Abstract
Bone marrow-derived mesenchymal stem cells (MSCs) are pluripotent stem cells that show a vital potential in the clinical application for cell transplantation. In the present paper, proteomic techniques were used to approach the protein profiles associated with porcine bone marrow MSCs and investigate the regulation of MSC proteins on the effect of 5-azacytidine (5-aza). Over 1,700 protein species were separated from MSCs according to gel analysis. Compared with the expression profiling of control MSCs, there were 11 protein spots up-regulated and 26 down-regulated in the protein pattern of 5-aza-treated cells. A total of 21 proteins were successfully identified by MALDI-TOF-MS analysis, among which some interesting proteins, such as alpha B-crystallin, annexin A2, and stathmin 1, had been reported to involve in cell proliferation and differentiation through different signaling pathways. Our data should be useful for the future study of MSC differentiation and apoptosis.
Key words: bone marrow mesenchymal stem cells, 5-azacytidine, two-dimensional gel electrophoresis, mass spectrometry, proteome
Introduction
The interest in the research of bone marrow mesenchymal stem cells (MSCs) has recently been increased by reports suggesting that MSCs seem to be pluripotent 1., 2. and may play a future therapeutic role in cellular replacement strategies for clinical applications. It is reported that MSCs could differentiate into cardiomyocytes after exposure to 5-azacytidine (5-aza) (3), and the MSC-derived cardiomyocytes could provide a useful and powerful tool for cardiomyocyte transplantation (4). Researches have shown that MSCs are capable of the stable cardiac engraftment and site-specific differentiation in myocardial scar tissues in the animal model of infarction 5., 6.. Recent study of mononuclear bone marrow cells demonstrate that the selectively intracoronary transplantation of autologous bone marrow cells is safe and seems to be effective for acute myocardial infarction patients under clinical conditions (7).
As an animal model, pig is often used in experiments because of several distinct advantages (8). Some reports have been published on the porcine MSCs, including their isolation (9), cardiac differentiation 10., 11., 12., induced osteoclastogenesis (13), and pluripotency (14). Despite this promising progress, the detailed molecular events that occur during the MSC differentiation induced by 5-aza still need to be fully understood, and some researchers have proposed a different view on this issue (15).
With the development of mass spectrometry (MS) technology, proteomics has been widely used for the analysis of cell proliferation and differentiation under different conditions 16., 17.. It has a potential for the definition of stem cell populations (18) and differentiation 19., 20., 21.. However, compared with the gene study of MSCs 22., 23., 24., 25., only a few proteomic reports on MSCs can be found in literature 26., 27..
In the present study, we analyzed the protein pattern of porcine bone marrow MSCs by using two-dimensional (2D) gel electrophoresis to investigate the changes (increase or decrease) of protein expression on the effect of 5-aza. Protein spots at different expression levels were examined by MS, and 21 proteins were successfully identified by peptide mass fingerprinting (PMF) with MS analysis. The results suggest that the molecular mechanisms of MSCs treated by 5-aza in vitro might be associated with these functional proteins.
Results and Discussion
Isolation of MSCs and treatment by 5-aza
The future use of adult MSCs for human therapies depends on the establishment of preclinical studies with other mammals such as rat and pig. The isolation of MSCs from porcine bone marrow was successfully completed (9). Two major types of cells, MSCs and hematopoietic stem cells (HSCs), were prepared from bone marrow. MSCs were spindle-shaped, attached to the culture dish, and proliferated in the culture medium. HSCs were rowed, not attached to the culture dish, and were removed with the culture medium change. According to the previous report (5), MSCs were cultured at the Dulbecco’s modified Eagles’ medium (DMEM), which was changed twice every week.
