Abstract
Outer membrane protein X (OmpX) and its homologues have been proposed to contribute to the virulence in various bacterial species. But, their role in virulence of extraintestinal pathogenic Escherichia coli (ExPEC) is yet to be determined. This study evaluates the role of OmpX in ExPEC virulence in vitro and in vivo using a clinical strain PPECC42 of porcine origin. The ompX deletion mutant exhibited increased swimming motility and decreased adhesion to, and invasion of pulmonary epithelial A549 cell, compared to the wild-type strain. A mild increase in LD50 and distinct decrease in bacterial load in such organs as heart, liver, spleen, lung and kidney were observed in mice infected with the ompX mutant. Complementation of the complete ompX gene in trans restored the virulence of mutant strain to the level of wild-type strain. Our results reveal that OmpX contributes to ExPEC virulence, but may be not an indispensable virulence determinant.
Keywords: extraintestinal pathogenic Escherichia coli, outer membrane protein X, swimming motility, virulence
Extraintestinal pathogenic Escherichia coli (ExPEC) can cause a variety of extraintestinal infections and diseases in humans and animals, typically characterized by multi-organ infections including urinary tract infections, meningitis, polyserositis and septicemia. ExPEC strains represent a large economic burden due to both medical costs and lost productivity and attract more attention than ever before [9, 18, 28, 34]. Various virulence factors have been recognized in ExPEC, including pathogenicity-associated islands (PAIs), adhesins (e.g., P fimbriae, Afa/Dr adhesins and type 1 fimbriae), invasins (e.g., IbeA), toxins (e.g., hemolysin, cytotoxic necrotizing factor, etc.), surface antigens (e.g., capsule and lipopolysaccharide), iron acquisition system (e.g., aerobactin system) and secretion systems (e.g., type III secretion systems) [2, 9, 12, 16, 21, 32]. Recently, ExPEC was frequently discovered in clinical samples of the pig industry [10]. Moreover, it was also widely found in retail chicken, beef, pork and ready-to-eat foods, which means a potential threat on public health [1, 6, 23, 24, 26, 28, 33].
Outer membrane protein X (OmpX), as well as its biological and molecular characteristics, was initially described for Enterobacter cloacae [34,35,36], but its homologues had been identified in other Gram-negative bacteria like Salmonella enterica serovar Typhimurium (PagC, Rck) [13], Yersinia pestis (OmpX/Ail) [19, 20], Y. enterocolitica and Y.pseudotuberculosis (Ail) [4], E. coli (OmpX, Lom) [3, 25, 30] and Klebsiella pneumoniae (OmpK17) [7]. These proteins display small size (from 15 to 18 kDa) and fold in eight-stranded antiparallel β-barrel structure with membrane-spanning domains, protruding from the cell surface [5, 38], and thus influence the binding to external proteins [37] and participate in channeling, antibiotic resistance and signal transduction, as well as cell adhesion and invasion, resistance to complement-mediated killing, survival within macrophages and virulence [4, 13, 14, 17, 19, 20, 27, 31]. OmpX and its homologues have been proposed to be potential bacterial vaccine candidates [11]. However, conflicting studies that deletion of ompX did not reduce the adhesion by different E. coli strains [25] or invasiveness by E. cloacae [8] had also been reported. ExPEC strains are significantly different from other E. coli strains causing intestinal infections in virulence and in phylogenic background [22]. Thus, the present study was aimed at characterizing the role of OmpX in regulating the virulence in vitro and in vivo of ExPEC strain PPECC42, isolated by our research group from China and probably related to pig’s lung disease [15].
MATERIALS AND METHODS
Bacterial strains, plasmids, media and growth conditions: The bacterial strains and plasmids used in this study are listed in Table 1. The wild-type (WT) ExPEC strain PPECC42 (O11 serogroup) was isolated from the lung of a diseased pig, and its complete genome sequence had been submitted to NCBI (Genbank Accession No. LRGE00000000). Plasmid pRE112 was used as a suicide vector for homologous recombination to construct the mutant. E. coli χ7213 was a host for pRE112 to conjugally transfer [39]. E. coli DH5α and plasmid pHSG396 were purchased from Takara Bio (Otsu, Japan). All strains were routinely cultivated either in lysogeny broth (LB) medium or on tryptic soy agar (TSA) plate (Difco Laboratories, Sparks, MD, U.S.A.).
