Abstract
Altered infant human gut microbiome composition and metabolic activity are implicated in childhood atopy and asthma1. We hypothesized that compositionally distinct neonatal human gut microbiota exist and are differentially related to relative–risk (RR) of childhood atopy and asthma. Using stool samples (n = 298; aged 1–11 months) from a US birth cohort and 16S rRNA sequencing, neonates (median age 35 days) were divisible into three microbiota–composition states (NGM1–3). Each incurred significantly different RR for multi–sensitized atopy at age–two years and doctor–diagnosed asthma at age–four years. The highest risk group, NGM3, showed lower relative abundance of certain bacteria (e.g. Bifidobacterium, Akkermansia and Faecalibacterium), higher relative abundance of particular fungi (Candida and Rhodotorula), and a distinct fecal metabolome enriched for pro-inflammatory metabolites. Ex vivo culture of adult human peripheral T–cells with sterile fecal water from NGM3 subjects increased the proportion of CD4+ cells producing interleukin–4 and reduced the relative abundance of Foxp3+CD25+CD4+ cells. 12,13 DiHOME which discriminated NGM3 from lower–risk NGMs, recapitulated the effect of NGM3 fecal water on Foxp3+CD25+CD4+ cell relative abundance. These findings suggest that neonatal gut microbiome dysbiosis drives CD4+ T–cell dysfunction associated with childhood atopy.
Atopy, the propensity to produce IgE antibodies in response to allergens, is one of the most common chronic health issues2, considered a significant risk factor for childhood asthma development3 and has recently been linked to bacterial taxonomic depletions in the gut microbiota at 3, but not 12 months of age1. We therefore hypothesized that distinct neonatal (~1 month of age) gut microbiota exist and idiosyncratically influence CD4+ populations in a manner that differentially relates to RR of childhood atopy and asthma. Independent fecal samples collected during a study visit targeting 1–month of age (median age 35 days; range 16–138 days; n = 130; “neonates”), or 6–months of age (median age 201 days; range 170–322 days; n = 168; “infants”) from participants in the racially and socio–economically diverse Wayne County Health, Environment, Allergy and Asthma Longitudinal Study birth–cohort4 were studied. Atopic status at age–two years was defined using latent–class analysis, an unsupervised statistical algorithm that clusters subjects based on their pattern of serum IgE responses to a panel of ten food and aeroallergens5 (Supplementary Table 1; Supplementary Fig. 1).
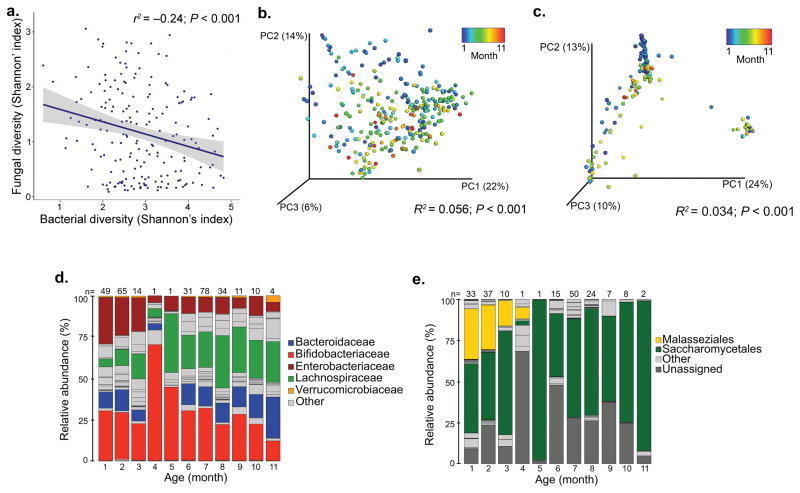
At the population level (independent of atopy status) bacterial community α–diversity (taxon number and distribution) expanded with increasing age (Pearson’s correlation r = 0.47, P < 0.001; Supplementary Fig. 2a). In parallel fungal α–diversity contracted (Pearson’s correlation, r = −0.23, P = 0.0014; Supplementary Fig. 2b), and a reciprocal relationship between these microbial kingdoms existed (Shannon’s Index; Pearson’s correlation r = −0.24, P < 0.001; Fig. 1a). Both bacterial and fungal β–diversity (inter–personal taxonomic composition), were related to participant age (PERMANOVA; R2 = 0.056, P < 0.001 and R2 = 0.034, P < 0.001, respectively; Fig. 1b–c). Neonatal fecal microbiota were typically dominated by Bifidobacteriaceae, Enterobacteriaceae (Fig. 1d), Malasseziales (Malassezia) and Saccharomycetales (Saccharomyces; Fig. 1e). Infant participants exhibited sustained presence, though diminished relative abundance of Bifidobacteriaceae and Enterobacteriaceae, an expansion of Lachnospiraceae (Blautia and Ruminococcus) and fungal communities characteristically dominated by Saccharomycetales (Saccharomyces and Candida; Fig. 1e), the dominant fungal order in healthy adults6. These findings indicate an inter–kingdom gut microbial co–evolution along an age–associated developmental gradient, over the first year of life.
Fig. 1. Bacterial and fungal α– and β–diversity are related to age of participant at the time of fecal sample collection.