Protein separation by 2D gel electrophoresis
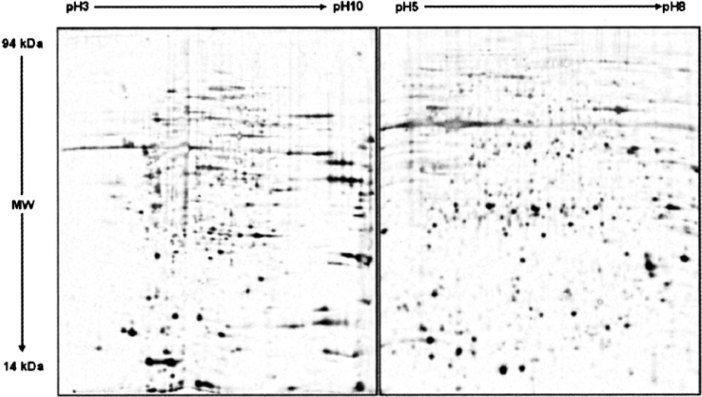
For the purpose of initially scanning the changes in the protein levels of MSCs treated by 5-aza, we used IPG strips with a wide range of linear IPG (pH 3–10) in the first dimension. In the wide-range 2D maps, there was a loss of resolution in the region pH 5–8. Most probably the fact is due to that the pI values of many proteins occur in this range. Therefore, we also performed electrophoresis on pH 5–8 to achieve a better protein separation. These narrow pH gels allowed a higher resolution and more protein spots in the relative pH zones. Figure 1 shows the analytical silver-stained 2D maps of MSCs using different pH-range IPG strips.
Fig. 1.
Effect of IPG strips with different pH range on the protein expression of MSCs. The protein lysates (100 μg) were subjected to 2D gel electrophoresis, followed by silver staining and image analysis. Results were quantified from three sets of 2D gel electrophoresis. IEF (pH 3–10 and pH 5–8, linear gradient) is in the horizontal direction. SDS-PAGE (12% gel) is in the vertical direction.
Comparison of protein patterns between control and 5-aza-treated MSCs
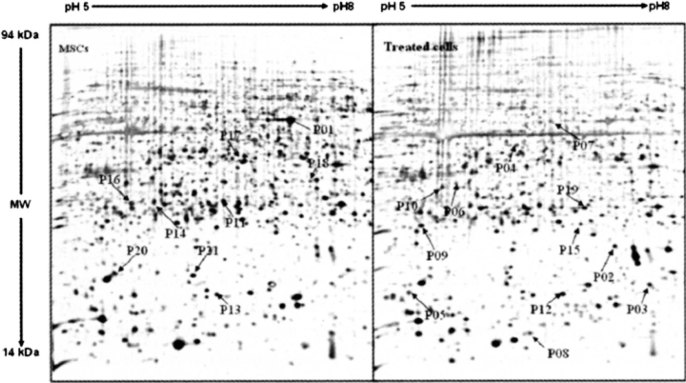
To investigate the effect of 5-aza on the expression of proteins, we compared the protein expression profiles between control and 5-aza-treated MSCs (Figure 2). Three analytical gels were run for each sample. Proteins were considered to be significantly up- or down-regulated when the corresponding spot volumes showed an increase (>two folds) or decrease (<50%). According to the analysis, about 1,700 protein spots were detected from the silver-stained gels of control and treated MSCs. Compared with the profiling of control MSCs, there were 11 protein spots up-regulated (>two folds) and 26 down-regulated in the protein pattern of treated cells according to the density of protein spots. These differently expressed proteins that can be detected in CBB R-250 stained gels were chosen for further MS analysis.
Fig. 2.
The 2D profiles of control and treated MSCs. Upward arrows indicate up-regulation, and downward arrows indicate down-regulation. The spots of interest were excised from the gels and digested with trypsin. The extracted peptides were subjected to MALDI-TOF-MS, and the spectra of peptides were used for protein identification. The marked spots were successfully identified and the information of proteins was listed in Table 1.
Protein identification
Based on the comparison of protein profiles of control and treated MSCs, we chose the protein spots differently expressed for the protein identification by MS. The protein spots in different sets of the gels were identified by the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis on the basis of peptide mass matching with the theoretical peptide masses in tryptic digests of all known proteins from the NCBI nonredundant (NCBInr) database and the Swiss-Prot database. Table 1 lists the identified proteins in our study.
Table 1.