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| ExPEC strain PPECC42 | Wild-type (WT), porcine origin, serotype O11, CmS | [15] |
| PPECC42ΔompX | Mutant deleted a 345 bp fragment from whole ORF of ompX gene in PPECC42, CmS | This study |
| ompX+/ΔompX | PPECC42ΔompX mutant complemented with a copy of the ompX gene via pHSG396, CmR | This study |
| E. coli χ7213 | Thi-1 thr-1 leuB6 fhuA21 lacY1 glnV44 ΔasdA4 recA1 RP4 2-Tc::Mu[λpir] KmR | [39] |
| χ7213ΔompX | E. coli strain χ7213 containing suicide vector pRE112ΔompX | This study |
| E. coli DH5α | Used for recombinant DNA method | Takara |
| Plasmids | ||
| pRE112 | oriT oriV Δasd CmR SacB, suicide vector | [39] |
| pREΔompX | pRE112 vector inserted disrupted ompX gene in Kpn I and Sac I sites | This study |
| pHSG396 | ori lacZ CmR | Takara |
| pHSG-ompX | pHSG396 vector containing the whole ompX gene, CmR | This study |
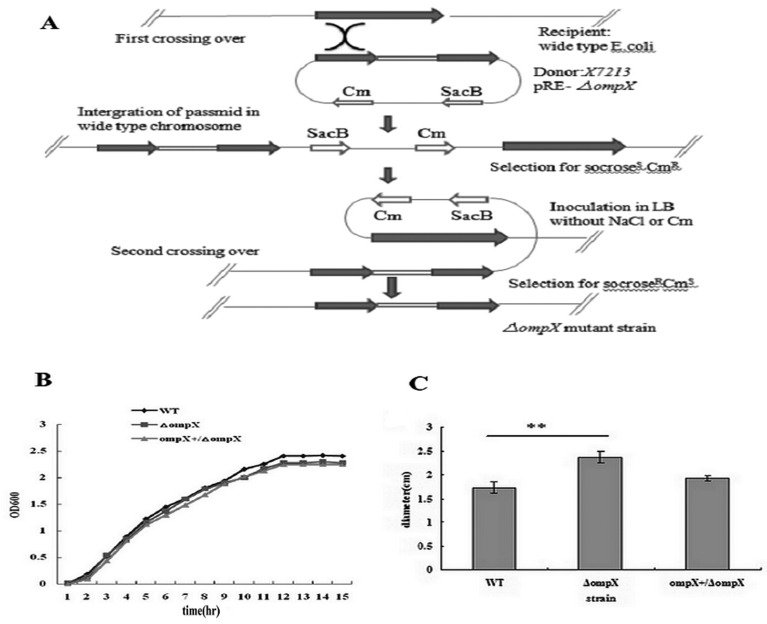
Construction of the ompX mutant and complemented strains: Deletion of ompX was performed by allelic exchange method with a constitutive chloramphenicol resistance (CmR) expression cassette, as described by Hou et al. [15]. Briefly, the upstream and downstream regions of ompX gene were PCR amplified from the WT strain’s genomic DNA using primer sets P1/P2 and P3/P4, respectively, and mixed to perform the overlapping PCR. Then, the primer set P1/P4 was added in order to amplify the disrupted ompX gene. The plasmid pREΔompX with the disrupted ompX gene was introduced into E. coli strain χ7213, followed by co-culturing with the WT strain. Transformants resistant to sucrose and sensitive to chloramphenicol were selected and used to perform colony PCR to confirm the deletion of ompX gene using primer set P5/P6. The mutant strain finally selected was named PPECC42ΔompX (Fig.1A).
Fig.1.
Construction of the ompX mutant (A), and growth curves (B) and swimming motility (C) of ExPEC strains. (A) Protocol of construction of the ompX mutant. (B) The WT strain (♦), ompX mutant (■) and complemented strain (▲) were grown in LB medium at 37°C, respectively, and their optical densities at 600 nm were measured each hour. The graph is representative of three independent experiments. (C) The overnight culture of each strain was stabbed on 0.3% LB agar plates. After incubation for 6 hr, the diameters of the swimming rings were measured. All values of the bar graphs are presented as the mean ± SD. Asterisks indicate significant differences between the values of the mutant and the WT strains (**P<0.01).
