Abstract
The absence of a gold standard to determine when antibiotics have induced sterilizing cure confounds the development of new approaches to treat pulmonary tuberculosis (PTB). We detected PET-CT imaging response patterns consistent with active disease along with the presence of Mycobacterium tuberculosis mRNA in sputum and bronchoalveolar lavage samples in a substantial proportion of adult, HIV-negative PTB patients after standard 6-month treatment plus one year follow-up, including patients with a durable cure and others who later developed recurrent disease. The presence of MTB mRNA in the context of non-resolving and intensifying lesions on PET-CT might indicate ongoing transcription, suggesting that even apparently curative PTB treatment may not eradicate all organisms in most patients. This suggests an important complementary role for the immune response in maintaining a disease-free state. Sterilizing drugs or host-directed therapies and better treatment response markers are likely needed for the successful development of improved and shortened PTB treatment strategies.
Keywords: tuberculosis, treatment response, PET-CT, mRNA, persistence
Introduction
Currently, there is no absolute measure of sterilizing cure of pulmonary tuberculosis (PTB). The current standard treatment period of 6 months was determined by acceptable rates of treatment failure and disease recurrence after discontinuation of chemotherapy.1 The global rate of unsuccessful treatment in HIV-negative patients with drug-sensitive (DS) Tuberculosis (TB) is 12%.2 In recently published multi-national trials, relapse rates within 12 – 24 months after successful standard treatment ranged from 2.6%, to 9.7%.3–7 Shortening treatment is also a major aim of anti-tuberculosis drug development.8 Three trials3,4,6 introduced an experimental 4-month treatment arm, which yielded significantly increased relapse rates, even though sputum culture conversion rates were higher than in the 6 month arm within the first 2 months of treatment.3,4,6,9,10 Thus Mycobacterium tuberculosis (MTB) may persist in lung tissue for months to years after culture negativity has been achieved through antibiotics.
In contrast to sputum culture conversion, lung lesions seen on chest X-ray (CXR) or computed tomography (CT) often persist long after the end of successful treatment.11,12 Post tuberculosis lung impairment, which contributes the majority of TB-related disability burden,13 commonly causes persistent symptoms and signs after microbiologically confirmed cure.14–20 These factors contribute to uncertainty when defining sterilizing cure and greatly increase the complexity of developing new treatments.
Some sub-populations of slow-replicating MTB are naturally tolerant to all TB drugs,21 which can lead to persistent, viable, non-culturable MTB. This has been demonstrated in mice after 14 weeks of isoniazid and pyrazinamide treatment when MTB mRNA transcripts (shown to be absent in dead bacteria in vitro) were detected in culture negative tissue from all mice necropsied at this time-point. High-dose steroid treatment induced bacteriological relapse in 21 of 23 mice that were followed up after treatment22. The presence of mRNA could suggest transcriptionally active bacteria due to its very short half-life (on average 9.5 minutes for MTB mRNA in vitro), although stabilization of transcripts occurs when intact MTB is exposed to stress such as cold shock or hypoxia and therefore the in vivo half-life is not known.23 The retention of low-abundance mRNA and a relative increase of non-coding RNA in dormant MTB have been shown during in vitro persistence induced by potassium depletion and Rifampicin exposure and might play a role in early reactivation.24 To date, however, persistent bacteria have not been observed in humans.
Positron Emission Tomography and Computerized Tomography (PET-CT) is well established in the management of cancer. 18F-fluorodeoxyglucose (FDG) is the most widely used PET tracer and reflects glucose metabolism, showing increased uptake in areas of inflammation.25–27 PET images are viewed alongside corresponding CT images, which reflect tissue density and provide anatomical information.
In mouse, rabbit and primate models, FDG PET-CT has been used to accurately display the progression of MTB disease. These studies suggest that relative lesion uptake correlates approximately with bacterial numbers in lesions.28–32 Individual lesions can improve or deteriorate in apparent contrast to the overall trend of disease progression in the host, with or without antibiotic treatment, pointing to a dynamic and localized nature of TB vs host interactions.28–31 Human PET-CT studies have shown promise for monitoring the effects of different anti-TB drug regimens and seem to be superior to CT alone.33–39 More PET-CT background is provided in Supplementary Note 1.
The present study was conducted to track changes in PET-CT imaging at baseline, during and after antibiotic treatment and to correlate radiologic findings with microbiologic data in South-African and South-Korean PTB patients. We report a striking heterogeneity in radiological responses amongst patients, with ongoing inflammation after curative treatment in the majority. In the South-African cohort, we also detected the presence of several different mycobacterial mRNA transcripts in sputum and bronchoalveolar lavage (BAL) samples through qRT-PCR at the end of successful treatment outcomes.
Results
Patient demographics and treatment
We recruited ninety-nine HIV-uninfected, non-diabetic, adult patients with PTB at diagnosis (Dx) and followed them during treatment in Cape Town, South Africa. Ninety-five patients had DS strains. Of these, 72 received a standard, 6 month regimen, while 23 received an extended regimen of 8 months [22 due to previous episode(s) of TB and one due to delayed smear conversion]. Two patients had INH mono-resistant TB and were treated for 6 and 12 months respectively and two patients with multidrug-resistant (MDR) TB were treated for 2 years on individualized regimens. Month 6 (M6) was selected as a fixed time-point for PET-CT and sputum collection, consistent with current WHO treatment guidelines.40
Of the 99 patients, eight remained culture positive at M6 and failed treatment, 76 converted to MTB-negative sputum culture and maintained cure and 12 became sputum culture negative, but were diagnosed with recurrent PTB within 2 years after treatment completion (EOT + 2y). Supplementary Dataset 1 shows a summary of time to re-diagnosis and the supporting results. Initial treatment outcome was un-evaluable (UE) for three patients due to contaminated cultures. All extended treatment patients were culture negative from M6 to EOT. Fourteen patients took fewer than approximately 80% of their treatment dosages during the 6-month period, which is regarded as poor adherence in most clinical trial designs.3,6 The failed treatment group included four patients with poor treatment adherence and one with MDR disease. Most patients (96%) reported improvement during treatment, but residual chronic symptoms were often encountered. The most frequent were coughing (found in 47 patients), dyspnoea (21) and low body- mass index (24). Table 1 provides a summary of patient demographics, treatment adherence, as well as culture and GeneXpert MTB/Rif (Xpert) results.
Table 1.
Summary of median (range) or number (%) of clinical and microbiological parameters for PTB patients. The asterisk (*) in the Failed and Recurrence columns indicate a significant difference from cured values (two-tailed, non-paired, Student’s T-test; P-value < 0.05). Poor adherence refers to patients that missed more than 20% of their treatment (note: only three of these patients met WHO criteria of treatment default).
| All (n=99) | Cured (76) | Failed (8) | Recurrence (12) | |
|---|---|---|---|---|
| Age | 31 (17–66) | 32 (17–64) | 29 (18–66) | 34 (21–52) |
| Male (n) | 60 (60%) | 49 (82%) | 5 (63%) | 8 (67%) |
| Previous PTB | 39 (39%) | 20 (27%) | 4 (50%) | 4 (33%) |
| Poor adherence | 14 (10%) | 9 (12%) | 4* (50%) | 1 (8%) |
| MDR | 2 (2%) | 1 (1%) | 1 (12.5%) | 0 (0%) |
| M6 Xpert positive (n) | 30 (30%) | 16 (22%) | 7 (88%) | 6 (50%) |
| Smoker (n) | 77 (78%) | 56 (74%) | 6 (75%) | 12 (100%) |
| Dx Body Mass Index | 18 (13.1–42) | 18 (13.1–23) | 17 (16.7–23) | 19* (16.3–42) |
| M6 Body Mass Index | 18 (14.5–43) | 19 (14.5–25) | 19 (17.2–26) | 19* (17.5–43) |
| Time to Negativity (weeks) | 8 (0.2–24) | 8 (1–24) | >24* | 8 (0.3–12) |
We also recruited 14 HIV-negative patients with PTB in Masan, South-Korea, including three MDR cases and two with mono-resistance to rifampicin. Twelve were cured (culture converted within 6 months), one had un-evaluable outcome and one had culture confirmed recurrence after initial cure. Study design and research procedures are detailed in Figure 1.
