Abstract
The H6N1 avian influenza virus has circulated in Taiwan for more than 40 years. The sporadic activity of low pathogenic H5N2 virus has been noted since 2003, and highly pathogenic H5N2 avian influenza virus has been detected since 2008. Ressortant viruses between H6N1 and H5N2 viruses have become established and enzootic in chickens throughout Taiwan. Outbreaks caused by Novel highly pathogenic H5 avian influenza viruses whose HA genes were closely related to that of the H5N8 virus isolated from ducks in Korea in 2014 were isolated from outbreaks in Taiwan since early 2015. The avian influenza virus infection status is becoming much more complicated in chickens in Taiwan. This necessitates a rapid and simple approach to detect and differentiate the viruses that prevail. H6N1, H5N2 and novel H5 viruses were simultaneously subtyped and pathotyped in this study using reverse transcription loop-mediated isothermal amplification and microarray, with detection limits of 10°, 101 and 10° viral copy numbers, respectively. The microarray signals were read by the naked eye with no expensive equipment needed. The method developed in this study could greatly improve avian influenza virus surveillance efficiency.
Keywords: avian influenza virus, microarray, pathotyping, reverse transcription loop-mediated isothermal amplification, subtyping
Avian influenza (AI) is a highly contagious disease caused by the type A influenza virus, which has many subtypes with respect to two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) [15]. The H6N1 virus was first isolated in Taiwan in 1972 [18] and subsequently isolated frequently from chickens. Although Taiwanese H6N1 viruses were considered to be low pathogenic, significant loss may occur when these viruses are associated with other infectious agents [22]. Sporadic H5N2 virus activity has been observed in chickens in Taiwan from 2003 to 2012 [16].
It is generally accepted that H5 highly pathogenic avian influenza viruses (HPAIVs) were derived from low pathogenic avian influenza viruses (LPAIVs) introduced to terrestrial birds from aquatic birds [2, 14]. After consecutive passages in the new hosts, LPAIVs may acquire additional basic amino acids at the connecting peptide of the cleavage site of the HA [12, 20, 21]. The number of these basic amino acids is directly associated with the virulence of the viruses in chickens and other terrestrial poultry [10, 11]. In general, LPAIVs have one to three basic amino acids, while HPAIVs have four or more basic amino acids at the cleavage sites [9, 17]. H5N2 LPAIVs whose amino acid residues at HA cleavage site (CS) was PQREKR*GLF (Asterisk means cleavage point by host proteases) caused outbreaks in Taiwan in late 2003 [6, 16]. A potentially highly pathogenic H5N2 avian influenza virus with PQRKKR*GLF at HA CS was first isolated in 2008 [6]. A H5N2 HPAIV with PQRRKR*GLF was also isolated in 2012 [16]. Lee et al. proved that these H6N1 and H5N2 viruses cocirculated in chickens in Taiwan [16].
Outbreaks caused by novel H5 HPAIVs (novel H5N2, H5N3 and H5N8) with PLRERRRKR*GLF were found in Taiwan in early 2015. According to the official website data of the Bureau of Animal and Plant Health Inspection and Quarantine (BAPHIQ), the HA gene of novel H5 HPAIVs was closely related to that of the H5N8 virus isolated from ducks in Korea in 2014, and the other genes were close to avian influenza viruses (AIVs) isolated in China, Japan and Korea [4]. These novel H5 viruses were therefore verified as from abroad and not from the evolution of local viruses in Taiwan.
The surveillance of AIVs depends mostly on reverse transcription polymerase chain reaction (RT-PCR) and sequencing, which are time- and labor-intensive. A number of methods have been developed, including real-time RT-PCR (rRT-PCR) [1, 9, 19, 24], reverse transcription loop-mediated isothermal amplification (RT-LAMP) [5, 13] and microarray [7, 23]. However, combination of RT-LAMP and microarray for simultaneous subtyping and pathotyping of AIVs has not been reported, and the detection limit with one viral RNA copy was neither achieved.
AIVs have become much more diversified over the past decade in Taiwan, and the surveillance workload has exhausted official staff. The huge economic loss has also caused a great impact on this country. The purpose of this study was to develop a precise, low-cost method to rapidly subtype and pathotype AIVs in chickens in Taiwan. This proposed approach could significantly improve the sensitivity of the detection and greatly contribute to the control of avian influenza.