Proteins Identified by MALDI-TOF-MS
| No. | Protein/Gene name | Accession No. | MW (kDa)/pI | Score | Matched/ all peptides | Seqence coverage | Regulation |
|---|---|---|---|---|---|---|---|
| P01 | Phosphopyruvate hydratase (Fragment) | Q29568 | 16.08/4.6 | 119 | 11/42 | 71% | High expression |
| P02 | Alpha B-crystallin (Cryab) | gi|75063982, Q7M2W6 | 20.13/6.8 | 116 | 4/19 | 21% | 4.14 |
| P03 | Peptidyl-prolyl isomerase A (Ppia) | gi|51702768, P62936 | 17.96/8.4 | 104 | 12/42 | 57% | 2.26 |
| P04 | Moesin (Msn) | gi|127236, P26042 | 67.66/6.3 | 77 | 8/28 | 11% | 2.07 |
| P05 | Cardiac alpha tropomyosin (Tpml) | gi|1927, P42639 | 32.72/4.7 | 138 | 24/77 | 59% | 1.83 |
| P06 | Heat shock 27 kDa protein 1 (Hspb1) | gi|55926209 | 22.98/6.2 | 82 | 9/40 | 35% | 1.77 |
| P07 | Phosphopyruvate hydratase (Enolase) | Q29568 | 16.08/4.6 | 63 | 8/31 | 51% | 1.59 |
| P08 | Beta-catenin (Ctnnb1) | gi|47523792, Q8WNW4 | 85.51/5.5 | 83 | 5/10 | 7% | 1.48 |
| P09 | Peroxiredoxin 2 (Prdx2) | gi|1717797, P52552 | 14.18/4.7 | 129 | 5/7 | 36% | 1.33 |
| P10 | MHC class I antigen (Sla-3) | gi|33468760, Q7YNZ2 | 43.68/5.4 | 71 | 4/28 | 10% | 1.19 |
| P11 | Heat shock 27 kDa protein 1 (Hspb1) | gi|55926209 | 22.94/6.2 | 122 | 5/31 | 22% | 1.11 |
| P12 | Superoxide dismutase 1 (Sod1) | gi|15082144, Q95ME5 | 15.25/6.0 | 87 | 8/28 | 44% | 1.10 |
| P13 | Guanosine diphosphate dissociation inhibitor 2 (Gdi2) | gi|75043802, Q6Q7J2 | 50.27/6.3 | 66 | 4/14 | 8% | 1.09 |
| P14 | Cytoskeletal beta actin (Actb) | gi|45269029, Q6QAQ1 | 44.79/5.5 | 67 | 7/23 | 19% | 1.08 |
| P15 | Guanosine diphosphate dissociation inhibitor 2 (Gdi2) | gi|48675953, Q6Q7J2 | 50.27/6.3 | 54 | 6/21 | 22% | 1.06 |
| P16 | Peroxiredoxin 6 (Prdx6) | gi|47523870, Q9TSX9 | 25.04/5.7 | 151 | 8/11 | 40% | 0.96 |
| P17 | Aldose reductase (Akr1b1) | gi|48374071, P80276 | 35.87/5.9 | 183 | 10/12 | 38% | 0.95 |
| P18 | Esterase D (Esd) | gi|7288152, Q9GM98 | 31.41/6.8 | 62 | 4/14 | 13% | 0.84 |
| P19 | Heat shock 27 kDa protein 1 (Hspb1) | gi|55926209 | 22.98/6.2 | 153 | 13/34 | 42% | 0.80 |
| P20 | Annexin A2 (Anxa2) | gi|54020966, Q5Y2C7 | 38.54/6.5 | 85 | 5/38 | 16% | 0.60 |
| P21 | Stathmin 1 (Stmn1) | gi|57528035, Q6DUB7 | 17.29/5.8 | 84 | 6/11 | 36% | 0.53 |
Functional analysis of the identified proteins
After protein identification, we analyzed the information of these identified proteins from the NCBInr and Swiss-Prot databases. Some interesting proteins, such as HSP27 (a heat shock protein), alpha B-crystallin (an HSP), annexin A2 (a calcium-binding protein), and stathmin 1 (a structural protein), were regulated when MSCs were treated with 5-aza.
