals. Both EG and PVP, however, did not show significant improvement in the crystal quality. The crystals soaked in EG often showed cracks on their surfaces. While PVP seemed to be less invasive, diffraction images from crystals soaked in the PVP solution showed severe ice rings. We therefore applied a multistep soaking method with EG and PVP. The best result was obtained by two-step soaking: first a crystal was soaked in the less invasive PVP and then EG was supplemented to the soaking solution. This multistep soaking significantly improved the average resolution of the PI5P4Kβ crystals. The resolutions of the final structures were 2.60, 2.15, 2.45, 2.70, and 2.60 Å for the apo, AMP, GMP, Adenylyl-imidodiphosphate (AMPPNP), Guanylyl imidodiphosphate (GMPPNP) complexes, respectively. These structures demonstrated clear differences in their GTP- and ATP-binding modes.
Unexpected guanine nucleotide recognition via a ‘Tetris-spin’ mechanism
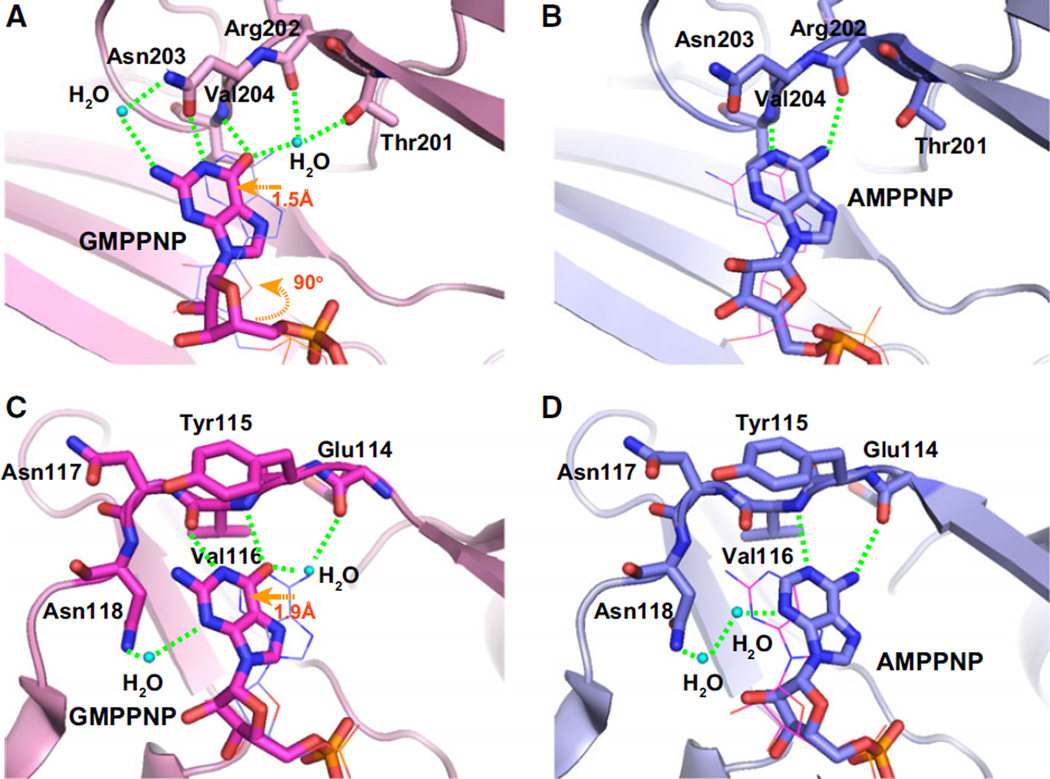
In the nucleotide-bound PI5P4Kβ, the bases of both the ATP and GTP analogs occupy a hydrophobic groove formed by Phe-139, Val-148, Val-204, Phe-205, Leu-282, and Ile-368 (Fig. 2A,B). In the deep end of the nucleotide-binding groove, there are three hydrogen donors and two hydrogen acceptors available for nucleotide base binding (Fig. 2C–F). The N1 atom in the adenine nucleotide is a hydrogen acceptor and coordinates with the Val-204 main chain HN of PI5P4Kβ (Fig. 2D). However, as the corresponding position in the guanine nucleotide is a hydrogen, it cannot form a hydrogen bond with the Val-204 main chain HN in the same way. Thus, the guanine nucleotide instead utilizes the neighboring O6 atom to coordinate with the Val-204 main chain HN (Fig. 2C). This shifts the guanine nucleotide base by approximately 1.5 Å to a deeper position (Figs 2E and 3A). After shifting, the N1 atom of the guanine nucleotide base also finds its partner (Asn-203 side-chain CO) to form an additional hydrogen bond. This mode of interaction is similar to the ‘hydrogen-bonding frame-shift’ in the dual specific kinase, casein kinase II (CKII), to ATP and GTP [18] (Fig. 3). In both kinases, ATP/GTP dual recognitions are enabled by the alternate emergence of the hydrogen donors and acceptors in the binding pocket and both nucleotides find their positions by shifting to match the hydrogen donor/acceptor positions in the binding pocket. While PI5P4Kβ forms a more extensive hydrogen-bonding network with GTP than with ATP, CKII has an equivalent hydrogen bond network with both nucleotides. This would be one of the reasons why PI5P4Kβ binds more strongly to GTP than to ATP.