MATERIALS AND METHODS
Viral reference strains and field samples: The viruses used in this study are listed in Table 1. H5N2 LPAIV (H5l), chicken H6N1 virus (H6c), New castle disease virus (NDV) and infectious bronchitis virus (IBV) were obtained from the Poultry Disease Laboratory, School of Veterinary Medicine, National Taiwan University. The viral nucleotides of H5N2 HPAIVs (H5ha and H5hb) and novel H5 HPAIVs (H5c, H5d and H5e) were obtained from the Epidemiology Division of the Animal Health Research Institute, Council of Agriculture, Tamsui, Taiwan. The viral nucleotide of human H6N1 virus (H6m) was from the Centers for Disease Control, Ministry of Health and Welfare, Taiwan. Seventy-two field samples, including oral swab, cloacal swab, brain, heart, trachea, lung and kidney, were collected from free-range, broiler and layer chickens at slaughterhouses in the north, central, south and east Taiwan, from November 2013 to March 2015.
Table 1. Viruses used in this study.
| Designation | Strain | Virus type | Amino acids at CSa) | Accession no. |
|---|---|---|---|---|
| H6c | A/chicken/Taiwan/2838V/00 | chicken H6N1 | EF681878 (Genbank) | |
| H6mb) | A/Taiwan/2/2013 | human H6N1 | EPI459855 (GISAID) | |
| H5l | A/chicken/Taiwan/1209/03 | H5N2 LPAIV | PQREKR*GLF | AY573917 (Genbank) |
| H5ha | A/chicken/Changhua/0101/12 | H5N2 HPAIV | PQRKKR*GLF | KF193386 (Genbank) |
| H5hb | A/chicken/Yunlin/0502/12 | H5N2 HPAIV | PQRRKR*GLF | |
| H5hc | A/chicken/Taiwan/a288/15 | novel H5N2 HPAIV | PLRERRRKR*GLF | |
| H5hd | A/chicken/Taiwan/a174/15 | novel H5N3 HPAIV | PLRERRRKR*GLF | |
| H5he | A/chicken/Taiwan/b214/15 | novel H5N8 HPAIV | PLRERRRKR*GLF | |
| NDV | 2209 | New castle disease virus | ||
| IBV | 3263 | infectious bronchitis virus |
a) Basic amino acids at CS are marked with bottom line (Asterisk means actual HA0 cleavage point by host proteases). b) Human infected H6N1 influenza virus.
RNA extraction: Viral RNA was extracted using QIAamp viral RNA kit (Qiagen, Valencia, CA, U.S.A.). Field tissue samples were ground in liquid nitrogen, and RNA was extracted using QIAamp RNeasy Mini Kit (Qiagen, Hilden, Germany). The RNA was immediately stored at −80°C until used for the RT-LAMP reaction.
Primer design: The HA gene highly conserved sequences of H6 and H5 AIVs in chickens in Taiwan were chosen for the primers. Primers flanking the HA CS of H5 viruses were selected. Sequences from Genbank or GISAID (The Global Initiative on Sharing All Influenza Data) were aligned using the MegAlign program (DNASTAR Lasergene 7.2.1, Madison, WI, U.S.A.). Primers (Table 2) were designed using the PrimerExplorer V4 software (http://primerexplorer.jp/elamp4.0.0/index.html) and were 5′ end-biotinylated.
Table 2. LAMP primers used to amplify H5 and H6 AIVs in Taiwan.
| Virus | Primer | Sequence (5′–3′) |
|---|---|---|
| H6 AIVs | F3 | CACAATTYGAAGCTGTHGAY |
| B3 | CCTAGRTCATTAGCRTTGTCY | |
| FIP | CAAACRTCYAGAAAYCCATCTTCCATTTTCACGAATTTTCAAAYCTDGAGAG | |
| BIP | AGAACAYTAGAYCTGCATGAYGCRTTTTTTAGYTGYGAYTTGACCTT | |
| H5 AIVs | F3 | GTATGCCTTTCCACAATGTY |
| B3 | TGCAGCATATCCRCTACC | |
| FIP | TCTYAGYCCTGTTGCWAGGATTTTCCCTTYRCCATTGGGGAG | |
| BIP | GCCGGATTYATAGAAGGAGGRTTTTCTYATTGCTRTGATGGTATCC | |
| Novel H5 HPAIVs | F3 | GGGCGATAAACTCTAGCA |
| B3 | TACCATTCCCTGCCATCC | |
| FIP | GCAAGGACTAATTTGTTTGACTTCATTTTCCACAATATACACCCTCTCAC | |
| BIP | CTGGGCTCAGAAATAGTCCTCTTTTTTCCCTCTATAAACCCTGCTA |
Multiplex RT-LAMP and uniplex RT-LAMP assays: RT-LAMP assays were carried out using Loopamp RNA Amplification Kit (Eiken, Tokyo, Japan). The H6N1 and H5N2 AIVs were amplified together using multiplex RT-LAMP. Each 25 µl reaction contained 20 pmole of each H6N1 inner primer (FIP and BIP), 2.5 pmole of each H6N1 outer primer (F3 and B3), 70 pmole of each H5N2 inner primer (FIP and BIP), 8.75 pmole of each H5N2 outer primer (F3 and B3) and 2 µl of each template RNA. The novel H5 HPAIVs were amplified using uniplex RT-LAMP. Each 25 µl reaction contained 4 pmole of each inner primer (FIP and BIP), 0.5 pmole of each outer primer (F3 and B3) and 2 µl of template RNA. The RT-LAMP reaction was performed at 61°C for 90 min, and the products were then visualized by 1.5% agarose gel electrophoresis.