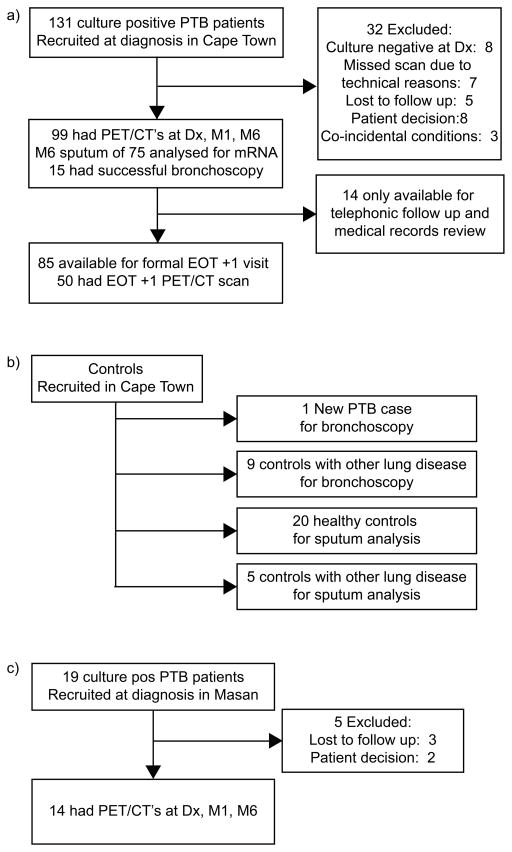
Figure 1.
Flow chart of study design. (a) In, South-Africa, 99 sputum culture positive Pulmonary Tuberculosis (PTB) patients underwent 18F-FDG PET-CT scans at diagnosis (Dx), month 1 (M1) and month 6 (M6) of treatment. Fifty patients also had PET-CT scans 1 year after the end of treatment (EOT + 1y). QRT-PCR assays for MTB mRNA were performed on month 6 sputum from 75 patients and end of treatment bronchoalveolar lavage (BAL) samples of 15 patients. (b) QRT-PCR assays were repeated on sputum from 20 community controls, and five controls with lung disease other than PTB; as well as BAL samples of 10 controls undergoing diagnostic bronchoscopies for suspected lung cancer. (c) PET-CT scans were also performed on 14 sputum culture positive PTB patients from South-Korea at Dx, M1 and M6.
Month 6 PET-CT findings in 99 South African patients
All patients had 18F-FDG PET-CT lung scans at Dx, month 1 (M1), and M6 of TB treatment
Comparing M6 to Dx scans we identified three distinct response patterns: 1) Resolved, 2) Improved and 3) Mixed. A resolved response pattern refers to minimal or no increased FDG uptake compared to surrounding healthy tissue, regardless of structural abnormalities on CT. Improved scans showed decreased intensity of all lesions compared to the baseline scan, but had one or more lesion(s) with increased uptake compared to background and reference structures. Mixed responses showed at least one new FDG-avid lesion or at least one lesion with increased FDG uptake (intensified) compared to the baseline scan. Representative cases are shown in Figure 2.
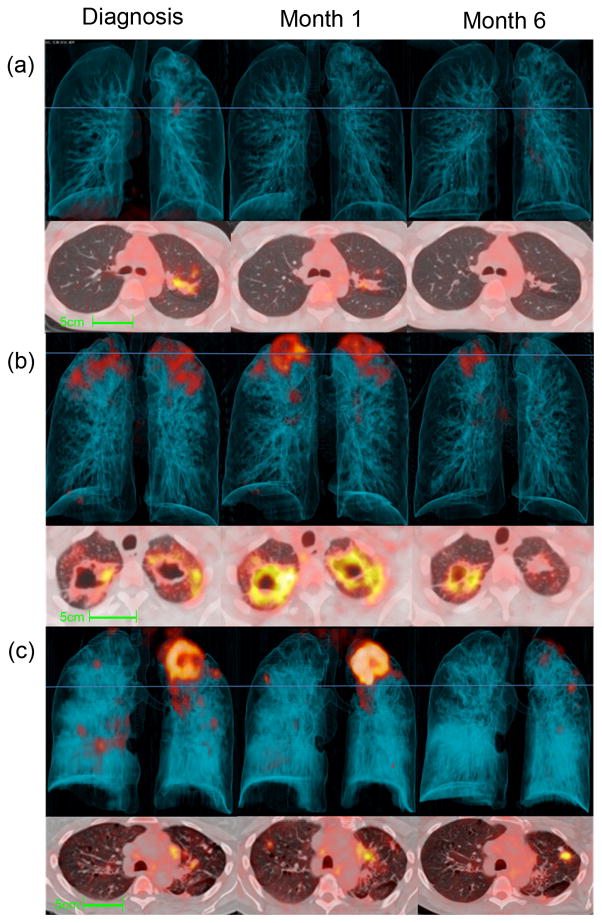
Figure 2.
Representative Dx, M1 and M6 PET-CT images for cured patients after 6 months of standard treatment. 3-dimensional anterior (top panels) and transverse views, at the level of blue lines, (lower panels). (a) Resolved scan, without residual abnormal FDG uptake, some structural abnormalities remain on CT scan. (b) Improved scan, where all lesions have improved since diagnosis, but abnormal CT lesions and increased uptake persist. This example includes thick-walled cavities with high FDG uptake. (c) Mixed scan response with either new FDG-avid lesions or increased intensity of some lesions. This example shows a large new FDG-avid nodule with high intensity uptake in the left lung upper lobe.
Surprisingly, only 14 (14%) patients had a resolved pattern on their M6 scan (Fig 2a) and in 51 (52%) we found an improved response on M6 scan (Fig 2b). A mixed response (Fig 2c) was seen in 34 (34%) patients, of which 14 had both new and intensified lesion(s), 16 had only intensified lesion(s) and four had only new FDG-avid lesion(s).
The morphology associated with the most intense lesion of each mixed and improved M6 scan included features suggestive of active PTB, such as cavities (found in 26 cases), patchy consolidation (in 22 cases), complex lesions involving consolidation with cavitation (16), nodular infiltrates (17), enlarged hilar lymph nodes (3) and pleural based infiltrates (1). Treatment outcome was associated with scan response pattern, (Figure 3a; P < 0.01) with a mixed response found in all failed patients. Neither a mixed response nor a high maximum lesion intensity, however, was specific to a poor outcome and 21 (28%) of cured patients had a mixed response, while 55 (72%) still had M6 lesions with moderate to very high intensity, which would be compatible with the intensity range found at Dx.
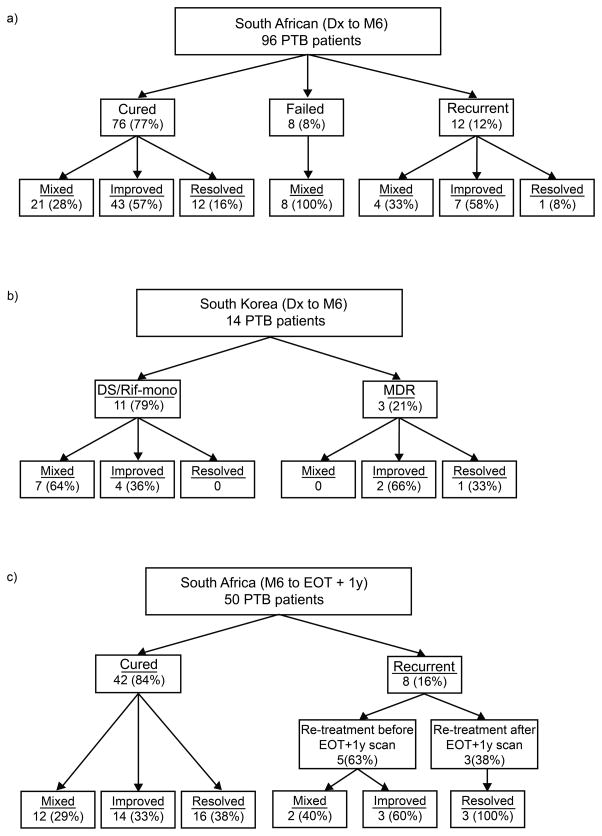
Figure 3.