To construct the ompX complemented strain, the complete ompX gene was PCR amplified from the WT strain’s genomic DNA using primer set P7/P8 and inserted into the pHSG396 vector. The plasmid pHSG-ompX was transformed into the ompX mutant by electroporation. Clones that reacquired the complete ompX gene were checked by PCR using primer set P5/P6. The complemented strain was named ompX+/ΔompX. A complete list of the primers used in this study is shown in Table 2.
Table 2. Primers used in this study.
| Primer | Sequence (5΄-3΄) | Application |
|---|---|---|
| P1 | CGGGGTACCAGACATCCAGCGATGCTAG KpnI | Mutant |
| P2 | ACTTATGCCCGTCTCGGCATCGTCGCAACGGGTATT | Mutant |
| P3 | AATACCCGTTGCGACGAT GCCGAGACGGGCATAAGT | Mutant |
| P4 | TCCGAGCTCGGCGGTGACAATGGGTTA Sal I | Mutant |
| P5 | GCCGTACTGCAAGCTCTG | Checking PCR |
| P6 | AGTCGCTGGTGTCGTGT | Checking PCR |
| P7 | GGCGTCGACATGAAAAAAATTGCAT Sal I | Complementation |
| P8 | CGCGAATTCTTAGAAGCGGTAACCAACA Ecol I | Complementation |
Growth analysis: Growth curve profile was constructed in order to determine the significance of OmpX on ExPEC growth. An equal number of cells from the WT, ompX mutant and complemented strains were inoculated in LB medium and incubated at 37°C with shaking (200 rpm). Samples were obtained hourly, and the optical densities were measured at 600 nm (OD600 nm) using a BioPhotometer (Eppendorf AG, Hamburg, Germany). The data were acquired from three independent experiments, each having three replicates.
Swimming motility assays: Semi-solid agar plates (LB medium with 0.3% agar) were used to characterize the bacterial swimming motility phenotype [29]. The plates were spotted with 2 µl of an overnight bacterial culture and then incubated at 37°C for 6 hr. Subsequently, bacterial motility was qualitatively assessed by measuring the diameter of swimming ring.
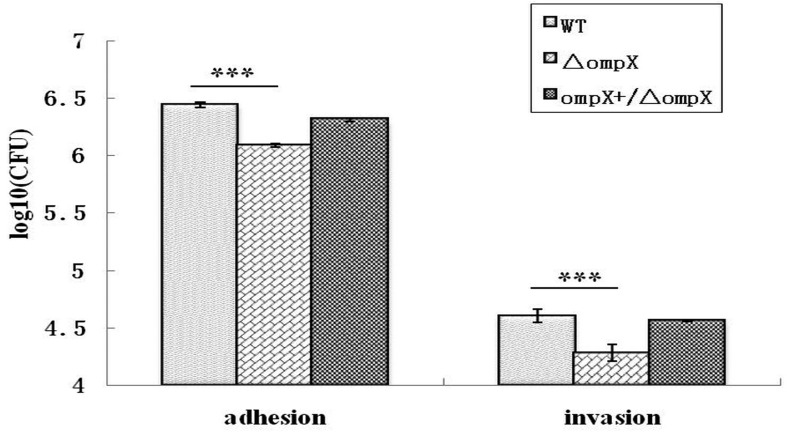
Adhesion and invasion assays: The bacterial adhesion and invasion assays were preformed in human pulmonary epithelial A549 cells. Briefly, A549 cell monolayers were seeded in 24-well tissue culture plates (Nunc, Roskilde, Denmark) with about 105 cells/well and incubated for 48 hr, and then infected with bacterial cells at a multiplicity of infection (MOI) of 10 per cell in 1 ml of internalization medium (IM) (low-glucose modified RMPI-1640 medium supplemented with heat-inactivated fetal calf serum (Gibco, Grand Island, NY, U.S.A.)). The infected plates were centrifuged for 10 min at 1,000 × g and further incubated at 37°C for 2 hr. After washed six times with phosphate-buffered saline (PBS), the infected cells were lysed with 0.1% Triton X-100 (Sigma-Aldrich, St.Louis, MO, U.S.A.). Samples were diluted and spread on TSA plates to determine the number of colony-forming units (CFU) corresponding to the total number of cell-associated bacteria (adherent and intracellular bacteria). In order to measure invasion, a set of wells were washed with PBS after initial 2 hr incubation and further incubated for 2 hr at 37°C with 20 µg/ml gentamicin (Sigma-Aldrich) to eliminate extracellular bacteria. Monolayers were washed with PBS, lysed with 0.1% Triton X-100 and plated to enumerate the invading bacteria. The bacteria were quantified as described above. Each assay was repeated a minimum of three times, with each repeat including four technical replicates per bacterial strain.