HSP27 and alpha B-crystallin are a group of stress-related proteins that are synthesized by cells in response to heat and other environmental stress. Their expression in cells may have important physiological and pathological implications such as protection, cell proliferation, migration and differentiation, and disease prognosis. Increasing evidence suggests that the HSP family of chaperones may play a role in the formation and function of cytoskeleton (28). The small HSP alpha B-crystallin is a molecular chaperone whose function is to stabilize the proteins damaged by inflammatory stress. In our experiment, alpha B-crystallin (P02, gi|75063982) showed up-regulation (~4 folds). HSP27 is part of a signaling pathway regulating the dynamics of actin filaments (29). Interestingly, HSP27 has been shown to be essential for the differentiation of mouse embryonic stem cells (30). The different expression of HSP27 (P06, P11, and P19, gi|55926209) showed that it is also involved in the treatment process of MSCs.
About two-fold down-regulation expression of annexin A2 (P20, gi|54020966) was found in our study. Annexins are phospholipid-binding proteins that may cross-link plasma membrane phospholipids, actins, and cytoskeletons to regulate proliferation, exocytosis, and membrane fusion. The annexin A2 gene is a growth-regulated gene in mammalian cells, and the annexin A2 protein links the plasma membrane to cytoskeleton in a calcium-dependent manner. Gilmore et al. (31) reported that annexin A2 is an important player in the cellular differentiation of myeloid cell lines.
Peroxiredoxins (Prdxs) are a family of antioxidant proteins that reduce peroxide levels by using reducing agents such as thioredoxin. In addition to their antioxidant activity, Prdxs have been implicated in a number of cellular functions such as cell proliferation and differentiation (32), enhancement of the natural killer cell activity, protection of radical-sensitive proteins, metabolism, and intracellular signaling (33). Our data showed that two isoforms were found in the control and treated MSCs. However, Prdx2 (P09, gi|1717797) and Prdx6 (P16, gi|47523870) showed inversely regulation between the proliferating and treating stages, which may contribute to the using of different reducing agents. Therefore, they may be involved in different signaling pathways.
Beta-catenin is a multifunctional protein that is known to participate in two well-defined cellular processes, cell-cell adhesion and Wnt-stimulated transcriptional activation 34., 35.. Mbalaviele et al. (36) demonstrated that beta-catenin, in synergy with BMP-2, promotes the MSC commitment to the oteoblast pathway, inhibits adipogenic differentiation, and induces bone formation in vivo. Furthermore, the Wnt/beta-catenin pathway regulates the cardiac valve formation (37). Nakamura et al. (38) tested the involvement of Wnt and beta-catenin in mammalian cardiac myogenesis by using a pluripotent mouse cell line (P19CL6) that recapitulates the early steps for cardiac specification. Their findings suggested that Wnt/beta-catenin signaling is activated at the inception of mammalian cardiac myogenesis and is indispensable for cardiac differentiation, at least in this pluripotent model system. From our data, the beta-catenin (P08, gi|47523792) showed up-regulation by the 5-aza treatment; it may be involved in the cell process through the Wnt signaling pathway.
Three closely related proteins, namely ezrin, radixin, and moesin, which compose the ERM family, are thought to work as cross-linkers between plasma membranes and actin-based cytoskeletons. In our study, the moesin (P04, gi|127236) expressed up-regulation by the 5-aza treatment. Real-time PCR confirmed a significant reduction of moesin gene expression following the dexamethasone treatment. The decrease of expression for this protein, which involves in the cytoskeletal organization, could explain the effects of the dexamethasone treatment on bone marrow stromal cell differentiation (39).
Stathmin 1 (P21, gi|57528035) showed down-regulation by the 5-aza treatment. It is a ubiquitous cytosolic protein that has been independently studied in various cellular systems under different names, including Oncoprotein 18, metablastin phosphoprotein 19, and LAP18. Stathmin 1 is proposed to act as a relay for a diverse array of cell signaling pathways, which influences cell growth and differentiation 40., 41., 42., 43.. The function of stathmin 1 during the MSC differentiation needs to be investigated in further research.