Clinical outcome and associated PET-CT findings (due to rounding of percentage values totals may equal to 99% or 101%; Lesion intensity rankings summary in Supplementary Dataset 5). (a) South-African: Clinical outcome and associated M6 scan response pattern. Mixed scan response was more likely to have an unfavourable outcome (Fisher exact test, two-sided: P = 0.002). Source data in Supplementary Dataset 6. (b) South-Korea: Patients grouped into those diagnosed with MDR strains or with drug-sensitive or rifampicin mono-resistant (Rif-mono) strains. All MDR cases culture converted within 6 months, while the DS/Rif-mono group includes nine cured, one unevaluable and one recurrent patient. Source data in Supplementary Dataset 2. (c) South-Africa: Clinical outcome and associated EOT + 1y scan response pattern in comparison to M6. Source data in Supplementary Dataset 7.
PET-CT findings in 14 South-Korean patients
All 14 patients had PET-CT scans at Dx, M1 and M6. We applied the same criteria as above and found seven mixed (50%), six improved and only one resolved M6 scans. Eleven had moderate to very high intensity lesions at M6. Figure 3b shows a summary of scan findings; detailed demographical data in Supplementary Dataset 2 and representative images in Supplementary Note 2.
PET-CT findings 1 year after completion of treatment
In view of the findings in the M6 scans we added a fourth scan, 1 year after the end of treatment (EOT + 1y) for 50 South-African patients who had not yet passed this time-point and who were cured at M6. Eight of these 50 patients were diagnosed with recurrent disease by healthcare providers within two years of treatment completion, five before the EOT + 1y scan and three subsequently. The other 42 maintained cure.
When we compared the EOT + 1y scans to the M6 scans, there was improvement in size and intensity of most residual lesions. However, only 32% of EOT + 1y scans were completely resolved. The remaining 68% had significant residual lesion(s), half of which had improvement of all lesions and the other 34% a mixed lesion response compared to the M6 scan. Morphology of new FDG-avid lesions at EOT + 1y included nodular infiltrates (found in four cases), hilar lymph-nodes (one case), cavitation (two), consolidation (two) or lesions with combined morphology (three). Residual M6 lesions showing similar or more intense FDG uptake at EOT + 1y included consolidation (found in two cases), cavitation (four) and nodules (two). Examples of EOT + 1y scan progression can be seen in Figure 4. All three patients who developed recurrent PTB after EOT + 1y had mixed scan outcomes at this time-point, while none of the resolved scans were diagnosed with recurrence (Figure 3c).
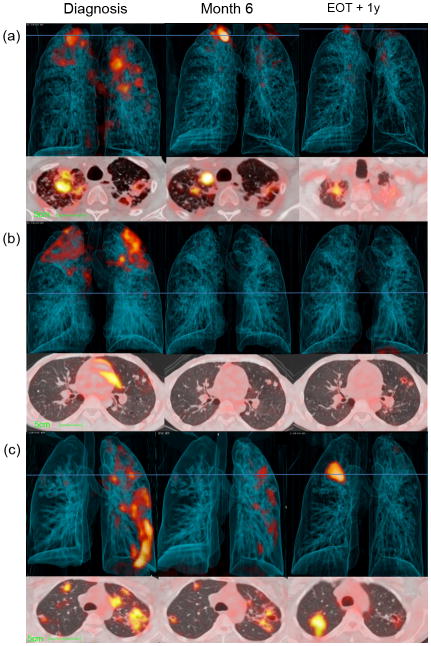
Figure 4.
Representative Dx, M6 and EOT + 1y, PET-CT images for patients after 6 months of standard treatment. 3-dimensional anterior (top) and transverse views (lower) at the level of horizontal blue line. (a) A large new FDG-avid nodule in right lung apex lesion develops by M6, but improves over the next year, with some residual increased FDG uptake. (b) Bilateral upper lobe cavities improve during treatment, but nodules with normal FDG-uptake at M6 develop new cavitation and increased uptake by EOT + 1y. (c) Multiple lesions with continuing reduction in FDG-avidity at M6 and EOT + 1y but with a large new area of consolidation at EOT + 1y. This patient was Gene Xpert positive, but culture negative at the time of EOT + 1y scan, but diagnosed with recurrent disease 6 months later (18 months after treatment completion), while patients (a) and (b) maintained cure.
DNA and mRNA in M6 sputum
Patients with an Xpert positive M6 sputum were more likely to have an unfavourable clinical outcome (7/8 failed treatment, 6/12 recurrent and 16/65 cured; P < 0.001) and M6 PET-CT lesions with high to very high intensity (P = 0.04). [Shenai et al. in press]
We performed mRNA detection assays on M6 sputum from 75 consecutive participants (60 cured, four failed treatment, nine recurrent and two un-evaluable) who produced adequate volume sputum, as well as on sputum from 20 community controls and five controls with lung disease other than TB (OLD). We assayed the sputum samples for 13 MTB-specific mRNA targets [85B, dosR, hspX (acr), pstS1, carD, nuoB, tgS1, TB8.4, TB31.7, acpM, icL, prcA and rrnAP1].
Among the 75 patients’ M6 sputum, at least one MTB mRNA target was detected in 29 patients (39%): 22 (37%) of the cured, four (100%) of the failed treatment, two (22%) of the recurrent PTB and one of the un-evaluable outcome patients. The most frequently detected transcripts were hspX (Acr), acpM and rrn, present in 20, 17 and 12 patients’ M6 sputum respectively. Tgs1 and acpM transcripts were detectable at low levels in one community control each.
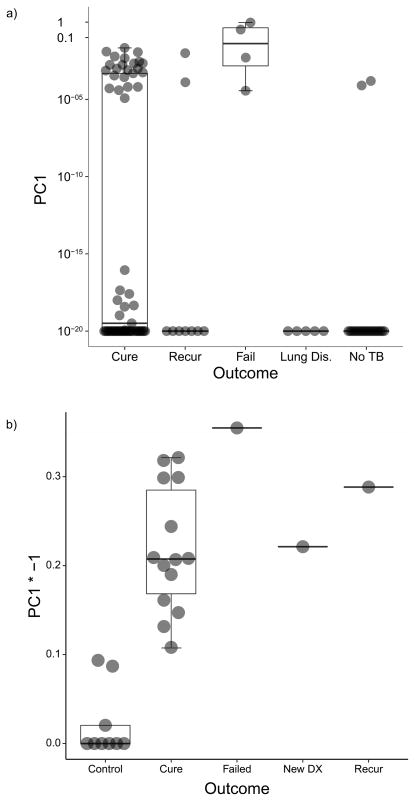
We performed principal component analysis (PCA) to reduce the dimensions of all transcript levels into one variable that indicated mRNA presence. PCA results in Figure 5a illustrate that all cases with failed treatment and 22 cured patients still had detectable MTB mRNA in their M6 sputum (transcript values in Supplementary Dataset 3). Two community controls and no controls with other lung disease had low levels of mRNA transcripts (M6 vs controls; Fisher Exact test P = 0.001). We found a trend for collapsed categories of M6 PET-CT lesion intensity to be correlated with MTB mRNA detection in M6 sputum (P = 0.08).
Figure 5.
Transformed (arcsinh) principal component one of mRNA transcript PCR data, demonstrating separation by detectable MTB mRNA as captured in PC1. Box plots represent median (middle), 75th percentile (top) and 25th percentile (bottom); whiskers represent range. (a) M6 sputum from 75 PTB cases by clinical category; 22 of 60 cured cases, all four failed treatment cases and two of the nine recurrent cases in the group have detectable MTB mRNA in their sputum. None of the other lung disease participants and two of 20 healthy community controls have detectable MTB mRNA (Fisher Exact Test, two-sided: P < 0.01). Source data in Supplementary Dataset 3. (b) EOT bronchoalveolar lavage samples by clinical category. The new PTB case and 15 EOT cases (one failed treatment, one recurrent and 13 cases that maintained cure), all still have significantly detectable mRNA. Three of the six Quantiferon® positive controls show inconclusive levels of MTB mRNA, whereas the Quantiferon® negative controls have no detectable MTB mRNA (Fisher Exact test, two-sided: P < 0.001). Source data in Supplementary Dataset 4.