(a) Bacterial and fungal α–diversities are inversely correlated (Shannon’s index; n = 188; Pearson’ correlation r2 = −0.24; P < 0.001). (b) Age of participant is associated with bacterial β–diversity (n = 298; PERMANOVA R2 = 0.056; P < 0.001). (c) Age of participant is related to fungal β–diversity (n = 188; PERMANOVA R2 = 0.034; P < 0.001). (d) Age-stratified taxa summaries (presented at the family level) of bacterial relative abundance (n = 298; number of participants per age-group is provided above bars). (e) Age-stratified taxa summaries (presented at the order level) of fungal relative abundance (n = 188; number of participants per age-group is provided above bars).
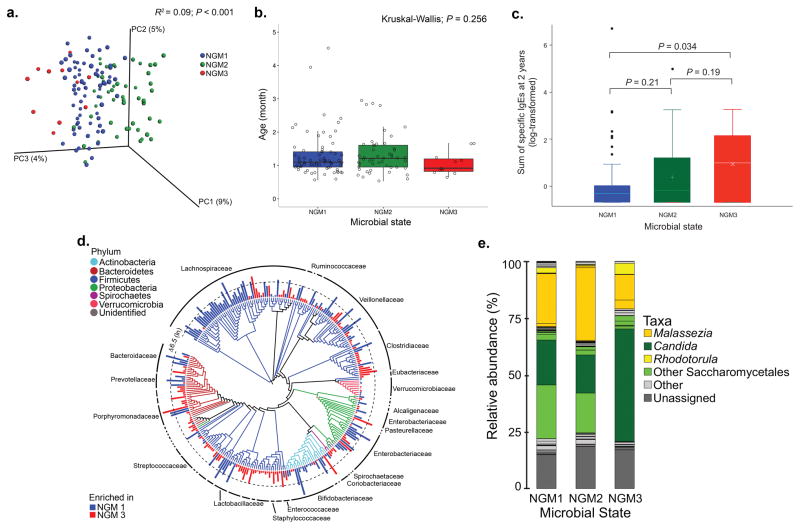
To address our primary hypothesis a Dirichlet Multinomial Mixture (DMM) model was used to group participants based on bacterial community composition7; three distinct neonatal gut microbiota–states (NGM1, 2 and 3) represented the best model fit (Supplementary Fig. 3). PERMANOVA confirmed that NGM designation explained a small but non–trivial proportion of bacterial β–diversity (PERMANOVA; R2 = 0.09, P < 0.001; Fig. 2a), indicating that these compositional states (which did not differ in age [Kruskal–Wallis; P = 0.256; Fig. 2b]) may represent a gradient of neonatal gut microbiota compositions. NGMs trended towards a significant relationship with fungal β–diversity (Bray Curtis; PERMANOVA, R2 = 0.037, P = 0.068), signifying that each co–associates with a mycobiota that primarily varies in the relative abundance of the dominant fungal taxa present. Infant samples were divisible into two compositionally distinct gut microbiota–states, IGM1 (Bifidobacteriaceae–dominated) and IGM2 (Lachnospiraceae–dominated (unweighted UniFrac; PERMANOVA, R2 = 0.032, P = 0.001), which did differ in age (Wilcoxon rank sum, P = 0.0257); IGM1 participants were younger. IGMs were not related to fungal community β–diversity (Bray–Curtis; PERMANOVA, R2 = 0.011, P = 0.33), presumably because infant subjects were consistently enriched for Saccharomycetales. Using the conventional atopy definition (IgE > 0.35 IU ml−1), no significant difference in RR between NGMs was observed (Table 1). However when the asthma predictive5 PM–atopy definition was used, NGM3 participants incurred a higher RR of atopy at age two-years compared with either NGM1 (RR = 2.94; 95% CI 1.42–6.09 P = 0.004; Table 1) or NGM2 groups (RR = 2.06; 95% CI 1.01–4.19 P = 0.048; Table 1). Even larger effect sizes for NGM3 were observed for parental reported doctor–diagnosed asthma at age–four years (Table 1). NGM–associated RR of PM–atopy was supported by the sum of specific IgE responses at age–two years (Fig. 2c). As hypothesized, IGMs did not exhibit different RR for PM–atopy (RR = 1.02; 95% CI 0.59–1.75, P = 0.94; Supplementary Table 2) or asthma (RR = 0.51; 95% CI 0.22–1.17 P = 0.11). Using available early–life characteristics, we identified maternal education, season of birth, gestational age, age at sample collection and breastfeeding to be significantly distinct across IGMs (Supplementary Table 3). Only detectable dog allergen (Can f 1) concentrations in the home during the neonatal study visit (lowest in the NGM3 group), and baby gender (NGM3 was almost entirely male) were significantly associated with NGM state (Supplementary Table 4). Despite adjustment for these and other early–life factors commonly related to allergic disease, the relationship between NGM and atopy or asthma remained (Supplementary Table 5). Only one other large pediatric gut microbiota–atopy study exists1, the youngest participants of which were substantially older (~100 days) than neonates in our cohort (median age 35 days). Application of our DMM model parameters to this dataset identified two compositionally distinct groups (Bifidobacteria-dominated NGM1 and Lachnospiraceae-dominated IGM2 [Supplementary information]), indicating that neonatal examination of gut microbiota is necessary to identify distinct pioneer microbiota related to differential RR.