Fig. 2.
Recognition of GTP and ATP by PI5P4Kβ. (A–D) Hydrophobic and hydrogen bond interactions in the human PI5P4Kβ–nucleotide complexes. (A) Guanine and (B) adenine nucleotides are tightly fitted into the slit formed by the hydrophobic residues shown in the stick representation. Aromatic interactions between the protein and nucleotide bases are highlighted by orange dotted lines. The observed hydrogen-bonding networks between the protein and (C) guanine and (D) adenine nucleotides are highlighted by green dotted lines. (E, F) Schematic representation of (E) GTP and (F) ATP recognition by PI5P4Kβ. The red and blue circles indicate the positions of proton donors and acceptors in the nucleotides, respectively. The red and blue squares indicate those in PI5P4Kβ. Hydrogen bonds are depicted as dashed green lines. For comparison, the superimposed ATP structure is represented as light gray in the GTP scheme. The figure was reproduced from [15].
Fig. 3.
Comparison of nucleotide recognition by PI5P4Kβ and CKIIα. (A and B) PI5P4Kβ in complex with (A) GMPPNP and (B) AMPPNP. (C and D) CKIIα in complex with (C) GMPPNP and (D) AMPPNP. The observed hydrogen-bonding networks between proteins and (A, C) guanine and (B, D) adenine nucleotides are highlighted by green dotted lines. In each panel, the positions of counterpart nucleotides are shown in line representation for comparisons. The panels A and B were reproduced from [15].
In addition, the guanine nucleotide base is located close to Phe-205, promoting formation of an aromatic–aromatic interaction (Fig. 2A,E). The aromatic– aromatic interaction of the adenine nucleotide base to Phe-205 is less significant due to its separation from the phenyl ring (Fig. 2B,F). The characteristic interaction with GTP is also associated with ~ 90° rotation of the ribose group relative to the ATP analog (Fig. 2). Thus, GTP binds to its designated position in PI5P4Kβ by shifting and rotating its position relative to ATP, which could be analogous to the ‘Tetris-spin’ in the tile-matching video game, Tetris.
Development of the GTP-insensitive mutant of PI5P4Kβ to define the significance of the GTP-sensing activity in vivo
The crystal structures indicated that Thr-201, Asn-203, and Phe-205 are specialized for guanine base recognition in PI5P4Kβ (Fig. 2). To analyze the effect of these residues on nucleotide recognition, Thr-201, Asn-203, and Phe-205 in PI5P4Kβ were mutated to methionine, alanine, and leucine, respectively. We confirmed that the individual structures of the PI5P4Kβ mutants were nearly the same as that of the wild-type (WT) PI5P4Kβ in nucleotide-bound forms (root-mean-square deviation values for all Cα atoms are less than 0.4 Å). Among them, only the PI5P4KβF205L mutant showed a decrease in GTP-dependent kinase activity, without having any perturbation of the ATP-dependent activity.
Isogenic cell lines that express the PI5P4KβF205L mutant and the WT PI5P4Kβ demonstrated that PI5P4Kβ’s GTP-sensing activity regulates cellular PI (5)P levels in response to the changes in GTP concentration [15]. WT PI5P4Kβ cells displayed concerted metabolic changes in response to the GTP concentration change, while the PI5P4KβF205L mutant could not faithfully change the metabolism under the same conditions [15]. Furthermore, the PI5P4KβF205L mutant-expressing cells abolished the tumorigenic activity observed for WT PI5P4Kβ cells in mouse xenograft models [15]. These results from the structural reverse genetics approach clearly indicate that the GTP-sensing activity of PI5P4Kβ regulates cellular PI (5)P signaling and metabolism, and provides an advantage in tumorigenesis in vivo.
Structural reverse genetics and future perspective
The conventional reverse genetics strategy based on knockout or knockdown of a specific gene requires the prior knowledge of the associating signaling networks to be able to reach an eureka moment by solving ‘simultaneous equations’ derived from a combination of experiments. Energy sensors for ATP, such as AMP-activated protein kinase [5] and possibly including mammalian target of rapamycin have been established [6]. Identification of these ATP energy sensors has been accomplished by extensive biological and biochemical studies on their upstream and downstream signaling pathways. As for PI5P4Kβ, we exploited the structural reverse genetics strategy to dissect the link between the GTP-dependent activity and its significance in GTP-sensing function. Despite the limited biological knowledge on PI5P4Kβ, this approach revealed the hidden function of PI5P4Kβ as described above.