RNA in end of treatment bronchoalveolar lavage samples
We obtained bronchoalveolar lavage (BAL) samples from the most affected lung segment identified on PET-CT as well as post-bronchoscopy sputa from 15 consecutive, consenting patients with DS PTB within 3 months after the end of treatment (compared to PET-CT scans and sputum at fixed month 6 time-point). Treatment duration was 6 months for this group. We also collected BAL samples in 10 control patients undergoing bronchoscopy as part of clinical workup for suspected lung cancer. The 15 EOT patients included 13 cured (M6 sputum culture negative), one recurrent (M6 culture negative, but became positive again 3 months after treatment) and one failed treatment (M6 culture positive) case. The 10 controls included one patient in whom new active PTB was subsequently diagnosed based on the BAL culture (although sputum culture was negative). All other BAL and post-bronchoscopy sputa were negative for MTB in liquid culture. Cancer was confirmed in five controls, while three controls had bacterial infections and one had interstitial fibrosis. Five of these controls had a positive Quantiferon® blood test (suggesting latent TB infection). Xpert was positive on BAL or post-bronchoscopy sputum at EOT for 10 of the 15 PTB cases and the new PTB case, and MTB DNA was detected in all EOT BAL samples by qRT-PCR.
We performed blinded mRNA detection assays on all BAL samples. 19 MTB-specific mRNA targets (recF, eccD3, menA, iprB, Rv1421, gabD2, Rv1910c, hspX, fadD9, tgs1, fadE34, Rv3675, sodA, trxB2, rpsJ, rplV, sigI, sigH and rpsK) previously shown to be differentially regulated in stress or persistence41,42 were assayed.
A median of eight mRNA transcripts were positive in the BAL from EOT cases (range 1–19); one MTB mRNA transcript was detected in BAL from three of the five Quantiferon® positive control cases (EOT vs controls P < 0.001). The target most frequently detected was hspX, present in the treatment failure and new diagnosis cases, as well as in 14 of the cured patients, but none of the controls. HspX have previously been linked to long-term persistence. 43 In BAL from the newly diagnosed PTB from the control group, we detected five transcripts. We used PCA to summarize mRNA positivity (Figure 5b; transcript values per patient in Supplementary Dataset 4).
Discussion
This study shows ongoing inflammation, as detected by PET-CT, in the majority and the presence of MTB mRNA and DNA in respiratory samples from a substantial number of culture negative South African PTB patients at the end of treatment. Similar PET-CT response patterns were seen in a smaller South-Korean cohort, for whom RNA data were unavailable.
Our study confirms the persistence of TB lung lesions previously described on CT and CXR.11,12 We also frequently found residual symptoms or signs after treatment, similar to other reports on post-tuberculosis lung sequelae.14–20 In addition, although most lesions improved and scan response patterns showed an association with clinical outcomes, we found that the majority of patients still had lesions with an ongoing inflammatory response on PET and roughly a third of patients showed intensified or new lesions. These dynamic patterns were found after anti-TB treatment and even a year later, regardless of drug sensitivity, sustained culture conversion or clinical cure. A European study of 35 patients with pulmonary or extra-pulmonary TB reported persistent PET-CT lesions in 15 and progressive lesions in four patients after a mean treatment duration of 16.1 months,38 while a Japanese study reported resolution of all lesions in eight patients with DS-PTB after 12 months of treatment.35 It’s not clear whether the favourable scan outcome in the Japanese cohort is related to extent of disease, treatment duration, lower infection pressure or better living standards and health.
FDG uptake only reflects the increased metabolic activity associated with inflammation and does not necessarily imply active MTB infection. After the successful treatment of bacterial or pneumocystis pneumonia, however, radiological lesions typically resolve within 6 weeks44 and relative intensity on PET has shown correlation with MTB bacterial load in animal lesions.29 PTB related pathology is the most concordant with our PET-CT findings, considering the underlying morphology of individual lesions and the absence of clinical findings supporting other diagnoses. Persistent antigens, associated with dead MTB, might lead to increased FDG-avidity after treatment, but it is unlikely to cause very high intensity, intensified or new lesions. PET-CT imaging provides insight into the local host response and further research is required to unravel the interaction with MTB.
Our results confirm the high Xpert positivity rate previously reported in culture-negative sputum samples and noted an even higher rate in bronchoscopy samples.45,46 In addition, we detected MTB mRNA in 35% of culture-negative sputum samples at M6 and in all BAL samples taken from 15 PTB patients at EOT. Alpha-crytallin (hspX), linked to long-term MTB persistence,43 was present in 20 (27%) of the M6 sputum samples and 14 (93%) of the EOT BAL samples, while absent in M6 sputum and BAL of all controls. The detection of MTB mRNA in two separate types of respiratory samples suggests either ongoing transcription, based on the short half-life of mRNA,14,15 or persistence of stabilized mRNA as seen in non-replicating MTB in vitro.24 Although persistent MTB DNA in intact, non-viable bacteria may explain Xpert positivity, the same is unlikely to apply to MTB mRNA.
This is the largest study examining PET-CT scans and mRNA after treatment for pulmonary TB; however, it is a relatively small study, with a small number of unfavourable outcomes and without differentiation between relapse and re-infection. Other studies in Cape Town reported similar recurrence rates and if onset of recurrence occurred within 2 years of EOT, found roughly 66% due to relapse.47,48 We could not establish whether the positivity of some mRNA transcripts in the sputum of two controls could be the result of latent or subclinical MTB infection or transient MTB excretion after recent exposure;49,50 or eliminate the possibility that even pre-validated probes could show cross-reactivity. However, in a blinded test, the EOT patients scored more MTB transcriptional signals, more frequently than the community controls and most probes did not show any positivity in the controls. The extent to which the stochastic detection of low levels of MTB mRNA in individuals in communities with a high TB-burden represents true detection of MTB in individuals without manifestations of clinical TB has yet to be determined.
The paradox that some samples were Xpert negative, but mRNA positive could be explained by the different sensitivities of the assays: the mRNA isolation procedures concentrate specimens prior to the assays and usually there are many copies of an mRNA species per genome copy. We also observed Xpert-positive, mRNA-negative specimens, possibly due to MTB DNA’s greater stability in both dead and alive cells. In BAL samples, however, we observed a good concordance between mRNA and DNA detection, since the BAL samples were from targeted sampling of the affected lobe, processed immediately and concentrated. The presence of DNA and mRNA in all BAL samples, suggest that both are usually present in parenchyma, but may be variable in sputum. We found an excellent correlation between MTB mRNA in BAL and clinical TB status (P < 0.001; Supplementary Note 3).
Our study was exploratory and the patients were investigated intensively, allowing complementary results from different tests. In summary, we describe a marked heterogeneity in treatment responses of individual PET-CT lesions and the presence of MTB mRNA in sputum and BAL of a substantial number of cured patients at end of treatment. Although MTB mRNA stability remains a possible explanation for its presence in intact bacteria, we suggest that viable MTB, with the potential to elicit a host response, often persist even after clinically curative treatment. The higher relapse rates in patient groups with impaired immunity7,51–53 support the concept that a competent immune response plays an important complementary role in the ultimate control of residual bacteria after antibiotic treatment. The nature of host-pathogen interaction and association with poor treatment outcome and post-tuberculosis lung impairment need to be investigated in future studies. Sterilizing drug or host-directed therapies and improved treatment response markers are likely needed for the successful development of improved or shortened PTB treatment strategies.
Online methods
Recruitment and follow-up, South-Africa
Ethical approval was obtained from the Stellenbosch University Human Research Ethics Committee (registration number N10/01/013). In total, 131 HIV-uninfected adults with newly diagnosed PTB, confirmed by sputum culture, were recruited after informed consent between April 2010 and April 2013 at Primary Healthcare Clinics (PHC’s) in Cape Town. Of these patients, 32 were excluded for: missed scan visits due to technical reasons (7), patient decision (8), sputum culture negativity at diagnosis (8) and severe co-morbidity (3).
Sputum specimens for MGIT culture with speciation and additional clinical information were collected at Day 0, week 1, 4, 8, 12, and 24 for all participants and an additional culture, collected at week 20 for the last 64 patients who completed the study. Of the 99 completing study time-points (including 60 males and 39 females), 85 patients were also available for a formal follow-up visit one year after the end of treatment, while the other cured patients were screened by telephonic interview and review of medical records and results. The screen was repeated for all participants, 2 years after the end of treatment. Patients’ clinical outcomes were classified as cured if they proved and maintained sputum culture negativity by month 6 (M6), failed if they’re M6 culture was still positive, un-evaluable if contamination caused uncertainty in outcome, and recurrent if they were restarted on TB treatment by healthcare providers in the 2 years after initial treatment completion. Of the recurrent patients, two were culture confirmed; five were confirmed by both Xpert with supporting acid fast bacillus (AFB) testing for smear positivity by direct microscopy; three were Xpert negative at month 6, but converted back to positive; and two remained Xpert positive for more than 6 months, but complained of increased symptoms. None of the un-evaluable group was diagnosed by healthcare providers with recurrent PTB before EOT + 2y.