Fig. 2. Compositionally distinct, age-independent bacterial gut microbiota–states (NGMs) exist in neonates, exhibit significant differences in fungal taxonomy and are related to relative risk of atopy at age–2 years.
(a) NGM designation significantly explains observed variation (n = 130; PERMANOVA with unweighted UniFrac R2 = 0.09; P < 0.001) in bacterial β–diversity. (b) NGM participants do not differ significantly in age (n = 130; Kruskal–Wallis; P = 0.256). Boxplots are defined by the 25th and 75th percentiles with the center line representing the median (50th percentile). Lines that extend from the box are defined as 1.5 times the interquartile range (IQR, 75th–25th percentile), plus or minus the 75th and 25th percentiles, respectively. (c) The sum of allergen–specific serum IgE concentrations measures at two–years of age (n = 130) is significantly higher in NGM3 versus NGM1 participants (Welch’s t–test; P = 0.034). Boxplots are constructed as defined in (b). (d) Taxonomic comparison of NGM3 with NGM1 subjects; taxa exhibiting significant difference (ZINB; Benjamini–Hochberg, q < 0.05) in mean relative abundance (natural log transformed for purposes of illustration) are shown. Bar height indicates the magnitude of between–group relative abundance delta. (e) Relative abundance of fungal genera differs across NGMs.
Table 1. Neonatal gut microbiota–states exhibit significantly different RR of PM–atopy development at age–two and parental report of doctor diagnosed of asthma at age–four.
Significance of risk ratios between microbiota–states were calculated based on log–binomial regression.
| DMM Community Types | RR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| NGM1 (n = 70) | NGM2 (n = 49) | NGM3 (n = 11) | NGM2 versus NGM1 | NGM3 versus NGM1 | NGM3 versus NGM2 | Overall P value | |
| Atopy (PM) | 13 (18.6%) | 13 (26.5%) | 6 (54.5%) | 1.43 (0.73–2.81) P = 0.30 |
2.94 (1.42–6.09) P = 0.004 |
2.06 (1.01–4.19) P = 0.048 |
0.034 |
| Parental report of doctor diagnosed asthmaa, b | 8 (13.6%) | 5 (11.9%) | 4 (40.0%) | 0.87 (0.31–2.50) P = 0.81 |
2.95 (1.09–7.98) P = 0.033 |
3.36 (1.10–10.3) P = 0.034 |
0.13 |
| Atopy (IgE > 0.35 IU ml−1) | 29 (41.4%) | 25 (51%) | 7 (63.6%) | 1.23 (0.83–1.82) P = 0.30 |
1.54 (0.91–2.60) P = 0.11 |
1.25 (0.74–2.11) P = 0.41 |
0.30 |
n = 19 missing doctor diagnosis of asthma information;
average age at interview 4.1 years (standard deviation of 0.84)
NGM3 microbiota were characteristically depleted of bacterial taxa including Bifidobacteria (Bifidobacteriaceae), Lactobacillus (Lactobacillaceae), Faecalibacterium (Clostridiaceae) and Akkermansia (Verrucomicrobiaceae) when compared with NGM1 (zero–inflated negative binomial regression (ZINB), Benjamini–Hochberg q < 0.05; Fig. 2d; Supplementary Table 6). These observations were consistent when NGM3 was compared to NGM2 (Supplementary Fig. 4; Supplementary Table 7) and with previously described atopy–associated taxonomic depletions1. Mycologically, NGM3 subjects were consistently depleted of multiple Malassezia taxa (Fig. 2e; ZINB; Benjamini–Hochberg q < 0.05; Supplementary Tables 8–9), striking given our population–based observation that this genus is characteristically enriched in the neonatal gut microbiota. Fungal taxonomic enrichments in the NGM3 group were also consistent when compared to either of the lower–risk groups, and included members of the Saccharomycetales, Rhodotorula, Pleosporales and Candida (Supplementary Table 8–9). Hence neonatal inter–kingdom microbiota dysbiosis is characteristic of PM–atopy and asthma development in childhood.
NGM3 microbiota were predicted8 to lack an extensive range of bacterial amino acid, lipid and xenobiotic metabolism pathways (Supplementary Fig. 5). Un–targeted liquid chromatography mass spectrometry identified fecal metabolites present in a subset (n = 28) of the representative subjects from each NGM (those with the highest posterior probability of NGM membership). A significant relationship existed between 16S rRNA profile, predicted metagenome and the metabolome of the NGMs (Procrustes; Supplementary Table 10), indicating a deterministic relationship between community composition and the metabolic microenvironment of the neonatal gut. Between–group comparisons identified specific metabolites enriched in each NGM (Welch’s t–test; P < 0.05; Supplementary Fig. 6, Supplementary Table 11–12). As previously reported in the urine of atopic subjects1, NGM3 participants exhibited fecal enrichment of primary and secondary bile metabolites. However more expansive metabolic dysfunction involving lipid, amino acid, carbohydrate, peptide, xenobiotic, nucleotide, vitamin and energy metabolism pathways, essentially those bacterial pathways predicted to be deficient in NGM3, was evident. Though NGM1 and NGM2 exhibited distinct metabolic programs, a common subset of metabolites differentiated them from NGM3. These included anti–inflammatory polyunsaturated fatty acids, docosapentaenoate (n3 DPA; 22 n5) and dihomo–γ–linolenate9,10 (DGLA; 20:3n3 or n6), succinate, and the breast–milk oligosaccharides, 3–fucosyllactose and lacto–N–fucopentaose II known to influence gut epithelial colonization11,12 (Supplementary Fig. 6, Supplementary Table 11–12). In contrast, NGM3 participants were consistently enriched for 12,13 DiHOME, stigma– and sito–sterols, 8–hydroxyoctanoate, α–CEHC and γ–tocopherol.