Murine infection model: All animal experiments were carried out according to the International Guiding Principles for Biomedical Research Involving Animals (1985). Five-week-old male BALB/c mice, provided by Laboratory Animal Center of Zhongnan Hospital of Wuhan University, were used to determine the 50% lethal dose (LD50) values and systemic bacterial counts. The experimental strains were grown overnight at 28°C, washed, resuspended in 25% glycerol and frozen at −80°C. On the day of the experiment, the cultures were thawed, aliquoted and diluted in PBS to the desired concentration. In the LD50 experiment, a total of 80 mice were randomly divided into 15 experimental groups and 1 control group (5 mice/group). The mice in experimental groups received intraperitoneal injection with various doses of each strain (104 to 108 CFU), respectively. The control mice were injected with sterile PBS. All mice were observed for mortality for a 14-day period.
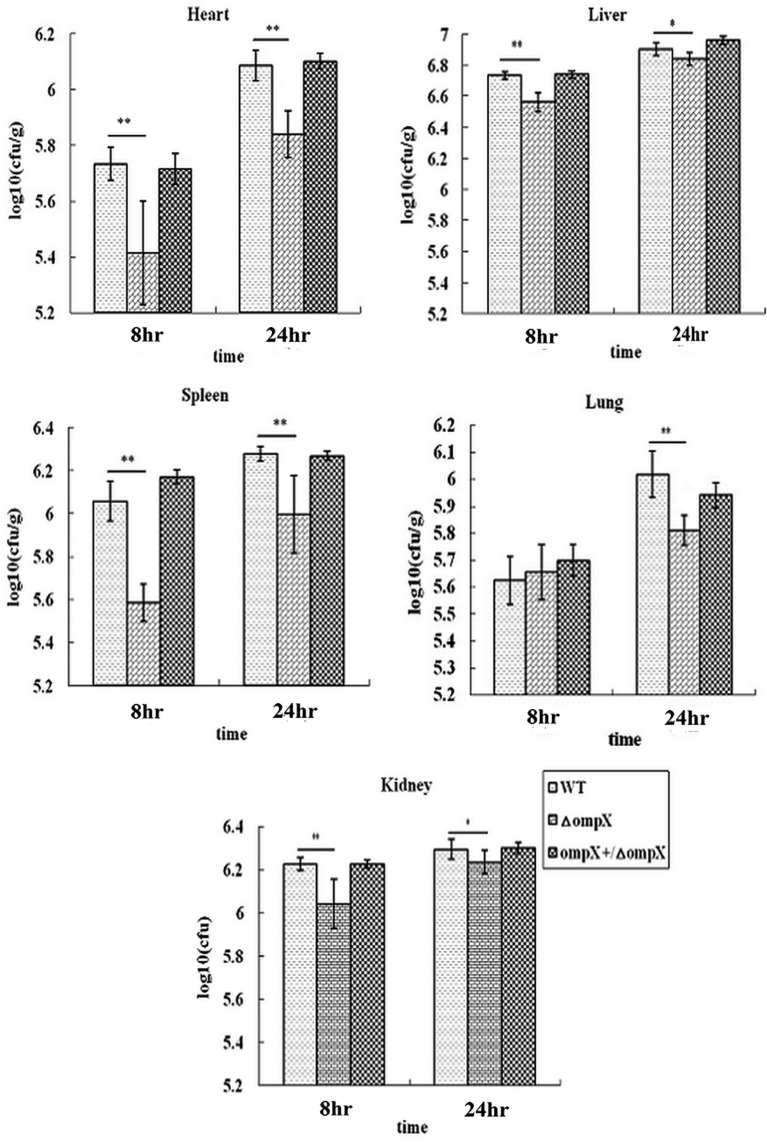
To assay the systemic bacterial counts, a total of 36 mice were randomly divided into 3 experimental groups and 1 control group (12 mice/group). The experimental mice were inoculated by intramuscular injection with 2 × 106 CFU of each strain, respectively. The control mice were injected with sterile PBS. Six mice from each group were euthanized at 8 hr and 24 hr post-inoculation. The heart, liver, spleen, lung and kidney were aseptically collected, weighed and homogenized in sterile PBS. Bacterial loads were determined by plating serial dilutions of the homogenates on TSA plates.