Spoligotyping was performed on sputum cultures from Dx and M6 for patients that failed treatment. In six of eight cases identical strains were found, while the remaining two patients had different strains at Dx and M6. This may be due to reinfection54 during the treatment phase, or concurrent infection with different strains from Dx. 55,56
Sixty-six asymptomatic community controls and 13 controls diagnosed with infective or inflammatory lung diseases other than PTB (other lung disease; OLD) were recruited from the same high TB- incidence communities. Out of this group, 20 community (9 males and 11 females) and five OLD (2 males and 3 females) consecutively enrolled controls were included in sputum MTB RNA analysis. OLD controls included four cases of pneumonia and one asthma exacerbation case. Active tuberculosis was excluded for all controls based on negative sputum culture and a chest X-ray not suggestive of PTB. All controls were HIV-negative and not on any systemic corticosteroid treatment.
Treatment regimen
All patients were subjected to directly observed treatment (DOT) at local healthcare clinics.
Seventy-two patients received a standard treatment regimen: two months intensive phase with daily fixed-dose combination tablets (HRZE): isoniazid (INH), rifampicin (RIF), ethambutol (EMB) and pyrazinamide (PZA) followed by 4 months continuation phase of daily INH and RIF (currently the standard of care according to the WHO guidelines).40 Twenty-three patients received an extended regimen: a 3 month initiation phase of HRZE, with additional streptomycin in the first month (19) or without (4), and a 5 month continuation phase. In 22 patients, the indication for the extended regimen was previous episode(s) of PTB and for the remaining patient, delayed smear conversion. The local guidelines were amended during the study period to standard 6 months of HRZE treatment for all drug-susceptible cases, resulting in seven patients with previous episode(s) not receiving extended treatment.
Two patients had INH mono-resistance and received 6 months of HRZE with added ofloxacin and 12 months of HRZE respectively. Two patients with MDR received individualised regimens, containing kanamycin and terizidone among others.
Recruitment and follow-up, South-Korea
All participants signed informed consent (Ethical approval from National Medical Centre Institutional Review board: IRB00008343). 19 culture positive PTB patients were recruited at diagnosis in Masan, South-Korea. Three patients were lost to follow-up; two decided to leave the study. The 14 patients (including 12 males and 2 females) that completed Dx, M1 and M6 scans included three with MDR strains and two with Rifampicin mono-resistance. Patients with DS-PTB were treated with standard HRZE for 6 months, Rifampicin resistant cases were treated for 12 months and MDR cases were treated for 18 months after culture conversion, on individualized regimens. Patients were followed for 2 years after treatment; one patient was diagnosed with recurrent PTB with a strain different than at diagnosis.
PET-CT
A one week window was allowed for the Dx and M1 scans; a 4 week window was allowed for M6 and EOT + 1y year scans. Patients fasted for at least 6 hours before FDG administration. According to body weight 185–259 MBq of 18F-FDG was administered intravenously 60 min before scan. Scans at baseline were performed from the base of skull to the upper thigh and from the neck to the upper abdomen (whole lung) at M1, M6 and EOT + 1 yr. The CT scan parameters were set at 120kV, 100mAs (no dose modulation) with a 3 mm slice thickness.
CT and PET images were evaluated in collaboration and consensus with a radiologist, a nuclear physician and a pulmonologist. Patient clinical outcomes were not known at the time of reading. Lesion morphology was characterised and measured based on CT images. PET uptake intensity of prominent lesions was measured using the mean and maximum standardized uptake value (SUV) and compared across time-points, using the uptake in the right lobe of the liver and the mediastinal aortic blood pool, as reference values to account for variability in background uptake.25,57–63
The most intense lesion of each scan was ranked based on its mean standard uptake value (SUVmean). The method was adapted from the Deauville grading system (low, moderate, high intensity) that used the mediastinal blood pool and liver as references.57,64,65 None: no increased uptake compared to surrounding lung tissue. Minimal: Uptake more intense than surrounding lung tissue, but less intense than mean (SUV) of 10 mm sphere in mediastinal blood pool. Mild: Uptake more intense than mediastinal blood pool but less than one standard deviation (SD) below the mean SUV of a 30 mm sphere in right liver lobe. Moderate: Uptake similar to liver (between 1 SD below mean SUV and 2 SD above mean SUV. High: Uptake more intense than 2 SD above mean liver SUV and less than double the mean liver SUV. Very high: Intensity higher than double the mean liver SUV.
Scans were grouped into response patterns by comparing each M6 lesion to the corresponding M1 lesion to summarize the central trend of lesion progression. Resolved response pattern refers to minimal or no increased FDG uptake compared to surrounding healthy tissue, regardless of structural abnormalities on CT. Improved scans had decreased intensity of all lesions compared to the baseline scan, but still one or more lesion(s) with increased uptake compared to surrounding lung tissue and reference structures. Mixed responses showed at least one new FDG-avid lesion or at least one intensified lesion compared to the baseline scan.
MTB RNA in M6 sputum
For transcription a QuantiFast Multiplex RT-PCR kit (QIAGEN) with a primer designed to be specific for each MTB target gene was used. We selected genes that were previously shown to have increased expression under conditions of stress and persistence.41,42 Two ml sputum specimens were collected and added to 2 ml of Trizol (Invitrogen), flash frozen in liquid nitrogen and stored at −80°C. For RNA extraction, samples were thawed and spun down at 3500 g for 25 min. The supernatant was discarded and the pellet was re-suspended in 1 ml Trizol reagent. The entire content of the tube was transferred into new 2 ml screw cap micro-centrifuge tubes containing 0.5 ml of zirconia-silica beads (diameter, 0.1 mm). Bacteria were disrupted in a Bead Beater (FastPrep Cell Disrupter, FP120) using two 45 second pulses at maximum speed of 6.5 m/sec, incubated on ice for 5 min, followed by the addition of 250 μl of chloroform and vigorous mixing for 15 s and then a 2–3 min room temperature incubation. Samples were then centrifuged at 12, 000 g for 5 min and the upper clear phase transferred carefully to a new tube and mixed with an equal volume of 70% ethanol, applied to an RNeasy mini column (QIAGEN) and processed according to the manufacturer’s recommendations.
Fourteen mRNA targets (85B, dosR, hspX (acr), pstS1, carD, nuoB, tgS1, TB8.4, TB31.7, acpM, icL, prcA sigA, rrnAP1) were reverse transcribed, using a QuantiFast Multiplex RT-PCR kit (QIAGEN) and a primer specific for each target gene. The conditions for Reverse Transcription PCR were 50°C for 50 min, 95 °C for 2 min. The amount of cDNA produced was quantified by real-time PCR with the corresponding molecular beacon. The 10 μl PCR reaction mixture consisted of 1X PCR buffer, 250 μM deoxynucleoside triphosphates, 4 mM MgCl2, 0.5 μM each primer, 4ng/μl molecular beacon and 0.03U/μl Jumpstart Taq polymerase (Sigma-Aldrich). To normalize the individual reactions 6-carboxy-x-rhodamin (ROX) was always included as passive reference dye. PCRs were performed in 384-well microtiter plates in an ABI 7900 prism (Applied Biosystems, Foster City, CA) according to the following parameters: initial denaturation 95°C for 1 min, then50 cycles of: denaturation 95°C for 30 seconds, annealing 58°C 30 seconds, and extension at 72°C, 15 seconds. PCR conditions were identical for all assays. The fluorescence was recorded during the annealing step of the assay. The quantity of specific target DNA was determined from the cycle threshold (CT) value with reference to a standard curve of genomic MTB DNA. The copy numbers of target standards used ranged from 1 to 106 genomic copies per reaction (i.e. 10 fg to 10 ng DNA from CDC 1551 strains). The lower limit of detection for each assay was 10 fg which is equivalent to 1–5 copies of cDNA. qRT-PCR reactions were performed in triplicate. The mean of each triplicate is used in calculations. The sigA target showed cross reactivity with non-tuberculosis mycobacterial RNA at high concentrations in addition to low level positivity in four community controls and one control with other lung disease and was subsequently dropped from further analysis.
Primer sequences are listed in Supplementary Note 4.