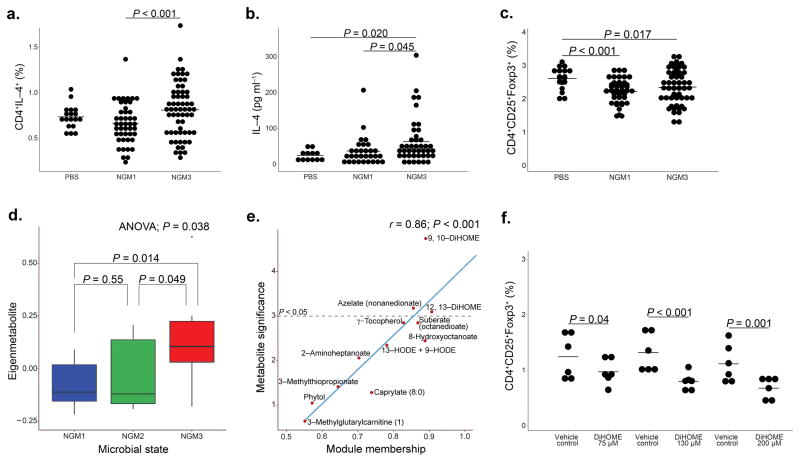
Compared with NGM1, sterile fecal water from NGM3 participants decreased the ratio of CD4+IFNγ+:CD4+IL–4+ cells (LME, P = 0.095; Supplementary Fig. 7), increased the proportion of CD4+IL–4+ cells (LME, P < 0.001; Fig. 3a), the concentration of IL–4 released (LME, P = 0.045; Fig. 3b) and reduced the percentage of CD4+CD25+Foxp3+ cells (compared to control; Fig. 3c) ex vivo, indicating that the NGM3 gut microenvironment promotes adaptive immune dysfunction associated with established atopic asthma. Weighted correlation network analysis identified 32 metabolic modules (Supplementary Fig. 8), one of which discriminated the NGMs (ANOVA; P = 0.038; Fig. 3d) and contained 12,13 DiHOME, which was identified both as a hub metabolite (MM = 0.91; Fig. 3e) and relatively enriched in NGM3 subjects compared with the other two NGMs (P < 0.05; Supplementary Fig. 6 and 9, Supplementary Table 11–12). All concentrations of 12,13 DiHOME examined reduced the percentage of CD4+CD25+Foxp3+ cells compared to vehicle–treatment (LME, P = 0.04, P < 0.001, P = 0.001 respectively; Fig 3f).
Fig. 3. NGMs significantly differ in CD4+ cell differentiation.
Dendritic cells and autologously purified naïve CD4+ cells from serum of two healthy adult donors (biological replicates), were incubated with sterile fecal water from NGM1 (n = 7; three biological replicates per sample) or NGM3 (n = 5; three biological replicates per sample) participants. NGM3 induced significantly increased (a) proportions of CD4+IL–4+ (LME, P < 0.001; center line represents mean) and (b) expression of IL–4 (LME; P = 0.045). (c) Both NGMs expressed significantly increased proportions of CD4+CD25+Foxp3+ cells (LME; P < 0.001 for NGM1 and P = 0.017 for NGM3) compared to control. (d) Weighted correlation network analysis identified a metabolic module that differentiated NGM3 from NGM 2 and NGM1 participants (n = 28; ANOVA; P = 0.038). Boxplots define the 25th and 75th percentiles, median represented by centerline. IQR (75th–25th percentile) represented by whiskers. (e) Scatterplot of metabolite significance versus module membership (MM) of the 12 metabolites in the NGM3 discriminating metabolic module. Metabolites with a value of P < 0.05, significantly discriminate NGM3 from other NGMs. MM value indicates the degree of inter-connectedness of a specific metabolite to other metabolites in the module (higher MM value indicates greater inter-connectedness). (f) Using the same ex vivo assay as performed in 3a–c, 12, 13 DiHOME significantly reduced the proportion of CD4+CD25+Foxp3+ cells at three different concentrations (LME; P = 0.04, P < 0.001, P = 0.001 for concentrations of 75, 130 and 200 μM respectively).
Thus these findings indicate that the neonatal gut microbiota influences susceptibility to childhood allergic asthma, via alterations in the gut microenvironment that influence CD4+ T–cell populations and function. These data suggest that very-early life interventions to manipulate the composition and function of the gut microbiome may offer a viable strategy for disease prevention.