According to the previous reports, these functional proteins may play an important role in the cell process of MSCs induced by 5-aza, and they should be highlighted during the MSC proliferation and differentiation research.
Conclusion
In conclusion, this study describes for the first time a proteomics approach employed to understand the biochemical processes related to porcine bone morrow MSCs treated by 5-aza. The expression level of proteins involved in cell proliferation and differentiation as well as cell defense was found to change when MSCs were treated by 5-aza. Our data should be useful for the future study of MSC differentiation and apoptosis.
Materials and Methods
Cell culture
The porcine sample was anesthetized by an injection of sodium pentobarbital (20 mg/kg). Under anesthesia, thighbone was excised by a syringe containing heparin with an 18-gauge needle. Mononuclear bone marrow cells were obtained by centrifugation. Cells were cultured in DMEM (Gibco BRL, Rockville, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, USA), penicillin, and streptomycin (Sigma, St. Louis, USA) at 37°C humidified atmosphere with 5% CO2 for three days before the first medium change, and then the medium was changed twice a week. At 90% confluence, the cells were trypsinized with 0.25% trypsin-EDTA and were passaged to flask at 1:3 ratio.
For the 5-aza treatment, the MSCs at passage 2 were cultured for 72 h, incubated with the medium containing 10 μM 5-aza for 24 h, and then cultured with DMEM without 5-aza. About 106 MSCs were harvested by gently trypsinization after incubation for three weeks, and the harvested cells were washed by cold PBS solution and centrifuged at 800 g for 10 min at 4°C for more than three times. The pipettes were stored at −80°C before protein lysis. Triplicate control and treated MSCs were cultured for the future protein preparation.
Sample preparation
The harvested cells were washed by cold PBS and the cellular pellets were dissolved in a lysis buffer containing 8 M urea (Promega, Madison, USA), 4% w/v CHAPS (Promega), 100 mM DTT (Promega), 0.5% ampholyte (Bio-Rad Laboratories, Hercules, USA), and 1 mM PMSF (Promega), and then sonicated at 4°C for 2 min. After that, the DNase and RNase (Roche, Basel, Switzerland) solution was added and laid at 4°C for 15 min. The sample was then centrifuged at 14,000 g for 30 min at 4°C to remove any insoluble cell debris. The total protein concentration was determined using the Bradford method and the remained protein solution was stored at −80°C for further proteomic analysis.
2D gel electrophoresis
For the first dimension, 350 μL of each sample (100 μg and 600 μg of proteins for analytic and preparative gels, respectively) was subjected to isoelectric focusing (IEF) on an IPG strip (pH 3–10 or pH 5–8, 17 cm, linear gradient, Bio-Rad). The amount of proteins applied in the samples from the different treatment groups (control and treated MSCs) was the same in each experimental repeat, allowing comparisons between the different groups. IPG Ready Strips were actively rehydrated at 50 V for 15 h to enhance protein uptake. IEF was carried out on a Protean IEF Cell (Bio-Rad) at 20°C with a maximum current setting of 50 μA per gel. Focusing was performed using a four-phase program in which the first phase was at 250 V for 30 min, the second at 1 kV for 1 h, the third at 10 kV for 5 h at a linear ramping mode, and the final phase at 10 kV for 6 h. After focusing, the strips were either stored at −80°C or used directly for 2D gel electrophoresis. Prior to the 2D SDS-PAGE, IPG strips were incubated for 10 min in the equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% v/v glycerol, 2% w/v SDS) supplemented with 10 mg/mL DTT (Promega), followed by a 10-minute incubation in the equilibration buffer supplemented with 25 mg/mL iodoacetamide (Bio-Rad), then rinsed once with the SDS-PAGE buffer (25 mM Tris, 192 mM glycine, pH 8.3, 0.1% SDS). The strips were then transferred on the 2D SDS-PAGE gel and sealed in place with 0.5% low melting point agarose (Promega). Proteins were resolved by 12% SDS-PAGE using a Protean II XL system (Bio-Rad). Electrophoresis was carried out at 10 mA per gel for 30 min, followed by 30 mA per gel until the bromophenol blue front reached the bottom of the gel, and the temperature of the electrophoresis system was kept at the range of 4°C–5°C.