Probe design: Probes located within the amplicon amplified by each specific primer set were designed. Seven differentiating probes were employed that specifically recognized and differentiated all of the H6N1 viruses isolated from chickens and human and all of the chicken H5 viruses, H5N2 LPAIVs, H5N2 HPAIVs and novel H5 HPAIVs in Taiwan (Table 3). Probe H5h could recognize both the PQRKKR*GLF and PQRRKR*GLF amino acid motifs at the CS of H5N2 HPAIVs. All probe designs were derived from the nucleotide sequence alignment and analyses retrieved from the GenBank or Global Initiative on Sharing Avian Influenza Data (GISAID), and conducted using the MegAlign program (DNASTAR Lasergene 7.2.1).
Table 3. Probes used to detect and differentiate H5 and H6 AIVs in Taiwan.
| Probe | Sequence (5′–3′) | Recognition viruses |
|---|---|---|
| H6u | CTTCTTGAARAYGAAAGAACAYTAGAYCTGCATG | All H6 AIVs (including both chicken and human) |
| H6c | AATGCTGAACTKYTGRT | Chicken infected H6N1 viruses |
| H6m | ATGCTGAGTTGTTG | Human infected H6N1 virus (A/Taiwan/2/2013) |
| H5u | AGA GGMCTWTTT GGAGCWATAGCM GG | All chicken H5 AIVs |
| H5l | MGAGAAAAAAGAGGHCTA | H5 chicken LPAIVs |
| H5h | CCYCAAAGRARRAAAAGAG | H5 chicken HPAIVsa) |
| H5n | ATAGTCCTCTAAGAGAAAGA | Novel H5 chicken HPAIVs |
a) H5h recognizes H5 HPAIVs in Taiwan, including both amino acid patterns at CS (PQRKKR*GLF, PQRRKR*GLF).
Microarray analysis: A tail composed of 19 T bases was added onto each 5′end of the oligonucleotide probe, including the positive control probe (an oligonucleotide from capsid protein VP1 of human enterovirus 71 gene, 5′-ATGAAGCATGTCAGGGCTTGGATACCTCG-3′). Twenty µM of each probe was then spotted to each specific position on the microarray polymer substrate using an automatic spotting machine and immobilized using a UV crosslinker (Stratagene UV Stratalinker 1,800, Stratagene, Santa Clara, CA, U.S.A.) with 0.48 J. The hybridization reaction between each DNA template and probe was carried out with the DR. Chip DIYTM Kit (Dr. Chip Biotech, Miao-Li, Taiwan). The RT-LAMP product was denatured at 95°C for 5 min and cooled in an ice bath for 5 min. To the microarray chamber was added 200 µl of Hybridization Buffer (containing the 5′ end-biotinylated oligonucleotide complementary to the positive control probe sequence). Two µl of denatured multiplex H6N1 and H5N2 virus RT-LAMP product was incubated at 48°C with vibration for 1 hr. Four µl of denatured novel H5 HPAIV single RT-LAMP product was incubated at 57°C with vibration for 1 hr. The sample was then washed three times with Washing Buffer at room temperature for multiplex RT-LAMP product or at 58°C for uniplex RT-LAMP product. The blocking reaction was then performed by mixing 0.2 µl of Streptavidin conjugate alkaline phosphatase and 200 µl of Blocking Reagent at room temperature for 30 min, followed with washing three times with Washing Buffer. The colorimetric reaction was then implemented by adding 4 µl of NBT/BCIP and 196 µl of Detection Buffer in the chamber, developing in the dark at room temperature for 20 min, and washing twice with distilled water. The hybridization result was indicated as the developed pattern on the microarray, which was read directly with the naked eye.