Methods
Study population
Pregnant women (N = 1,258) between the ages of 21–49 were recruited from August 2003–November 2007 as part of the Wayne County Health, Environment, Allergy and Asthma Longitudinal Study (WHEALS). WHEALS is a prospective birth cohort from southeastern Michigan designed to investigate early life risk factors for allergic diseases as previously described4. Briefly, women were considered eligible if they lived in a predefined cluster of contiguous zip codes in, and surrounding Detroit, MI, had no intention of moving out of the area and provided informed written consent. Five follow–up contacts were conducted at 1, 6, 12, 24, and 48–months after the birth of their child, with the 24–month appointment being at a standardized study clinic for the child to be evaluated by a board–certified allergist. Stool samples were collected from the child at the 1– and 6–month home visits. All aspects of this research were approved by the Henry Ford Hospital Institutional Review Board.
Sub–sample criteria of WHEALS subjects for stool microbiome analyses
Children who had completed their 24–month clinic visit, which included a blood draw for IgE measurements and had dust samples collected from their homes at the same time as their stool sample collection were selected for this study (n = 308). Stool samples from children ranging from 1–11 months were collected from field staff during home visits and stored at −80 °C. Samples were randomized prior to being shipped to UCSF on dry ice where they were also stored at −80 °C until processed.
PM–atopy and asthma definition
Blood drawn at the 2–year clinic visit was used to determine levels of total and 10 allergen–specific IgEs (sIgE): Alternaria (Alternaria alternata), German cockroach (Blattella germanica Bla g 2), dog (Canis lupus familiaris Can f 1), house dust mites (Dermatophagoides farinae Der f 1), hen’s egg (egg), cat (Felis domesticus Fel d 1), cow’s milk (milk), peanut (Arachis hypogaea), common ragweed (Ambrosia artemisiifolia), and Timothy grass (Phleum pratense). Specific IgEs were measured using the Pharmacia UniCAP system (ThermoFisher Scientific, Waltham, MA, USA). Latent class analysis was used to group participants into four discrete atopic–classes based on sensitization patterns of the 10 allergen sIgEs as with the entire WHEALS cohort3. Our subset was assigned to one of four latent classes: 1. Low–no sensitization (n = 226); 2. Highly sensitized (both food and inhalant allergens; n = 9); 3. Milk and egg dominated (n = 50), or 4. Peanut and inhalant(s) dominated (n = 13) sensitization. Due to sample size, latent classes 2–4 were collapsed and considered “predominately multi–sensitized” (PM–atopy; n = 72), remaining subjects represented the “low–no sensitization” group. The conventional definition of atopy (least one positive test (sIgE ≥ 0.35 IU ml−1) to any of the 10 allergens) was also used for comparative purposes. Children were defined as asthmatic using parental reported doctor diagnosis of asthma at the 4–year interview.
Bacterial and fungal community profiling, PICRUSt and metabolomic analyses
DNA Extraction
Stool samples from 308 infants were extracted using a modified cetyltrimethylammonium bromide (CTAB) buffer based protocol13. Briefly, 0.5 ml of modified CTAB extraction buffer were added to 25 mg of stool in a 2 ml Lysing Matrix E tube (MP Biomedicals, Santa Ana, CA) then incubated (65 °C, 15 min). Samples were bead–beaten (5.5 m s−1, 30 sec) in a Fastprep–24 (MP Biomedicals, Santa Ana, CA) followed by the addition of 0.5 ml of phenol:chloroform:isoamyl alcohol (25:24:1). Following centrifugation (14,000 rpm, 5 min), the supernatant was added to a heavy phase–lock gel tube (5 Prime, Gaithersburg, MD) and chloroform (v:v) was added. Samples were centrifuged (14,000 rpm, 5 min); resulting supernatants were added to fresh tubes, followed by addition of 1 μl of linear acrylamide prior to PEG–NaCl (2v:v). Samples were incubated (21 °C, 2 h), washed with 70% EtOH and resuspended in 10 mM Tris–Cl, pH 8.5.
Sequencing preparation
The V4 region of the 16S rRNA gene was amplified as designed in Caporaso et al.14. PCR reactions were performed in 25 μl reactions using 0.025 U Takara Hot Start ExTaq (Takara Mirus Bio Inc, Madison, WI), 1X Takara buffer with MgCl2, 0.4 pmol μl−1 of F515 and R806 primers, 0.56 mg ml−1 of bovine serum albumin (BSA; Roche Applied Science, Indianapolis, IN), 200 μM of dNTPs, and 10 ng of gDNA. Reactions were performed in triplicate with the following: initial denaturation (98 °C, 2 min), 30 cycles of 98 °C (20 sec), annealing at 50 °C (30 sec), extension at 72 °C (45 sec) and final extension at 72 °C (10 min). Amplicons were pooled and verified using a 2% TBE agarose e—gel (Life Technologies, Grand Island, NY), prior to purification using AMPure SPRI beads (Beckman Coulter, Brea, CA), quality checked with the Bioanalyzer DNA 1000 Kit (Agilent, Santa Clara, CA) and quantified using the Qubit 2.0 Fluorometer and the dsDNA HS Assay Kit (Life Technologies, Grand Island, NY). Samples were pooled and sequenced on the Illumina MiSeq platform as previously described15.