Statistical analysis: All data were expressed as the mean ± standard deviation (SD). Bacteria groups in all tests (growth rate, motility, adhesion, invasion and bacterial loads in different organs) were compared by using the Student’s t-test for independent samples. Differences were considered significant at a P value of <0.05. Statistical analyses were performed with SPSS17.0 software (SPSS inc., Chicago, IL, U.S.A.).
RESULTS
OmpX deletion did not affect ExPEC growth: Growth curves were constructed for the WT, ompX mutant and complemented strains. The growth rate of the ompX mutant during a period of 15 hr was similar to that of the WT and complemented strains (Fig. 1B), indicating that inactivation of OmpX had no obvious effect on ExPEC growth.
Deletion of ompX attenuated the virulence in vitro of ExPEC: A swimming motility assay was performed to determine if OmpX played an important role in ExPEC motility. A significant increase (P<0.01) in the diameter of bacterial colony was observed in the ompX mutant (2.37 ± 0.12 cm) when compared to the WT (1.73 ± 0.12 cm) and complemented (1.93 ± 0.06 cm) strains (Fig. 1C), indicating that deletion of ompX increased swimming motility in ExPEC.
Furthermore, the role of OmpX in ExPEC virulence was evaluated using in vitro cell adhesion and invasion assays. The ompX mutant exhibited a significant stronger reduction in adhesion to and invasion of A549 cells compared to the WT strain (P<0.05) (Fig. 2). The adhesion and invasion levels of the ompX mutant in A549 cells were decreased to 45% and 48% of those of the WT strain, respectively. Complementation of ompX gene in trans restored the adhesion and invasion abilities to a certain extent. These results showed that OmpX played an important role in cell adhesion and invasion by ExPEC.
Fig.2.
Effects of ompX deletion on ExPEC adhesion to (A) and invasion of (B) human pulmonary epithelial A549 cells. Adhesion and invasion levels of ExPEC strains were measured. Cell-associated bacteria (adherent + intracellular) were quantified after a 2-hr infection period. Invasion was determined after gentamicin treatment for an additional 2 hr. All values of the bar graphs are presented as the mean ± SD. Asterisks indicate significant differences between the values of the mutants and the WT strain (*P<0.05; ***P<0.001;****P<0.0001).
Inactivation of OmpX attenuated ExPEC virulence in mice: A murine infection model was used to determine the role of OmpX in ExPEC virulence in vivo. As shown in Table 3, the ompX mutant displayed a LD50 of 9.4 × 106 CFU, 6.27- and 3.86-fold increase when compared to the WT (1.5 × 106 CFU) and complemented strain (2.43 × 106 CFU), respectively, indicating that deletion of OmpX in ExPEC exhibited a slightly attenuated lethality to mice.
Table 3. Determination of LD50 of the experimental strains in BALB/c mice.
| dose (CFU) | WT | ΔompX | ompX+/ΔompX |
|---|---|---|---|
| 3 × 108 | 5 / 5 | 5 / 5 | 5 / 5 |
| 3 × 107 | 5 / 5 | 3 / 5 | 5 / 5 |
| 3 × 106 | 3 / 5 | 2 / 5 | 3 / 5 |
| 3 × 105 | 1 / 5 | 0 / 5 | 0 / 5 |
| 3 × 104 | 0 / 5 | 0 / 5 | 0 / 5 |
| LD50 | 1.5 × 106 | 9.4 × 106 | 2.43 × 106 |
Animals were inoculated by intraperitoneal injection and observed for a period of 14 days. The ratio indicated the number of dead mice per number of mice infected.