Detection limit tests on electrophoresis agarose gel and microarray: The RT-LAMP product detection limits on electrophoresis agarose gels and microarrays were tested and compared. AIV RNA standards of known copy numbers were prepared as follows. Reverse transcription was performed to produce AIV cDNA using Transcriptor High Fidelity cDNA Synthesis Kit (Roche, Mannheim, Germany) with uni12 primer [9]. PCR reactions were carried out using F3 and B3 primers at 94°C for 3 min, followed by 40 cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C for 40 sec, and then final elongation at 72°C for 7 min. The PCR products were purified using PCR cleanup kit (GeneMark, Taichung, Taiwan) and cloned into pGEM-T Easy Vector (Promega, Madison, WI, U.S.A.). The recombinant plasmid was linearized with SacI (New England Biolabs, Ipswich, MA, U.S.A.), and the 3′overhang was conversed with the DNA polymerase Klenow (Promega). In vitro transcription was performed using Riboprobe in vitro Transcription Systems (Promega) with T7 RNA Polymerase according to the manufacturer’s recommendations. DNase (Promega) was added to remove the remaining template DNA. The produced RNA was purified using RNeasy MiniElute Cleanup Kit (Qiagen) and verified by agarose gel. The RNA was quantified using spectrophotometer (WPA UV1101, Biochrom, Cambridge, U.K.), and the copy number was calculated. The RNA was 10-fold serially diluted (106 to 10° copies) in DEPC treated water and used as templates for the RT-LAMP reaction. The RT-LAMP detection limits visualized on electrophoresis agarose gels and microarrays were compared.
Virus combination test: Since H6N1 and H5N2 viruses were proven to be cocirculated in chickens in Taiwan [16], different copy numbers of H6N1 and H5N2 viral RNAs were combined to test the microarray detection efficiency in case of co-infection. A/chicken/Taiwan/2838V/00 (H6c) and A/chicken/Taiwan/1209/03 (H5l) were used as examples of H6N1 and H5N2 viruses, respectively. H6N1 RNA with 10° to106 copies was sequentially mixed with 106 to10° copies of H5N2 RNA as the co-temperate for the test.
One step RT-PCR: One-step reverse transcription-PCR (RT-PCR) was performed using One Step RT-PCR Kit (QIAGEN) to compare the detection effectiveness with the RT-LAMP-microarray assay for field samples. Outer primer set (F3 and B3) of H6N1 or H5N2 was used. For each reaction, 5 µl of RNA was mixed with a reaction mixture containing 5 µl of 5 × QIAGEN One-Step RT-PCR Buffer, 1.0 µl of dNTPs (10 mmol/l), 1.5 µl of forward and reverse primers (10 µmol/l), 1 µl of QIAGEN One-Step RT-PCR Enzyme Mix and distilled water in a final volume of 25 µl. One-step RT-PCR was performed using a thermo cycler (Biometra, Goettingen, Germany) with the following program: reverse transcription at 50°C for 30 min, initial denaturation at 95°C for15 min, amplification for 40 cycles at 94°C for 30 sec, 49°C for 30 sec, 72°C for 30 sec and followed by final extension at 72°C for 10 min. PCR product was visualized using electrophoresis in 1.5% agarose gel (Amresco, Solon, OH, U.S.A.) stained by 0.2 mg/ml ethidium bromide.
RESULTS
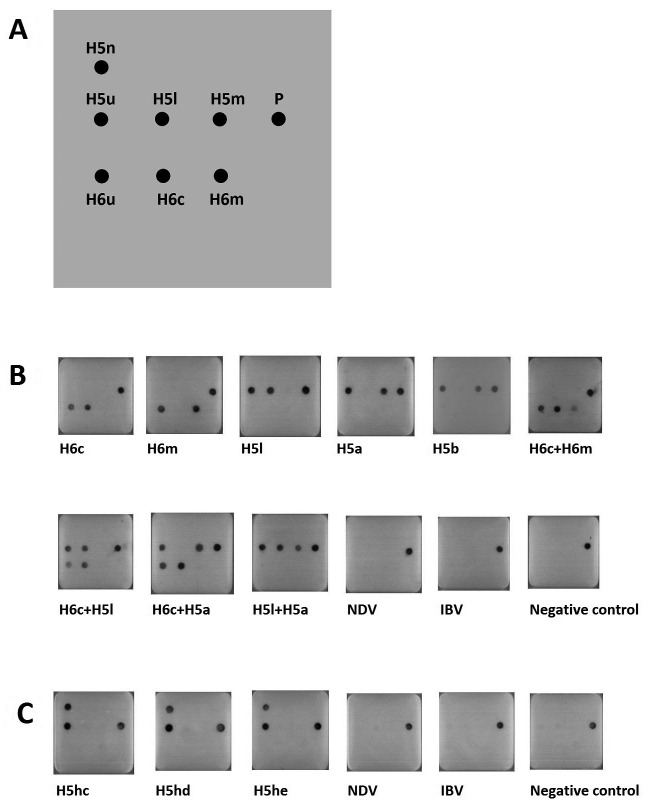
RT-LAMP product detection on microarray: The multiplex RT-LAMP products were differentiated and visualized on the microarrays. All influenza A viruses were clearly subtyped and pathotyped, including chicken H6 virus, human H6 virus, H5 LPAIV and H5 HPAIVs. Since H6 and H5 AIVs are co-circulating in Taiwan, various combinations of individual viruses were also tested on the microarrays. All showed clear signals without ambiguity (Fig. 1 B). Regarding the novel H5 HPAIVs, their HA genes were also identified using single RT-LAMP and microarray. The universal H5u probe for all chicken H5 viruses and the H5n probe for PLRERRRKR*GLF amino acid pattern at CS were both signalized on microarrays for the novel H5 HPAIVs (Fig. 1 C). No signals were displayed for other prevailing chicken respiratory viruses in Taiwan, including NDV and IBV (Fig. 1B and 1C). These results indicated that the developed microarray system had good detection and differentiation capacities for the complicated virus infection status in Taiwan.