The structural reverse genetics strategy can readily be applied to the proteins that interact with multiple partners using different interfaces and/or different domains [19]. As expected, it would be challenging to apply the strategy to proteins that express multiple biochemical activities using an overlapping site. The GTP- and ATP-dependent activities of PI5P4Kβ fall into this category as the binding sites to GTP and ATP are overlapping and deviated only by 1.5 Å (Figs 2 and 3). Even with such a small difference, we have successfully identified the mutant (F205L) that can specifically disrupt GTP-dependent activity. This is mainly due to the presence of additional interactions with GTP. In contrast, ATP do not have additional interactions; thus, specific disruption of ATP-dependent activity would be more challenging. It is also important to see if the mutation actually causes the expected effect to the protein activity by the structure determination and verification of the biochemical activities of the mutant, in order to make sure that the mutation does not cause protein misfolding and/or major structural changes.
The structural reverse genetics strategy can be applicable to any biomolecules with multiple biochemical activities, such as riboproteins, ribozymes, and noncoding RNA. Given the recent advances in the structure determination methods and genome editing technologies [20], the structural reverse genetics strategy will play a central role in understanding the in vivo role of a multifunctional biomolecules in the postgenomic era.
Acknowledgments
We acknowledge the Izayoi Meeting for stimulating discussions. We thank Emily Rose Kahoud for critical reading. The work was supported by Grants-in-Aid for Scientific Research (KAKENHI; grant numbers 25121743 to K. T.; 22121005 to T.S.) and Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology and Japan Society for the Promotion of Science. This work was also partly supported by JST, PRESTO to KT and by Kanae Foundation to SK. Funding was also provided by MTP UC-Brain Tumor Center grant, UC Internal Medicine grant, 1R03MH096575, and 1R01NS089815 (A.T.S.).
Abbreviations
- AMPPNP
adenylyl imidodiphosphate
- ATP
adenosine triphosphate
- CKII
casein kinase II
- EG
ethylene glycol
- GMPPNP
guanylyl imidodiphosphate
- GTP
guanosine triphosphate
- Km
Michaelis constant
- PI(4,5)P2
phosphatidylinositol 4,5-diphosphate
- PI(5)P
phosphatidylinositol 5-phosphate
- PTEN
phosphatase and tensin homolog
- PVP
polyvinylpyrrolidone K15
- TNFAIP3/A20
tumor necrosis factor alpha-induced protein 3
- WT
wild-type
Footnotes
Author contributions
KT, ATS, and TS designed the review. All authors contributed intellectually and to the writing of the manuscript.
References
- 1.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 2.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbloom FM, Henderson JF, Caldwell IC, Kelley WN, Seegmiller JE. Biochemical bases of accelerated purine biosynthesis de novo in human fibroblasts lacking hypoxanthine-guanine phosphoribosyltransferase. J Biol Chem. 1968;243:1166–1173. [PubMed] [Google Scholar]
- 5.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 7.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 10.Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB, Cantley LC. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol. 2004;24:5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keune WJ, Sims AH, Jones DR, Bultsma Y, Lynch JT, Jirströom K, Landberg G, Divecha N. Low PIP4K2B expression in human breast tumors correlates with reduced patient survival: a role for PIP4K2B in the regulation of E-cadherin expression. Cancer Res. 2013;73:6913–6925. doi: 10.1158/0008-5472.CAN-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, Bell EL, Shim HS, Lamia KA, Rameh LE, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155:844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke JH, Irvine RF. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem J. 2013;454:49–57. doi: 10.1042/BJ20130488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jude JG, Spencer GJ, Huang X, Somerville TDD, Jones DR, Divecha N, Somervaille TCP. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene. 2015;34:1253–1262. doi: 10.1038/onc.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumita K, Lo YH, Takeuchi K, Senda M, Kofuji S, Ikeda Y, Terakawa J, Sasaki M, Yoshino H, Majd N, et al. The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis. Mol Cell. 2016;61:187–198. doi: 10.1016/j.molcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- 17.Senda M, Hayashi T, Hatakeyama M, Takeuchi K, Sasaki AT, Senda T. Use of multiple cryoprotectants to improve diffraction quality from protein crystals. Cryst Growth Des. 2016;16:1565–1571. [Google Scholar]
- 18.Niefind K, Pütter M, Guerra B, Issinger OG, Schomburg D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Biol. 1999;6:1100–1103. doi: 10.1038/70033. [DOI] [PubMed] [Google Scholar]
- 19.Akai Y, Adachi N, Hayashi Y, Eitoku M, Sano N, Natsume R, Kudo N, Tanokura M, Senda T, Horikoshi M. Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction. Proc Natl Acad Sci. 2010;107:8153–8158. doi: 10.1073/pnas.0912509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]