Furthermore, we enumerated the bacterial load of the WT strain and its mutants in different internal organs of mice at 8 hr and 24 hr post-inoculations, as shown in Fig. 3. At 8 hr after infection, compared to the WT strain, the ompX mutant showed significant reduction in bacterial numbers in the heart, spleen, liver and kidney (P<0.01), but not in the lung (P>0.05). While at 24 hr after infection, there was a significant reduction in bacterial load of ompX mutant in all the tested internal organs, P<0.01 in the heart, spleen and lung, while P<0.05 in the liver and kidney. Complementation of the ompX gene in trans restored the bacterial load in all the internal organs to the levels of WT strain. Thus, it could be proposed that OmpX contributed to the ExPEC capacity of colonization and persistence in the internal organs.
Fig.3.
Determination of bacterial counts in heart, liver, spleen, lung and kidney. The mice were infected with 2 × 106 CFU of the WT ExPEC strain PPECC42, ompX mutant and complemented strain. Aliquots (0.1 g) of tissues were homogenized, serially diluted and spread on TSA plates to determine bacterial counts. All values of the bar graphs are presented as the mean ± SD. Asterisks indicate significant differences between the values of the mutants and the WT strain (*P<0.05; **P<0.01).
DISCUSSION
As an important zoonotic and foodborne pathogen, ExPEC can cause complex diseases and infections in humans and animals. It is essential to understand the virulence factors for the control of ExPEC infection. More recent studies had been performed on the role of OmpX and its homologues in the pathogenesis and virulence in different bacterial species, and conflicting results had been reported by different research groups [8, 20, 25, 27]. In this study, we firstly evaluated the relationship between OmpX and ExPEC virulence using a clinical strain PPECC42 of pig origin and its mutant strains. The ompX mutant displayed the same growth rate as the WT and complemented strains, but was defective in cell adhesion and invasion, in accompany with increased swimming motility. Furthermore, systemic bacterial counts were reduced in the mice infected by the ompX mutant. However, it was slightly that deletion of ompX attenuated ExPEC virulence in mice as indicated by LD50 determination.
Firstly, the effect of OmpX on ExPEC virulence was evaluated using a cell infection model. Deletion of ompX significantly impaired the ability to adhere to, and invade, human pulmonary epithelial A549 cells. Complementation of ompX gene in trans restored the adhesion and invasion to a certain extent (Fig. 2). The results supported that OmpX plays a role in ExPEC adhesiveness and invasiveness, in disagreement with the previous study reported by Mecsas et al. (1995) that deletion of ompX in E. coli K-12 didn’t affect the ability to adhere to Hep-2 cells [25]. The reason might be due to the significant difference between ExPEC and intestinal pathogenic E. coli strains like K-12 in virulence and phylogenic background [22, 25]. Perhaps, the increase of bacterial swimming motility in this study (Fig. 1C) can result in the decrease of cell association. OmpX may play the role in cell adhesiveness via mediating bacterial autoaggregation and interacting with the components of extracellular matrix, such as fibronectin and laminin [19], and its role in cell invasiveness may attribute to the receptor in the cell surface or the regulation of other internalization-related components [17, 19], which should be explored in the further study.
Then, we assessed the contribution of OmpX to ExPEC virulence using a murine infection model. The result showed that loss of OmpX led to the great reduction in bacterial counts in different organs tested of mice at 8 hr and 24 hr post-inoculation, including heart, liver, spleen, lung and kidney (Fig. 3), suggesting the reduced ability to colonize different organs and induce systemic infection. However, the mice infected with the ompX mutant displayed a slightly higher LD50 than the mice injected with the WT and complemented strains, indicating that OmpX might contribute to ExPEC lethality to mice, but might be not an indispensable virulence determinant. Kolodziejek et al. (2010) confirmed that the deletion of ompX (ail) in Yersinia Pestis had little consequence in mice with no change in LD50, while completely attenuated virulence in rats, the reason of which might be due to the different dependency of serum resistance on OmpX (Ail) expression between mice and rats [20]. However, Pulkkinen and Miller (1991) reported that PagC and Rck from S. typhimurium were important for virulence in mice [31]. The role of OmpX and its homologues in bacterial virulence might be determined by the strains and hosts used in the study. The identical difference of OmpX and its homologues among different bacterial species could also influence the results, as discussed by Mecsas et al. [25]. Nucleotide sequence analysis indicates that the ompX gene in ExPEC strain PPECC42 is highly conservative (99% identical) among E. coli strains (K12, ST2747, PCN033, etc.), while it exhibits relative lower identical varying from 81% to 87% to the homologues in other bacterial species, such as E. cloacae, S. enteric serovar typhimurium and Y. pestis.