The internal transcribed spacer region (ITS)2 of the rRNA gene was amplified using the primer pair fITS7 (5′–GTGARTCATCGAATCTTTG–3′) and ITS4 (5′–TCCTCCGCTTATTGATATGC–3′). Primers were designed for the Illumina MiSeq platform as described above. PCR reactions were performed in triplicate in 25 μl reaction with 1X Takara buffer (Takara Mirus Bio), 200 nM of each primer, 200 μM dNTPs, 2.75 mM of MgCl2, 0.56 mg ml−1 of BSA (Roche Applied Science, Indianapolis, IN), 0.025 U Takara Hot Start ExTaq and 50 ng of gDNA. Reactions were conducted under the following conditions: initial denaturation (94 °C, 5 min),30 cycles of 94 °C (30 sec), annealing at 54 °C (30 sec), extension at 72 °C (30 sec) and a final extension at 72 °C (7 min). PCR verification and purification were performed as described above. Samples were quantified using KAPA SYBR (KAPA Biosystems, Wilmington, MA) qPCR following manufacturer’s protocol. Samples were pooled in equal moles (50 ng) and prepped and denatured libraries with PhiX spike–in control, as described above, were loaded onto the Illumina MiSeq cartridge.
Sequence data processing and quality control
For bacterial sequences, paired–end sequences were assembled using FLASH16 v 1.2.7, demultiplexed by barcode, and low quality reads (Q–score < 30) were discarded in QIIME17 1.8. If three consecutive bases were < Q30, then the read was truncated and the resulting read retained in the data set only if it was at least 75% of the original length. Sequences were checked for chimeras using UCHIME18 and filtered from the dataset prior to OTU picking at 97% sequence identification using UCLUST19 against the Greengenes database20 version 13_5. Those sequence reads that failed to cluster with a reference sequence were clustered de novo. Sequences were aligned using PyNAST21, and taxonomy assigned using the RDP classifier and Greengenes reference database version20 13_5. To de–noise the OTU table, taxa with less than 5 total sequences across all samples were removed. A bacterial phylogenetic tree was built using FastTree22 2.1.3.
Fungal sequences were quality trimmed (Q–score < 25) and adapter sequences removed using cutadapt23 prior to assembling paired–end reads with FLASH16. Sequences were de–multiplexed by barcode and truncated to 150 bp prior to clustering using USEARCH vers. 7 pipeline, specifically the UPARSE24 function, and chimera–checked using UCHIME. Taxonomy was assigned using UNITE25 vers. 6.
To normalize variation in read depth across samples, data were rarefied to the minimum read depth of 202,367 sequences per sample for bacteria (n = 298) and 30,590 for fungi (n = 188). To ensure that a truly representative community was used for analysis for each sample, sequence sub–sampling at these defined depths was rarefied 100 times. The representative community composition for each sample was defined as that which exhibited the minimum average Euclidean distance to all other OTU vectors generated from all sub–samplings for that particular sample. Investigators at UCSF were blinded to sample identity until microbiota datasets underwent the aforementioned processing and were ready for statistical analyses.
Phylogenetic Reconstruction of Unobserved States (PICRUSt)
PICRUSt8 was used to predict pathways of those taxa significantly enriched in each neonatal gut microbiota–state based on zero–inflated negative binomial regression and corrected for multiple testing using the Benjamini-Hochberg false discovery rate26. These taxa were used to generate a new OTU table normalized in PICRUSt, and discriminatory pathways were illustrated in a heatmap constructed in R.
Metabolomic profiling
Stool samples (200 mg) from each of the three microbiota–states, 8 NGM3 subjects and 10 from each of NGM1 and NGM2, were provided to Metabolon (Durham, NC) for Ultrahigh Performance Liquid Chromatography–Tandem Mass Spectrometry (UPLC–MS/MS) and Gas Chromatography–Mass Spectrometry (GC–MS) using their standard protocol (http://www.metabolon.com/). These samples were chosen based on their exhibiting the highest posterior probability of belonging to a given NGM group and possessing sufficient sample volume for UPLC–MS/MS analysis. Compounds were compared to Metabolon’s in–house library of purified standards, including more than 3,300 commercially available compounds.
Ex vivo dendritic cell challenge and T–cell co–culture
Fecal samples (1 g ml−1) from five of ten NGM1 and seven of eight from NGM3 neonates that had undergone metabolically profiling were used (biological replicate). Samples were excluded due to lack of availability. Fecal samples were homogenized in w:v pre–warmed phosphate buffered saline (PBS) containing 20% fetal bovine serum (FBS). Samples were vortexed, incubated (37 °C, 10 min) and centrifuged (14,000 rpm, 30 min). Supernatant was filter–sterilized through a 0.2 μm filter, prior to use in the DC–T–cell assay described below. PBS was used as the negative control. Treatment conditions used for the DiHOME experiment included: 75 μM, 130 μM, and 200 μM 12,13 DiHOME (Cayman Chemical, Ann Arbor, MI) solubilized in 0.4%, 0.15% and 0.05% DMSO respectively. Controls included PBS and DMSO at corresponding percentages used to dissolve the different concentrations of DiHOME. Treatment group size was determined based on preliminary assays that demonstrated the effect size for suppression of CD4+CD25+Foxp3+ using 130 μM of 12, 13 DiHOME was approximately seven, indicating that least two samples per group was required to achieve a power > 0.80.