Fig. 1.
Subtyping and pathotyping of AIVs using oligonucleotide microarrays. (A) Microarray map. The meaning of each probe and its detecting virus are shown in Table 3. P: positive control. (B) The microarray detection results for multiplex RT-LAMP of H6 and H5 AIVs. (C) The microarray detection results for unique RT-LAMP of novel H5 AIVs. The designation of each virus is shown in Table 1.
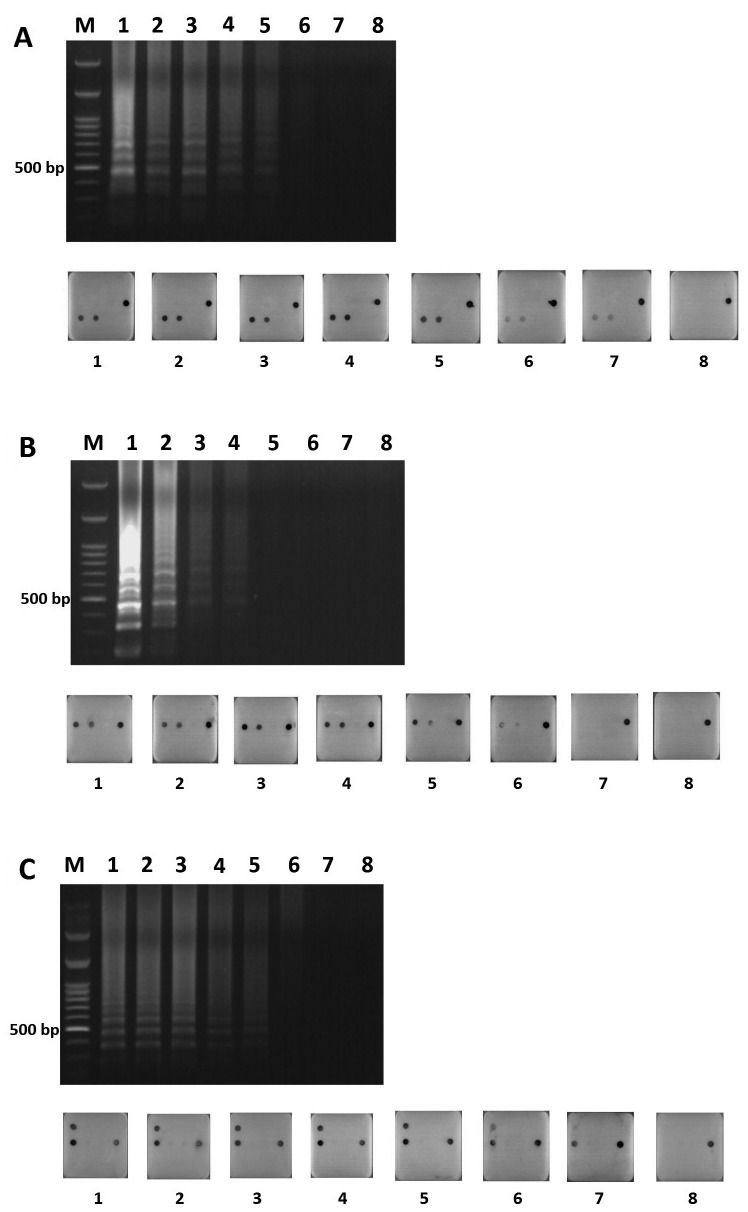
Detection limit comparison test: The detection limit comparison test between the agarose gel and microarray was performed for H6N1virus, H5N2 virus and novel H5 HPAIV using A/chicken/Taiwan/2838V/00 (H6c), A/chicken/Taiwan/1209/03 (H5l) and A/chicken/Taiwan/a288/15 (H5hc) as examples, respectively (Fig. 2). The viral RNA detection limit for H6N1 virus on agarose gel and microarray was 102 and 10° copies, respectively (Fig. 2A). The detection limit for H5N2 virus on agarose gel and microarray was 103 and 101 copies, respectively (Fig. 2B). The detection limit for H5 HPAIV on agarose gel and microarray was 101 and 10° copies, respectively (Fig. 2C). The results indicated that the RT-LAMP-agarose gel assay sensitivity was about 10–100 times lower than that for the RT-LAMP-microarray assay, which could even reach to one copy of viral RNA in this study.