Detection of proteins and image analysis
Analytical gels were silver-stained according to the protocol for compatibility with subsequent analysis of proteins by MS. Gels were fixed for 15 min in 40% v/v methanol, 10% v/v acetic acid, followed for another 15 min after changing fresh fixation solution. Gels were sensitized for 30 min in 0.02% w/v sodium thiosulfate, followed by three times of 5-minute washes with Milli-Q water, and then incubated in chilled (4°C) 0.25% w/v silver nitrate for 20 min, followed again by three times of 1-minute washes. Proteins were then visualized by several times of washes with the developing solution (2.5% sodium carbonate, 0.04% formaldehyde) until a desired level of staining was achieved, after which the development was stopped with 1.46% EDTA solution.
Preparative gels were incubated for 1 h in 50% v/v methanol/10% v/v acetic acid, 0.1% w/v CBB R-250, and the excess dye was removed with several changes of 20% v/v methanol, 10% v/v acetic acid. Gels were digitized at a resolution of 300 dpi using a Power-Look 2100XL scanner system (UMAX Technologies, Inc., Dallas, USA). Protein spots were then located, quantified, and matched to the spots on comparative gels using PDQuest 7.1 software (Bio-Rad). Analytical gels were analyzed to investigate the expression of proteins between control and treated MSCs. The corresponding spots in the preparative gels were matched with the spots of interest in analytical gels by the software.
In-gel digestion and protein identification
From the 2D gel analysis of control and treated samples, differentially expressed proteins were selected for the MS identification. These spots were manually excised from 2D gels, sliced into small pieces, and then destained three times with 25 mM NH4HCO3 buffer (pH 8.0) in 50% v/v acetonitrile (ACN) for 15 min. The gel pieces were dehydrated in ACN for 5 min and then dried for 20 min in a vacuum centrifuge. The dried gel volume was evaluated and three volumes of trypsin (12.5 ng/μL in 25 mM NH4HCO3) were added. The digestion was performed at 37°C overnight. The peptide fragments were extracted twice with 50 μL of 50% v/v ACN/0.1% v/v trifluoroacetic acid (TFA, Sigma). After removal of ACN by centrifugation in a vacuum centrifuge, the peptides were mixed with α-cyano-4-hydroxycinnamic acid (CHCA, 0.5 μL of 10 mg/mL) and then subjected to a gold-plated sample holder. The samples were introduced into the mass spectrometer after drying. Protein samples digested by trypsin were analyzed using a MALDI-TOF-MS instrument (Bruker Autoflex, Billerica, USA). The spectra were obtained in the reflection mode by summing 200 laser shots with the ion source 1 of 19 kV, ion source 2 of 16.27 kV, 100 ns delay, and the low mass gate at m/z 600. Peptide matching was carried out against the NCBInr and Swiss-Prot databases using the Mascot tool. The parameters were set to allow one possible missed cleavage for trypsin digestion with a peptide mass tolerance of 100 ppm. For an identity assignment, the minimum requirement number was four matching peptides. The considered modifications included carbamidomethylation of cysteine and oxidation of methionine.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 30271663 and No. 30500680).