Peripheral blood samples were obtained from healthy, de–identified human donors (PBMCs; Blood Centers of the Pacific, San Francisco, CA) through the cell–sourcing program that ensures donor confidentiality. Donors signed an agreement acknowledging that their blood may be used for research. PBMCs were isolated using Ficoll–Hypaque gradient centrifugation, washed twice with R10 media (RPMI 1640 with 10% heat–inactivated FBS [antigen activator] with 2 mM L–glutamine and 100 U ml−1 penicillin–streptomycin; Life Technologies) and incubated for 18 h. Dendritic cells were isolated from PBMCs using the EasySep™ Human Pan–DC Pre–Enrichment Kit (STEMCELL Technologies, Vancouver, BC). DCs (0.5 × 106 cells ml−1) from two donors (biological replication) were treated in 3–sets with either cell–free fecal water (treatment replicate; excluded if were < 50% of the T–cells were alive) or DiHOME treatment and cultured in R10 media supplemented with 10 ng ml−1 GM–CSF and 20 ng ml−1 IL–4 at 37 °C27 for 3 d for the fecal water assay, and 5 d for the DiHOME experiment. Treatment replicates were also considered biological replicates because the donor cells are not clonal. For the fecal water experiment, the assay was repeated twice on one donor (technical replicates) and once on donor B due to insufficient cells.
The DC treatment media was freshly prepared and replaced every 48 h. After 3 d (or 5 d for DiHOME treatment) of DC challenge, DC maturation was stimulated using DC growth mediators (10 ng ml−1 tumor nuclear factor–α [TNF–α], 10 ng ml−1 IL–1b, 10 ng ml−1 IL–6, and 1 mM prostaglandin E2 [PGE2]) which were added to the culture for 24 h prior to co–culture with T–cells. In preparation for co–culture, DCs were washed in fresh R10 media, counted via flow cytometry and plated in TexMACs Medium (Miltenyi Biotec, San Diego, CA) at 0.5 × 106 live CD45+ cells per well.
Autologous T lymphocytes were purified from the PBMCs using a Naïve CD4 T–cell isolation kit (Miltenyi Biotec). Following purification, naïve autologous CD4+ T–cells were suspended in the TexMACS Medium (Miltenyi Biotec) and added to the treated DCs at a ratio of 10:1 in the presence of soluble anti–CD28 and anti–CD49d (1 mg ml−1). T and DC cells were co–cultured for 5 d at 37 °C and replenished with fresh R10 media every 48 h. To assess cytokine production, the co–cultures were mixed with Phorbol Myristate Acetate–Ionomycin (SIGSa, St. Louis, MO) and GolgiPlug (BD Biosciences, San Jose, CA) for 16 h prior to flow cytometry. Cell–free media from the co–cultures were collected at 48 h and 5 d, prior to PMA–Gplug addition, to assess cytokine secretion. Cytokine secretion was evaluated by cytometric bead array following the manufacturer’s protocol (BD Biosciences).
For flow cytometry, single–cell suspensions were stained using a panel of antibodies including anti–CD3 (SP34–2), anti–CD4 (L200), anti–CD25 (M–A251), anti–IFNγ (B27; BD Biosciences); anti–CD8a (RPA–T8; BioLegend, San Diego, CA); anti–IL4 (7A3–3; Miltenyi Biotec); anti–IL–17A (64DEC17) and anti–FoxP3 (PCH101; Affymetrix eBioscience, Santa Clara, CA). Validation for each primary antibody is provided on the manufacturers’ websites. Dead cells were stained positive with LIVE–DEAD® Aqua Dead Cell Stain (Life Technologies). Permeabilization buffer (Affymetrix eBioscience) was used to permeabilize cell prior to staining for intracellular markers, IFNγ, IL–4, IL–17A, FoxP3. For flow analysis, live T–cells were gated as CD3+CD4+ cells. Among CD4+ T–cells subpopulations, Th1 were IFNγ+, Th2 were IL–4+, Th17 were IL–17A+, and T–regs were both CD25hi and FoxP3hi. Stained cells were assayed via flow cytometer on a BD LSR II (BD Biosciences).
Statistical analysis
Shannon’s diversity was calculated using QIIME. Pearson’s correlation was used to test for a relationship between bacterial and fungal Shannon’s diversity. Distance matrices (unweighted UniFrac28 and Bray–Curtis) were calculated in QIIME to assess compositional dissimilarity between samples and visualized using PCoA plots constructed in Emperor29. Permutational multivariate analysis of variance (PERMANOVA) was performed using Adonis in the R environment to determine factors that significantly explained variation in microbiota β–diversity.
To identify clusters of subjects based on bacterial taxon relative abundance, Dirichlet Multinomial Mixture (DMM) models were used, which implement an unsupervised Bayesian approach based on a Dirichlet prior7. The best–fitting DMM model was determined using the Laplace approximation to the negative log model evidence, testing up to 10 underlying microbiota–states. Each sample was assigned to a particular microbiota–state based upon the maximum posterior probability of microbiota–state membership. Kruskal–Wallis was used to test if age differentiated the microbiota states. Relative risk ratios (RR) and corresponding 95% confidence intervals were calculated using PROC GENMOD in SAS version 9.4 (Cary, NC). Unadjusted and adjusted RRs were calculated based on log–binomial regression using maximum likelihood estimation or robust Poisson regression when prevalence ratios were near one, or the log–binomial model did not converge. Two–tailed Welch’s t–test was used to test if sIgE concentrations (log–transformed) were significantly different between the three NGMs.