Fig. 2.
Detection limit tests on the agarose gel and the corresponding microarray. The viral RNA was serial diluted (indicated as copy numbers) for template of RT-LAMP. H6N1AIV (A), H5N2 AIV (B), novel H5 HPAIV (C) using A/chicken/Taiwan/2838V/00 (H6c), A/chicken/Taiwan/1209/03 (H5l) and A/chicken/Taiwan/a288/15 (H5hc) as examples, respectively. M: 100 bp ladder marker; 1: 106 copies; 2: 105 copies; 3: 104 copies; 4: 103 copies; 5: 102 copies; 6: 101 copies; 7: 10° copies; 8: negative control.
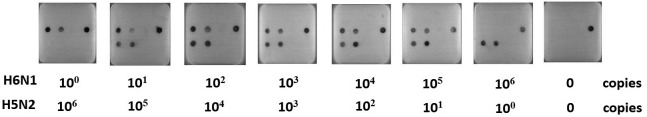
Detection limit test for co-infection cases: The virus combination test was performed using multiplex RT-LAMP and microarrays in case of chickens that were co-infected with H6N1 and H5N2 viruses, as H6N1 and H5N2 viruses were proven to be cocirculated in chickens in Taiwan [16]. The detection limit test using mixed viruses as templates showed that 101 to 105 copy mixtures of H6N1 and H5N2 viruses could ideally be identified simultaneously. However, one copy of H6N1 or H5N2 virus could not be detected when co-infection occurred (Fig 3).
Fig. 3.
H6N1 RNA with 10° to106 copies was sequentially mixed with 106 to10° copies of H5N2 RNA as the co-temperate for the detection limit test on microarrays. A/chicken/Taiwan/2838V/00 (H6c) and A/chicken/Taiwan/1209/03 (H5l) were used as examples of H6N1 and H5N2 viruses, respectively.
Field samples: Seventy-two field samples, including oral swabs, cloacal swabs, brain, heart, trachea, lung and kidney from chickens that showed respiratory signs, were collected in Taiwan for AIV detection. Forty-two samples showed RT-LAMP-microarray positive, including 14 H6N1 AIVs, 11 H5N2 LPAIVs, 1 H5N2 HPAIV and 16 novel H5 HPAIVs. Thirty-four samples showed one step RT-PCR positive, including 7 H6N1 AIVs, 12 H5N2 AIVs and 15 novel H5 HPAIVs (Table 4). The results indicated that RT-LAMP-microarray could subtype and pathotype AIVs simultaneously and was more sensitive than the traditional one step RT-PCR method. No H6N1 and H5N2 virus co-infections were found.
Table 4. AIV detection using RT-LAMP-microarray and one step RT-PCR for field samplesa).
| RT-LAMP-microarray assay | |||
|---|---|---|---|
| H6 N1+ | H5N2 + | Novel H5 + | |
| One step RT-PCR | |||
| H6N1 + | 7 | 0 | 0 |
| H6N1 − | 7 | 0 | 0 |
| H5N2 + | 0 | 12 b) | 0 |
| H5N2 − | 0 | 0 | 0 |
| Novel H5 + | 0 | 0 | 15 |
| Novel H5 − | 0 | 0 | 1 |
a) Forty-two samples showed AIV RT-LAMP-microarray positive among the total 72 field samples. b) Eleven showed H5 LAPIV (PQREKR*GLF), and one showed H5 HPAIV (PQRKKR*GLF/PQRRKR*GLF) within these 12 samples.