In summary, our study in vitro and in vivo confirmed that OmpX exhibited a weak effect on ExPEC lethality to mice, while it had a significant contribution to ExPEC adhesion to, and invasion, epithelial cells, as well as bacterial load in different organs. In order to clarify the mechanism of action, it would be interesting to further investigate the OmpX-associated regulation networks.
Acknowledgments
We thank Dr. Xiaoping LI at College of Animal Science, Huazhong Agricultural University for her help in statistics analysis to our data. This work was financially supported by the National Natural Science Foundation of China (NSFC) (Grant No. 31572539).
REFERENCES
- 1.Antão E. M., Wieler L. H., Ewers C.2009. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslam M., Toufeer M., Narvaez Bravo C., Lai V., Rempel H., Manges A., Diarra M. S.2014. Characterization of Extraintestinal Pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. Int. J. Food Microbiol. 177: 49–56. [DOI] [PubMed] [Google Scholar]
- 3.Barondess J. J., Beckwith J.1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346: 871–874. [DOI] [PubMed] [Google Scholar]
- 4.Bliska J. B., Falkow S.1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. U.S.A. 89: 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choutko A., Glättli A., Fernández C., Hilty C., Wüthrich K., van Gunsteren W. F.2011. Membrane protein dynamics in different environments: simulation study of the outer membrane protein X in a lipid bilayer and in a micelle. Eur. Biophys. J. 40: 39–58. [DOI] [PubMed] [Google Scholar]
- 6.Clermont O., Olier M., Hoede C., Diancourt L., Brisse S., Keroudean M., Glodt J., Picard B., Oswald E., Denamur E.2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11: 654–662. [DOI] [PubMed] [Google Scholar]
- 7.Climent N., Ferrer S., Rubires X., Merino S., Tomás J. M., Regué M.1997. Molecular characterization of a 17-kDa outer-membrane protein from Klebsiella pneumoniae. Res. Microbiol. 148: 133–143. [DOI] [PubMed] [Google Scholar]
- 8.de Kort G., Bolton A., Martin G., Stephen J., van de Klundert J. A.1994. Invasion of rabbit ileal tissue by Enterobacter cloacae varies with the concentration of OmpX in the outer membrane. Infect. Immun. 62: 4722–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dezfulian H., Batisson I., Fairbrother J. M., Lau P. C., Nassar A., Szatmari G., Harel J.2003. Presence and characterization of extraintestinal pathogenic Escherichia coli virulence genes in F165-positive E. coli strains isolated from diseased calves and pigs. J. Clin. Microbiol. 41: 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y., Tang X., Lu P., Wu B., Xu Z., Liu W., Zhang R., Bei W., Chen H., Tan C.2012. Clonal analysis and virulent traits of pathogenic extraintestinal Escherichia coli isolates from swine in China. BMC Vet. Res. 8: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erova T. E., Rosenzweig J. A., Sha J., Suarez G., Sierra J. C., Kirtley M. L., van Lier C. J., Telepnev M. V., Motin V. L., Chopra A. K.2013. Evaluation of protective potential of Yersinia pestis outer membrane protein antigens as possible candidates for a new-generation recombinant plague vaccine. Clin. Vaccine Immunol. 20: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia T. A., Ventura C. L., Smith M. A., Merrell D. S., O’Brien A. D.2013. Cytotoxic necrotizing factor 1 and hemolysin from uropathogenic Escherichia coli elicit different host responses in the murine bladder. Infect. Immun. 81: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffernan E. J., Harwood J., Fierer J., Guiney D.1992. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J. Bacteriol. 174: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffernan E. J., Wu L., Louie J., Okamoto S., Fierer J., Guiney D. G.1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 62: 5183–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou B., Meng X. R., Zhang L. Y., Tan C., Jin H., Zhou R., Gao J. F., Wu B., Li Z. L., Liu M., Chen H. C., Bi D. R., Li S. W.2014. TolC promotes ExPEC biofilm formation and curli production in response to medium osmolarity. Biomed. Res. Int. 2014: 574274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson J. R., Stell A. L.2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181: 261–272. [DOI] [PubMed] [Google Scholar]