To determine which OTUs were significantly different between NGMs, the zero–inflated negative binomial regression (pscl package) was used as a primary modeling strategy, appropriate for sequence count data; in cases where OTU distributions were not zero–inflated and the model failed to converge, the standard negative binomial was used as a secondary modeling strategy. These were corrected for multiple testing using the minimum positive false discovery rate26. Results were natural log transformed for illustration on phylogenetic trees using iTOL30 v3.0. When examining the association between early life factor and different microbiota states, P values were calculated based on covariate distribution by ANOVA (numerical, normally distributed), Kruskal–Wallis (numerical, skewed), chi–square (categorical), or Fisher’s exact (sparse categorical). Log–binomial regression model was used to test for confounding factors when assessing the relative risk of microbiota states developing atopy or asthma (PROC GENMOD in SAS version 9.4). Fisher’s exact 2–tailed test was conducted to test if breastfeeding was practiced significantly more in any of the microbiota–states.
Metabolites exhibiting significant different concentrations (log–transformed) between lower–risk NGMs and NGM3 were identified using 2–tailed Welch’s t–test. Shared and distinct super– and sub–pathway products amongst NGMs were illustrated using Cytoscape, vers. 3.2.131. Co–occurrence networks of metabolites were constructed using weighted correlation network analysis (WGCNA) with the R package WGCNA to find modules of highly interconnected, mutually exclusive metabolites. Pearson correlations were used to determine inter–metabolite relationships, where modules are composed of positively correlated metabolites. To avoid spurious modules, the minimum module size was set to five. Module “eigenmetabolites” (referred to as eigengenes), were defined as the first principal component of a given module and considered as a representative measure of the joint metabolic profile of that module. Each eigenmetabolite was used to test (ANOVA) the association between its respective module and NGM, Module membership was used to determine the interconnectedness of each metabolite to its assigned module and to identify “hub” metabolites: this was defined as the correlation between each metabolite and the eigenmetabolite (strong positive values indicate high interconnectedness).
Procrustes was used to test for concurrence between communities described by 16S phylogeny, PICRUSt and metabolomics datasets.
To test for T–cell and cytokine differences, linear mixed–effects model was used (R package lmerTest) and adjusted for donors. Except where indicated, all analyses were conducted in the R statistical programming language.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Health and National Institute of Allergy and Infectious Diseases P01 AI089473–01 (C.C.J., D.R.O., H.A.B., N.W.L., S.V.L., G.W., E.M.Z.) and the Alfred P. Sloan Foundation 2013–6–03 (S.V.L). We thank C. Arrieta and B. Finlay for graciously sharing sequence data from the CHILD study.
Footnotes
Accession codes
All sequence data related to this study are available in the European Nucleotide Archive (ENA), http://www.ebi.ac.uk/ena/data/view/PRJEB13896 (study accession no. PRJEB13896).
Author Contributions
S.V.L., C.C.J., D.R.O., H.A.B., N.W.L., G.W., and E.M.Z., designed research; C.C.J., D.R.O., K.E.F, D.F., B.L., D.L.L., S.L., A.R.P, E.R. and G.W., performed research; A.R.S., S.H., and A.M.L. contributed new analytic tools; K.E.F., A.R.S., S.H., S.L., A.M.L. and S.V.L. analyzed data; and K.E.F. and S.V.L. wrote the manuscript.
Competing Financial Interests Statement
We have no competing financial interests.
References
- 1.Arrieta M, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:1–14. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 2.Asher MI, Montefort S, Bjorksten B, Lai CK, Strachan DP, WS Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood. Int Study Asthma Allerg Child. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 3.Simpson A, et al. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181:1200–6. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- 4.Aichbhaumik N, et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin Exp Allergy. 2008;38:1787–1794. doi: 10.1111/j.1365-2222.2008.03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havstad S, et al. Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol. 2014;134:722–727. doi: 10.1016/j.jaci.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann C, et al. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:1–10. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin C, Blier PU, Fortin S. Eicosapentaenoic acid and docosapentaenoic acid monoglycerides are more potent than docosahexaenoic acid monoglyceride to resolve inflammation in a rheumatoid arthritis model. Arthritis Res Ther. 2015;17:142. doi: 10.1186/s13075-015-0653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amagai Y, et al. Dihomo-γ-linolenic acid prevents the development of atopic dermatitis through prostaglandin D1 production in NC/Tnd mice. J Dermatol Sci. 2015;79:30–7. doi: 10.1016/j.jdermsci.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weichert S, et al. Bioengineered 2′-fucosyllactose and 3-fucosyllactose inhibit the adhesion of Pseudomonas aeruginosa and enteric pathogens to human intestinal and respiratory cell lines. Nutr Res. 2013;33:831–838. doi: 10.1016/j.nutres.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 13.DeAngelis KM, et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 14.Werner JJ, et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magoc T, Salzberg SL. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- 24.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Meth. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 25.Abarenkov K, et al. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol. 2010;186:281–85. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 27.Obermaier B, et al. Development of a new protocol for 2-day generation of mature dendritic cells from human monocytes. Biol Proced Online. 2003;5:197–203. doi: 10.1251/bpo62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Env Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. 2003:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.