DISCUSSION
Detection limits of the present RT-LAMP microarray system were one RNA copy for H6N1 virus and novel H5 HPAIV and 10 copies for H5N2 virus. Such extremely low limits have not been achieved by using other molecular methods, such as100 copies of real time RT-PCR (rRT-PCR) [19, 24] and 10–100 copies of RT-LAMP [3, 5, 8]. The microarray hybridization effectiveness was approximately equal to sequencing, because of the specific matching reaction between the nucleotide bases. AIV subtyping and pathotyping could be achieved simultaneously using the multiplex RT-LAMP-microarray. The greater sensitivity and specificity of the RT-LAMP-microarray made it an excellent approach to AIV surveillance and studies. No expensive equipment was needed in this study, including PCR machines. A microarray imaging system was not necessary, because the results could be read with the naked eye. Low cost feature of the present RT-LAMP-microarray would be particularly attractive to regional diagnostic laboratories for large-scale AIV screening during outbreaks.
Novel H5 HPAIV was detected in this study using a uniplex RT-LAMP-microarray. Multiplex-LAMP was hardly performed for it to incorporate 12 primers at the same time, including 4 primers of H6N1 virus, 4 primers of H5N2 virus and 4 primers of novel H5 HPAIV. High temperature for microarray hybridization was also unique for novel H5 HPAIV. Lower temperature for hybridization made cross reaction to other unrelated H5 probes take place. During a suspected HPAIV outbreak, however, this uniplex RT-LAMP-microarray protocol could be conducted first to rule out the possibility of novel H5 virus infection.
Although H6N1 and H5N2 viruses co-circulate and reassortant viruses have become established and enzootic in chickens throughout Taiwan [16], H6N1 and H5N2 co-infection was not found in the field samples in this study. However, the development of multiplex RT-LAMP was important to make simultaneous H6N1 and H5N2 detection convenient and feasible. The detection limit was 10 copies for either H6N1 or H5N2 virus in virus combination test, indicating that the addition of extra viral template did not cause obvious impact on the detection sensitivity.
Since AIVs are constantly evolving into novel variants, it would be advantageous if the developed approach could be successful for the detection of future emerging AIVs. The microarray possesses such capacity because it can efficiently expend its efficacy by recruiting new probes, which would make this device more practical for meeting future needs.
Acknowledgments
Funding for this research was provided under the plan of Ministry of Science and Technology (grant no.: NSC 103–2622-B-002–012 -CC2), Taiwan.
References
- 1.Abolnik C.2008. A rapid and sensitive real-time reverse transcription PCR for the pathotyping of South African H5N2 avian influenza viruses. Onderstepoort J. Vet. Res. 75: 347–351. doi: 10.4102/ojvr.v75i4.110 [DOI] [PubMed] [Google Scholar]
- 2.Alexander D. J.2000. A review of avian influenza in different bird species. Vet. Microbiol. 74: 3–13. doi: 10.1016/S0378-1135(00)00160-7 [DOI] [PubMed] [Google Scholar]
- 3.Blomström A. L., Hakhverdyan M., Reid S. M., Dukes J. P., King D. P., Belák S., Berg M.2008. A one-step reverse transcriptase loop-mediated isothermal amplification assay for simple and rapid detection of swine vesicular disease virus. J. Virol. Methods 147: 188–193. doi: 10.1016/j.jviromet.2007.08.023 [DOI] [PubMed] [Google Scholar]
- 4.Bureau of Animal and Plant Health Inspection and Quarantine (BAPHIQ) 8 August 2015. Available at: http://ai.gov.tw/files/web_structure/324/file_1.pdf.