RNA in EOT bronchoalveolar lavage samples
Culture from BAL fluid is generally more sensitive than culture from sputum66,67, although the quality of the sample can be variable. An additional high-quality sputum sample is often taken after the procedure.68 Bronchoscopies were performed on 15 of the PTB patients within 3 months after the end of their standard treatment (EOT) and BAL fluid and post bronchoscopy sputa obtained. MGIT cultures on bronchoscopy samples were allowed extended growth periods (12 weeks). BAL fluid was also collected from 10 patients (3 males and 7 females) suspected of having cancer and undergoing diagnostics bronchoscopies, as controls. We added 15 ml of fresh BAL fluid to 30 ml Trizol and stored it at −80°C until RNA analysis.
Multiplex Real Time RT-PCR Transcriptional analysis of Sputum and BAL specimens from 14 recently cured and one failed treatment patient
Fluorescence-based qRT-PCR is accepted as a gold standard for gene expression profiling due to its high specificity, sensitivity, accuracy and large dynamic range.
Extraction of total RNA from BAL
BAL samples were sent, in a blinded fashion, for MTB RNA analysis via two-step qRT-PCR with validated TaqMan assays developed at Stanford.41,69 Total MTB RNA isolated from BAL was reverse transcribed using random primers and pre-amplified in a controlled multiplex PCR with 743 MTB-specific primer sets targeting corresponding mRNA transcripts as described previously.70 Finally, we used only 24 gene-specific TaqMan assays for individual gene transcript quantification of multiplex PCR products targeting the genes shown as differentially regulated in stress or persistence based on available literature. 41,42 These genes included recF, eccD3, menA, pabC, accD3, iprB, Rv1421, gabD2, Rv1910c, hspX, lipX, fadD9, tgs1, fadE34, Rv3675, sodA, trxB2, rpsJ, rplV, sigI, Rv1255c, sigB, sigH, rpsK.41
Samples were thawed and total RNA was extracted using a glass matrix tube for cell lysis (Lysing Matrix B, Q Biogene) in a FastPrep FP120 instrument (Bio 101, Thermo Savant) with a speed setting of six for three iterations of 30 s, with cooling on ice for 1 min after each iteration. After processing, chloroform was added, followed by separation of the aqueous and organic layers. The aqueous phase containing the RNA was removed to a fresh tube and the RNA was precipitated overnight at −20°C after addition of glycoblue (Ambion), 0.1 volume of 5 M ammonium acetate, and an equal volume of isopropanol were added. The resulting RNA pellet was re-suspended in 50 μl of RNase free H2O and cleaned by three rounds of the RNeasy® mini column system, interspersed with off-column DNase treatments (Promega RQ1 DNase). 41
Genome Expression Profiling of MTB was performed using Two-Step Multiplex RT-PCR. The protocol below consists of three parts: 1) First strand cDNA synthesis and Controlled Multiplex Pre-Amplification of cDNAs; 2) Preparation of Primer and Probe Sets; and 3) Individual qRT-PCR (Taqman) quantification of Amplified cDNAs using LightCycler480.
Each RNA sample (5 μl) was taken into two separate first strand cDNA synthesis reactions (RT+ and RT−) to control for DNA contamination. An additional water control (zero RNA) was also added. To each sample, 0.5 μl Exo-resistant Random Primer (Fermentas S0181), 1 μl 10mM dNTPs (Fermentas R0193) and 3.5 μl Nuclease Free Water (Ambion AM9938) were added for a total of 10 μl. This mix was incubated for 3 minutes at 70 °C in a thermal cycler, and then placed on ice.
During this incubation, two cocktails were prepared: one containing reverse transcriptase (RT+) and one without (RT−). Each RT+ cocktail contained 4 μl 5X Maxima RT Buffer (Fermentas EP0741), 0.5 μl Ribolock RNase-Inhibitor (Fermentas EO0382), 0.5 μl Maxima RT enzyme (Fermentas EP0741), and 3.0 μl Nuclease Free Water for a total of 10 μl. The RT− cocktail was exactly the same except water was substituted for the RT enzyme. These cocktails were scaled up for multiple samples and 10 μl aliquots were added to each RT+ or RT− sample prepared above. The samples were mixed gently and then gently centrifuged for 2 min. They were incubated at 50 °C 1 hour, 95 °C 2 minutes (to inactivate the reverse transcriptase), then stored at 4 °C. Maxima Reverse Transcriptase (RT) possesses an RNA and DNA-dependent polymerase activity as well as RNase H activity.
qRT-PCR on pre-amplified material: All outflanking primers and TaqMan probe sets had been validated in multiplex PCR pre-amplification for linearity of amplification using all the genes used in each pre-amplification cocktail. We also validated all individual TaqMan assays from our collection for sensitivity and linearity before we started using them in gene expression profiling. A complete database with all available validated TaqMan primer sets used for pre-amplification can be found at ftp://smd-ftp.stanford.edu/tbdb/rtpcr/taqman_oligos.fa. Sequences and design of specific PCR primer/probe sets can be found in Supplementary Note 4.
Primer and probe qRT-PCR sets for each gene consist of a forward primer (TMF), a FAM/BHQ-labelled probe (TMP), and a reverse primer (TMR). These were ordered from Biosearch at a 100 uM concentration and a Taqman Mix was prepared for each gene. Twenty-seven μl of the forward primer, 27 μl of the reverse primer, and 9 μl of the probe were mixed in 1737 μl of Nuclease Free water for a total of 1800 μl. This resulted in a dilution of the forward and reverse primer to 1500 nM and the probe to 500 nM. We used 2 μl of this mix in a 10 μl qRT-PCR, resulting in a final reaction concentration of 300 nM for each primer and 100 nM for the probe.
Two different water controls were run throughout the process: one starting at the cDNA step and one starting at the amplification step. All RT− controls were negative, but water controls for pabC, accD3, lipX, Rv1255c and sigB showed some reactivity and these targets were excluded from further analysis. Additionally, two samples of 104 gene copy number H37Rv genomic DNA were amplified in each amplification mix and run on each plate.
Genomic Equivalent DNA calculation
We isolated genomic DNA by buffer extraction from the interphase after MTB lysis with Trizol. DNA was pre- amplified with 24 genes and individual TaqMan assays quantified the amplicons. Median Ct was calculated for 24 genes for each sample and compared to genomic DNA reference to calculate GE.69
Statistical analysis
As an observational study, we did not apply randomization. The calculated sample size of the cohort was intended to provide a representative range of biological reactions during a favourable response to PTB treatment. Analysis was either performed blinded (BAL) or outcomes were not known at the time (M6 sputum mRNA and Xpert; M6 PET/CT scans).
Clinical and microbiological parameters of failed treatment and recurrent outcome groups were compared to the cured group using a two-tailed, unpaired Student’s t-test. Associations between categorical variables were evaluated using the Fisher Exact Test for independence, the Chi-squared Test for trend in proportions and the Asymptotic independence test, using Statistica Version 12 or R version 3.2.2. Values are shown in Supplementary Note 3.
Principal component analysis was performed using the singular value decomposition approach (prcomp in R version 3.2.2), to derive a limited set of variables (principle components; PC) that captured the variance in the mRNA values. Matrices of Ct values for each mRNA species in each patient were used as input for the analysis. The first PC captured more than 80% of the mRNA variance in the analyses and was used to represent the mRNA data for each individual with a single value. Figure 5 contains box and whisker plots of the arcsinh-transformed first principal component of mRNA transcript PCR data.
Supplementary Material
Acknowledgments
Funding was provided by the Catalysis Foundation for Health (grant to G.W., C.E.B., D.A.) the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (grant to C.E.B), the South African National Research Fund and the Medical Research Council’s Clinician Scholarship Program (bursary to S.T.M.), as well as the National Institute of Allergy and Infectious Diseases, International Collaborations in Infectious Disease Research (grant to G.W.). We thank our participants for their willingness to take part in this study. Further, we acknowledge Rich Thayer and Mickey Urdea of CFH, the staff at the Stellenbosch University Immunology Research Group, the International TB Research Center, Seoul, the Western Cape Academic PET-CT Centre, Ithemba LABS, Tygerberg Academic Hospital’s Nuclear Medicine Department and Pulmonology Unit, as well as managers and health care providers from the City of Cape Town Health Department.
Footnotes
Competing financial interests statement.
We have no competing financial interest to declare.