- 17.Kim K., Kim K. P., Choi J., Lim J. A., Lee J., Hwang S., Ryu S.2010. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76: 5188–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köhler C. D., Dobrindt U.2011. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 301: 642–647. [DOI] [PubMed] [Google Scholar]
- 19.Kolodziejek A. M., Hovde C. J., Minnich S. A.2012. Yersinia pestis Ail: multiple roles of a single protein. Front Cell Infect. Microbiol. 2: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodziejek A. M., Schnider D. R., Rohde H. N., Wojtowicz A. J., Bohach G. A., Minnich S. A., Hovde C. J.2010. Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect. Immun. 78: 5233–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G., Kariyawasam S., Tivendale K. A., Wannemuehler Y., Ewers C., Wieler L. H., Logue C. M., Nolan L. K.2012. tkt1, located on a novel pathogenicity island, is prevalent in avian and human extraintestinal pathogenic Escherichia coli. BMC Microbiol. 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., Zheng H., Yang M., Xu Z., Wang X., Wei L., Tang B., Liu F., Zhang Y., Ding Y., Tang X., Wu B., Johnson T. J., Chen H., Tan C.2015. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics 16: 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyhs U., Ikonen I., Pohjanvirta T., Raninen K., Perko-Mäkelä P., Pelkonen S.2012. Extraintestinal pathogenic Escherichia coli in poultry meat products on the Finnish retail market. Acta Vet. Scand. 54: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manges A. R., Johnson J. R.2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 55: 712–719. [DOI] [PubMed] [Google Scholar]
- 25.Mecsas J., Welch R., Erickson J. W., Gross C. A.1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177: 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellata M.2013. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10: 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller V. L., Beer K. B., Loomis W. P., Olson J. A., Miller S. I.1992. An unusual pagC:TnphoA mutation leads to an invasion- and virulence-defective phenotype in Salmonellae. Infect. Immun. 60: 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell N. M., Johnson J. R., Johnston B., Curtiss R., 3rd, Mellata M.2015. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 81: 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto K., Hermansson M.2004. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J. Bacteriol. 186: 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vica Pacheco S., García González O., Paniagua Contreras G. L.1997. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 156: 129–132. [DOI] [PubMed] [Google Scholar]
- 31.Pulkkinen W. S., Miller S. I.1991. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 173: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ron E. Z.2010. Distribution and evolution of virulence factors in septicemic Escherichia coli. Int. J. Med. Microbiol. 300: 367–370. [DOI] [PubMed] [Google Scholar]
- 33.Smith J. L., Fratamico P. M., Gunther N. W.2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4: 134–163. [DOI] [PubMed] [Google Scholar]
- 34.Stoorvogel J., van Bussel M. J., van de Klundert J. A.1987. Cloning of a beta-lactam resistance determinant of Enterobacter cloacae affecting outer membrane proteins of Enterobacteriaceae. FEMS Microbiol. Lett. 48: 277–281. [Google Scholar]
- 35.Stoorvogel J., van Bussel M. J., van de Klundert J. A.1991. Biological characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoorvogel J., van Bussel M. J., Tommassen J., van de Klundert J. A.1991. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsang T. M., Wiese J. S., Felek S., Kronshage M., Krukonis E. S.2013. Ail proteins of Yersinia pestis and Y. pseudotuberculosis have different cell binding and invasion activities. PLoS ONE 8: e83621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogt J., Schulz G. E.1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 7: 1301–1309. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z., Xue Y., Wu B., Tang X., Hu R., Xu Y., Guo A., Chen H.2008. Subcutaneous vaccination with attenuated Salmonella enterica serovar Choleraesuis C500 expressing recombinant filamentous hemagglutinin and pertactin antigens protects mice against fatal infections with both S. enterica serovar Choleraesuis and Bordetella bronchiseptica. Infect. Immun. 76: 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]