- 5.Chen H. T., Zhang J., Sun D. H., Ma L. N., Liu X. T., Cai X. P., Liu Y. S.2008. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 151: 200–203. doi: 10.1016/j.jviromet.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 6.Cheng M. C., Soda K., Lee M. S., Lee S. H., Sakoda Y., Kida H., Wang C. H.2010. Isolation and characterization of potentially pathogenic H5N2 influenza virus from a chicken in Taiwan in 2008. Avian Dis. 54: 885–893. doi: 10.1637/9208-120609-Reg.1 [DOI] [PubMed] [Google Scholar]
- 7.Gall A., Hoffmann B., Harder T., Grund C., Ehricht R., Beer M.2009. Rapid haemagglutinin subtyping and pathotyping of avian influenza viruses by a DNA microarray. J. Virol. Methods 160: 200–205. doi: 10.1016/j.jviromet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 8.Higashimoto Y., Ihira M., Ohta A., Inoue S., Usui C., Asano Y., Yoshikawa T.2008. Discriminating between varicella-zoster virus vaccine and wild-type strains by loop-mediated isothermal amplification. J. Clin. Microbiol. 46: 2665–2670. doi: 10.1128/JCM.00216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann B., Harder T., Starick E., Depner K., Werner O., Beer M.2007. Rapid and highly sensitive pathotyping of avian influenza A H5N1 virus by using real-time reverse transcription-PCR. J. Clin. Microbiol. 45: 600–603. doi: 10.1128/JCM.01681-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horimoto T., Kawaoka Y.1997. Biologic effects of introducing additional basic amino acid residues into the hemagglutinin cleavage site of a virulent avian influenza virus. Virus Res. 50: 35–40. doi: 10.1016/S0168-1702(97)00050-6 [DOI] [PubMed] [Google Scholar]
- 11.Horimoto T., Kawaoka Y.1994. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 68: 3120–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horimoto T., Rivera E., Pearson J., Senne D., Krauss S., Kawaoka Y., Webster R. G.1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213: 223–230. doi: 10.1006/viro.1995.1562 [DOI] [PubMed] [Google Scholar]
- 13.Jayawardena S., Cheung C. Y., Barr I., Chan K. H., Chen H., Guan Y., Peiris J. S., Poon L. L.2007. Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerg. Infect. Dis. 13: 899–901. doi: 10.3201/eid1306.061572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaoka Y., Webster R. G.1985. Evolution of the A/Chicken/Pennsylvania/83 (H5N2) influenza virus. Virology 146: 130–137. doi: 10.1016/0042-6822(85)90059-5 [DOI] [PubMed] [Google Scholar]
- 15.Lamb R. A., Krug R. M.1996. Orthomyxoviridae: the viruses and their replication. pp. 605–648. In: Fundamental Virology. (Fields, B. N., Knipe, D. M., Howley, P. M., Chanock, R. M., Melnick, J. L., Momath, T. P. and Roizman, B. eds.), Lippincott-Raven Publisher, Philadelphia. [Google Scholar]
- 16.Lee C. C., Zhu H., Huang P. Y., Peng L., Chang Y. C., Yip C. H., Li Y. T., Cheung C. L., Compans R., Yang C., Smith D. K., Lam T. T., King C. C., Guan Y.2014. Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan. J. Virol. 88: 5677–5686. doi: 10.1128/JVI.00139-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leijon M., Ullman K., Thyselius S., Zohari S., Pedersen J. C., Hanna A., Mahmood S., Banks J., Slomka M. J., Belák S.2011. Rapid PCR-based molecular pathotyping of H5 and H7 avian influenza viruses. J. Clin. Microbiol. 49: 3860–3873. doi: 10.1128/JCM.01179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y. S., Sugimura T., Shieh H. K., Lee Y. L., Jong M. H.1985. Isolation and identification of an influenza A virus in ducks in Taiwan. Jour. Chinese Soc. Vet. Sci. 11: 23–34. [Google Scholar]
- 19.Payungporn S., Chutinimitkul S., Chaisingh A., Damrongwantanapokin S., Nuansrichay B., Pinyochon W., Amonsin A., Donis R. O., Theamboonlers A., Poovorawan Y.2006. Discrimination between highly pathogenic and low pathogenic H5 avian influenza A viruses. Emerg. Infect. Dis. 12: 700–701. doi: 10.3201/eid1204.051427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perdue M., Crawford J., Garcia M., Latimer J., Swayne D.2003. Occurrence and Possible Mechanisms of Cleavage-Site Insertions in the Avian Influenza Hemagglutinin Gene. Avian Dis. 47: 182–193. [Google Scholar]
- 21.Senne D. A., Panigrahy B., Kawaoka Y., Pearson J. E., Süss J., Lipkind M., Kida H., Webster R. G.1996. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40: 425–437. doi: 10.2307/1592241 [DOI] [PubMed] [Google Scholar]
- 22.Wang C. W., Wang C. H.2003. Experimental selection of virus derivatives with variations in virulence from a single low-pathogenicity H6N1 avian influenza virus field isolate. Avian Dis. 47: 1416–1422. doi: 10.1637/6093 [DOI] [PubMed] [Google Scholar]
- 23.Wang L. C., Huang D., Cheng M. C., Lee S. H., Wang C. H.2015. H5 avian influenza virus pathotyping using oligonucleotide microarray. J. Virol. Methods 220: 39–42. doi: 10.1016/j.jviromet.2015.04.006 [DOI] [PubMed] [Google Scholar]
- 24.Yacoub A., Kiss I., Zohari S., Hakhverdyan M., Czifra G., Mohamed N., Gyarmati P., Blomberg J., Belák S.2009. The rapid molecular subtyping and pathotyping of avian influenza viruses. J. Virol. Methods 156: 157–161. doi: 10.1016/j.jviromet.2008.10.019 [DOI] [PubMed] [Google Scholar]