Author contributions
S.T.M. managed patient recruitment, follow-up and sample collection, analysed PET-CT scans, drafted the manuscript. R.Y.C., J.War., L.E.V., D.A., C.E.B., J.Win., and G.W., designed the study, analysed results and edited the manuscript. G.T., lead statistical analysis and edited the manuscript. S.S, K.R., A.G.L., G.D., T.V., T.S. and G.S., designed and conducted experiments, analysed results and edited the manuscript. M.K., M.L., managed patient recruitment, follow-up and sample collection and edited the manuscript. Members of the Catalysis TB-Biomarker Consortium contributing to the work: L.L., L.T., B.S., N.B., C.G.G.B., processed samples and conducted experiments. G.vd.S. and K.S., managed data collection, database and sample storage. E.M. and L.E.D. analysed results. A.E. and S.G-R. analysed PET-CT scans. H.C. and J.H. managed patient recruitment and follow-up. B.A., M.V. and C.K. recruited patients and conducted bronchoscopies.
Main References
- 1.Cox HS, Morrow M, Deutschmann PW. Long term efficacy of DOTS regimens for tuberculosis: systematic review. BMJ. 2008;336:484–487. doi: 10.1136/bmj.39463.640787.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Global Tuberculosis Report. 2015;20 [Google Scholar]
- 3.Gillespie SH, et al. Four-Month Moxifloxacin-Based Regimens for Drug-Sensitive Tuberculosis. N Engl J Med. 2014;371:1577–1587. doi: 10.1056/NEJMoa1407426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jindani A, et al. High-Dose Rifapentine with Moxifloxacin for Pulmonary Tuberculosis. N Engl J Med. 2014;371:1599–1608. doi: 10.1056/NEJMoa1314210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luzze H, et al. Relapse more common than reinfection in recurrent tuberculosis 1–2 years post treatment in urban Uganda. Int J Tuberc Lung Dis. 2013;17:361–367. doi: 10.5588/ijtld.11.0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merle CS, et al. A Four-Month Gatifloxacin-Containing Regimen for Treating Tuberculosis. N Engl J Med. 2014;371:1588–1598. doi: 10.1056/NEJMoa1315817. [DOI] [PubMed] [Google Scholar]
- 7.Sonnenberg P, et al. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet. 2001;358:1687–1693. doi: 10.1016/S0140-6736(01)06712-5. [DOI] [PubMed] [Google Scholar]
- 8.Stop TB Partnership W. The Global Plan to Stop TB 2016 – 2020. 2016;2020:1–4. [Google Scholar]
- 9.Johnson JL, et al. Shortening treatment in adults with noncavitary tuberculosis and 2-month culture conversion. Am J Respir Crit Care Med. 2009;180:558–563. doi: 10.1164/rccm.200904-0536OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner DF, Mizrahi V. Shortening Treatment for Tuberculosis - Back to Basics. N Engl J Med. 2014;371:1642–1643. doi: 10.1056/NEJMe1410977. [DOI] [PubMed] [Google Scholar]
- 11.Seon HJ, Kim YI, Lim SC, Kim YH, Kwon YS. Clinical significance of residual lesions in chest computed tomography after anti-tuberculosis treatment. Int J Tuberc Lung Dis. 2014;18:341–346. doi: 10.5588/ijtld.13.0565. [DOI] [PubMed] [Google Scholar]
- 12.Ralph AP, et al. A simple, valid, numerical score for grading chest x-ray severity in adult smear-positive pulmonary tuberculosis. Thorax. 2010;65:863–869. doi: 10.1136/thx.2010.136242. [DOI] [PubMed] [Google Scholar]
- 13.Pasipanodya JG, et al. Pulmonary impairment after tuberculosis and its contribution to TB burden. BMC Public Health. 2010;10:259. doi: 10.1186/1471-2458-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wejse C, et al. TBscore: Signs and symptoms from tuberculosis patients in a low-resource setting have predictive value and may be used to assess clinical course. Scand J Infect Dis. 2008;40:111–120. doi: 10.1080/00365540701558698. [DOI] [PubMed] [Google Scholar]
- 15.Pasipanodya JG, et al. Pulmonary impairment after tuberculosis. Chest. 2007;131:1817–1824. doi: 10.1378/chest.06-2949. [DOI] [PubMed] [Google Scholar]
- 16.Pasipanodya JG, et al. Using the St. George Respiratory Questionnaire to ascertain health quality in persons with treated pulmonary tuberculosis. Chest. 2007;132:1591–1598. doi: 10.1378/chest.07-0755. [DOI] [PubMed] [Google Scholar]
- 17.Hnizdo E, Singh T, Churchyard G. Chronic pulmonary function impairment caused by initial and recurrent pulmonary tuberculosis following treatment. Thorax. 2000;55:32–38. doi: 10.1136/thorax.55.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos LMM, Sulmonett N, Ferreira CS, Henriques JF, de Miranda SS. Functional profile of patients with tuberculosis sequelae in a university hospital. J Bras Pneumol publicaçao Of da Soc Bras Pneumol e Tisilogia. 2006;32:43–7. doi: 10.1590/s1806-37132006000100010. [DOI] [PubMed] [Google Scholar]
- 19.de Nihues SSE, et al. Chronic symptoms and pulmonary dysfunction in post-tuberculosis Brazilian patients. Brazilian J Infect Dis. 2015;19:492–497. doi: 10.1016/j.bjid.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baig IM, Saeed W, Khalil KF. Post-tuberculous chronic obstructive pulmonary disease. J Coll Physicians Surg Pak. 2010;20:542–4. [PubMed] [Google Scholar]
- 21.Mitchison DA. Basic Mechanisms of Chemotherapy. Chest. 1979;76 doi: 10.1378/chest.76.6_supplement.771. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, et al. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000;182:6358–6365. doi: 10.1128/jb.182.22.6358-6365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rustad TR, et al. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nuclecic Acids Res. 2013;41:509–517. doi: 10.1093/nar/gks1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatov DV, et al. Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA. BMC Genomics. 2015;16:954. doi: 10.1186/s12864-015-2197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheebsumon P, et al. Effects of image characteristics on performance of tumor delineation methods: a test-retest assessment. J Nucl Med. 2011;52:1550–8. doi: 10.2967/jnumed.111.088914. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, et al. Standard uptake value and metabolic tumor volume of 18F-FDG PET/CT predict short-term outcome early in the course of chemoradiotherapy in advanced non-small cell lung cancer. Eur J Nucl Med Mol Imaging. 2011;38:1628–35. doi: 10.1007/s00259-011-1838-5. [DOI] [PubMed] [Google Scholar]
- 27.Dibble EH, et al. 18F-FDG metabolic tumor volume and total glycolytic activity of oral cavity and oropharyngeal squamous cell cancer: adding value to clinical staging. J Nucl Med. 2012;53:709–15. doi: 10.2967/jnumed.111.099531. [DOI] [PubMed] [Google Scholar]
- 28.Lin PL, et al. Sterilization of granulomas is common in both active and latent tuberculosis despite extensive within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Via LE, et al. Infection dynamics and response to chemotherapy in a rabbit model of tuberculosis using [18F]2-fluoro-deoxy-D-glucose positron emission tomography and computed tomography. Antimicrob Agents Chemother. 2012;56:4391–402. doi: 10.1128/AAC.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin PL, et al. Radiologic responses in cynomolgous macaques for assessing tuberculosis chemotherapy regimens. Antimicrob Agents Chemother. 2013;57:4237–4244. doi: 10.1128/AAC.00277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman MT, et al. Early changes by 18F-PET-CT predict outcome after M. tuberculosis infection in cynomolgus macaques. Infect Immun. 2014;82:2400–2404. doi: 10.1128/IAI.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis SL, et al. Noninvasive pulmonary [18F]-2-fluoro-deoxy-D-glucose positron emission tomography correlates with bactericidal activity of tuberculosis drug treatment. Antimicrob Agents Chemother. 2009;53:4879–4884. doi: 10.1128/AAC.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez V, et al. (18)F-FDG PET/CT in tuberculosis: an early non-invasive marker of therapeutic response. Int J Tuberc Lung Dis. 2012;16:1180–5. doi: 10.5588/ijtld.12.0010. [DOI] [PubMed] [Google Scholar]
- 34.Dureja S, Sen I, Acharya S. Potential role of F18 FDG PET-CT as an imaging biomarker for the noninvasive evaluation in uncomplicated skeletal tuberculosis: a prospective clinical observational. Eur Spine J. 2014;23:2449–2454. doi: 10.1007/s00586-014-3483-8. [DOI] [PubMed] [Google Scholar]
- 35.Demura Y, et al. Usefulness of 18F-fluorodeoxyglucose positron emission tomography for diagnosing disease activity and monitoring therapeutic response in patients with pulmonary mycobacteriosis. Eur J Nucl Med Mol Imaging. 2009;36:632–639. doi: 10.1007/s00259-008-1009-5. [DOI] [PubMed] [Google Scholar]
- 36.Coleman MT, et al. Early Changes by (18)Fluorodeoxyglucose positron emission tomography coregistered with computed tomography predict outcome after Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2014;82:2400–4. doi: 10.1128/IAI.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen RY, et al. PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Sci Transl Med. 2014;6:265ra166. doi: 10.1126/scitranslmed.3009501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stelzmueller I, et al. 18F-FDG PET/CT in the Initial Assessment and for Follow-up in Patients With Tuberculosis. Clin Nucl Med. 2016;41:187–194. doi: 10.1097/RLU.0000000000001102. [DOI] [PubMed] [Google Scholar]
- 39.Sathekge M, Maes A, Kgomo M, Stoltz A, Van de Wiele C. Use of 18F-FDG PET to predict response to first-line tuberculostatics in HIV-associated tuberculosis. J Nucl Med. 2011;52:880–885. doi: 10.2967/jnumed.110.083709. [DOI] [PubMed] [Google Scholar]
- 40.WHO. Treatment of tuberculosis guidelines. 2010;4 [PubMed] [Google Scholar]
- 41.Walter ND, et al. Transcriptional adaptation of drug-tolerant Mycobacterium tuberculosis during treatment of human tuberculosis. J Infect Dis. 2015;212:1–9. doi: 10.1093/infdis/jiv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lew JM, Kapopoulou A, Jones LMCS. TubercuList - TB Gene Database. at < http://tuberculist.epfl.ch/quicksearch.php?gene+name=Rv2031c>.
- 43.Yuan Y, Crane DD, Barry CE. Stationary phase-associated protein expression in Mycobacterium tuberculosis: Function of the mycobacterial alpha-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittl RL, et al. Radiographic Resolution of Community-acquired Pneumonia. Am J Respir Crit Care Med. 1994;149:630–635. doi: 10.1164/ajrccm.149.3.8118630. [DOI] [PubMed] [Google Scholar]
- 45.Friedrich SO, et al. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 2014;2600:1–9. doi: 10.1016/S2213-2600(13)70119-X. [DOI] [PubMed] [Google Scholar]
- 46.Lahtinen SJ, et al. Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett Appl Microbiol. 2008;46:693–698. doi: 10.1111/j.1472-765X.2008.02374.x. [DOI] [PubMed] [Google Scholar]
- 47.Marx FM, et al. The temporal dynamics of relapse and reinfection tuberculosis after successful treatment: A retrospective cohort study. Clin Infect Dis. 2014;58:1676–1683. doi: 10.1093/cid/ciu186. [DOI] [PubMed] [Google Scholar]
- 48.Middelkoop K, Bekker LG, Shashkina E, Kreiswirth B, Wood R. Retreatment tuberculosis in a South African community: the role of re-infection, HIV and antiretroviral treatment. Int J Tuberc Lung Dis. 2012;16:1510–6. doi: 10.5588/ijtld.12.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breen RAM, et al. How good are systemic symptoms and blood inflammatory markers at detecting individuals with tuberculosis? Int J Tuberc Lung Dis. 2008;12:44–49. [PubMed] [Google Scholar]
- 50.Marais BJ, Schaaf HS, Hesseling AC, GR Tuberculosis case definition_: time for critical reassessment? Int J Tuberc Lung Dis. 2008;12:1217–1218. [PubMed] [Google Scholar]
- 51.Dooley KE, et al. Risk factors for tuberculosis treatment failure, default, or relapse and outcomes of retreatment in Morocco. BMC Public Health. 2011;11:140. doi: 10.1186/1471-2458-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comstock GW, Golub JE, Panjabi R. Recurrent tuberculosis and its risk factors: Adequately treated patients are still at high risk. Int J Tuberc Lung Dis. 2007;11:828–837. [PubMed] [Google Scholar]
- 53.Hesseling aC, et al. Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int J Tuberc Lung Dis. 2010;14:560–570. [PubMed] [Google Scholar]
- 54.Warren RM, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 55.Kritzinger FE, et al. No decrease in annual risk of tuberculosis infection in endemic area in Cape Town, South Africa. Trop Med Int Health. 2009;14:136–142. doi: 10.1111/j.1365-3156.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 56.Shanaube K, et al. Annual risk of tuberculous infection using different methods in communities with a high prevalence of TB and HIV in Zambia and South Africa. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chien David, Lodge Martin, RW Reproducibility of liver and mediastinal blood pool F-18 activity as normal reference tissues. J Nucl Med. 2011;52 [Google Scholar]
- 58.Fletcher JW. PET/CT Standardized Uptake Values (SUVs) n Clinical Practice Assessing Response to Therapy. Semin Ultrasouns CT MRI. 2010;31:496–505. doi: 10.1053/j.sult.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fogh SE, et al. Pathological correlation of PET/CT based auto contouring for radiation planning of lung cancer. Bodine J. 2010;78:202–203. [Google Scholar]
- 60.Delbeke D, et al. Expert opinions on positron emission tomography and computed tomography imaging in lymphoma. Oncologist. 2009;14(Suppl 2):30–40. doi: 10.1634/theoncologist.2009-S2-30. [DOI] [PubMed] [Google Scholar]
- 61.Lowe VJ, Hoffman JM, DeLong DM, Patz EF, Coleman RE. Semiquantitative and visual analysis of FDG-PET images in pulmonary abnormalities. J Nucl Med. 1994;35:1771–6. [PubMed] [Google Scholar]
- 62.Hatt M, et al. Reproducibility of 18F-FDG and 3′-deoxy-3′-18F-fluorothymidine PET tumor volume measurements. J Nucl Med. 2010;51:1368–76. doi: 10.2967/jnumed.110.078501. [DOI] [PubMed] [Google Scholar]
- 63.Dann EJ, et al. A functional dynamic scoring model to elucidate the significanceof post-induction interim fluorine-18-fluorodeoxyglucose positron emission tomography findings in patients with hodgkin’s lymphoma. Haematologica. 2010;95:1198–1206. doi: 10.3324/haematol.2009.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higashi K, et al. 18F-FDG uptake by primary tumor as a predictor of intratumoral lymphatic vessel invasion and lymph node involvement in non-small cell lung cancer: analysis of a multicenter study. J Nucl Med. 2005;46:267–73. [PubMed] [Google Scholar]
- 65.Barrington SF, et al. Concordance between four European centres of PET reporting criteria designed for use in multicentre trials in Hodgkin lymphoma. Eur J Nucl Med Mol Imaging. 2010;37:1824–1833. doi: 10.1007/s00259-010-1490-5. [DOI] [PubMed] [Google Scholar]
- 66.Jacomelli M, et al. Bronchoscopy for the diagnosis of pulmonary tuberculosis in patients with negative sputum smear results. J Bras Pneum. 2012;38:167–173. doi: 10.1590/s1806-37132012000200004. [DOI] [PubMed] [Google Scholar]
- 67.Reingold AL, Daley CL, Kritski AL. Comparison of Sputum Induction with Fiberoptic Bronchoscopy in the Diagnosis of Tuberculosis Experience at an Acquired Immune Deficiency Syndrome Reference Center in Rio de Janeiro, Brazil. 2000;162:2238–2240. doi: 10.1164/ajrccm.162.6.2003125. [DOI] [PubMed] [Google Scholar]
- 68.George PM, et al. Post-bronchoscopy sputum: Improving the diagnostic yield in smear negative pulmonary TB. Respir Med. 2011;105:1726–1731. doi: 10.1016/j.rmed.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 69.Galagan JE, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–83. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Commandeur S, et al. An Unbiased Genome-Wide Mycobacterium tuberculosis Gene Expression Approach To Discover Antigens Targeted by Human T Cells Expressed during Pulmonary Infection. J Immunol. 2013;190:1659–1671. doi: 10.4049/jimmunol.1201593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.