References
- 1.Jiang Y. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 2.Deans R.J., Moseley A.B. Mesenchymal stem cells: biology and potential clinical uses. Exp. Hematol. 2000;28:875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 3.Wakitani S. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417–1426. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda K. Reprogramming of bone marrow mesenchymal stem cells into cardiomyocytes. C. R. Biol. 2002;325:1027–1038. doi: 10.1016/s1631-0691(02)01524-x. [DOI] [PubMed] [Google Scholar]
- 5.Tomita S. Autologous transplantation of bone marrow cells improves damaged heart function. Circulation. 1999;100:II247–II256. doi: 10.1161/01.cir.100.suppl_2.ii-247. [DOI] [PubMed] [Google Scholar]
- 6.Tomita S. Improved heart function with myogenesis and angiogenesis after autologous porcine bone marrow stromal cell transplantation. J. Thorac. Cardiovasc. Surg. 2002;123:1132–1140. doi: 10.1067/mtc.2002.120716. [DOI] [PubMed] [Google Scholar]
- 7.Strauer B.E. Repair of infarcted myocardium by autologous intracoronary mononuclear bone marrow cell transplantation in humans. Circulation. 2002;106:1913–1918. doi: 10.1161/01.cir.0000034046.87607.1c. [DOI] [PubMed] [Google Scholar]
- 8.Edge A.S. Xenogeneic cell therapy: current progress and future developments in porcine cell transplantation. Cell Transplant. 1998;7:525–539. doi: 10.1177/096368979800700603. [DOI] [PubMed] [Google Scholar]
- 9.Fang L.J. Isolation and culture of multipotent mesenchymal stem cells from porcine bone marrow. Chin. Crit. Care Med. 2003;15:606–608. [PubMed] [Google Scholar]
- 10.Moscoso I. Differentiation “in vitro” of primary and immortalized porcine mesenchymal stem cells into cardiomyocytes for cell transplantation. Transplant Proc. 2005;37:481–482. doi: 10.1016/j.transproceed.2004.12.247. [DOI] [PubMed] [Google Scholar]
- 11.Shake J.G. Mesenchymal stem cell implantation in a swine myocardial infarct model: engraftment and functional effects. Ann. Thorac. Surg. 2002;73:1919–1925. doi: 10.1016/s0003-4975(02)03517-8. [DOI] [PubMed] [Google Scholar]
- 12.Tomita S. Improved heart function with myogenesis and angiogenesis after autologous porcine bone marrow stromal cell transplantation. J. Thorac. Cardiovasc. Surg. 2002;123:1132–1140. doi: 10.1067/mtc.2002.120716. [DOI] [PubMed] [Google Scholar]
- 13.Rassi C.M. Modulation of osteoclastogenesis in porcine bone marrow cultures by quercetin and rutin. Cell Tissue Res. 2005;319:383–393. doi: 10.1007/s00441-004-1053-9. [DOI] [PubMed] [Google Scholar]
- 14.Ringe J. Porcine mesenchymal stem cells. Induction of distinct mesenchymal cell lineages. Cell Tissue Res. 2002;307:321–327. doi: 10.1007/s00441-002-0525-z. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y. Growth and differentiation of rat bone marrow stromal cells: does 5-azacytidine trigger their cardiomyogenic differentiation? Cardiovasc. Res. 2003;58:460–468. doi: 10.1016/s0008-6363(03)00265-7. [DOI] [PubMed] [Google Scholar]
- 16.Turck N. Proteomic analysis of nuclear proteins from proliferative and differentiated human colonic intestinal epithelial cells. Proteomics. 2004;4:93–105. doi: 10.1002/pmic.200300480. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G. Applying proteomic methodologies to analyze the effect of hexamethylene bisacetamide (HMBA) on proliferation and differentiation of human gastric carcinoma BGC-823 cells. Int. J. Biochem. Cell Biol. 2004;36:1613–1623. doi: 10.1016/j.biocel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Unwin R.D. The potential for proteomic definition of stem cell populations. Exp. Hematol. 2003;31:1147–1159. doi: 10.1016/j.exphem.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Guo X. Proteomic characterization of early-stage differentiation of mouse embryonic stem cells into neural cells induced by all-trans retinoic acid in vitro. Electrophoresis. 2001;22:3067–3075. doi: 10.1002/1522-2683(200108)22:14<3067::AID-ELPS3067>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 20.Hayman M.W., Przyborski S.A. Proteomic identification of biomarkers expressed by human pluripotent stem cells. Biochem. Biophys. Res. Commun. 2004;316:918–923. doi: 10.1016/j.bbrc.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 21.Maurer M.H. Comprehensive proteome expression profiling of undifferentiated versus differentiated neural stem cells from adult rat hippocampus. Neurochem. Res. 2004;29:1129–1144. doi: 10.1023/b:nere.0000023600.25994.11. [DOI] [PubMed] [Google Scholar]
- 22.Barry F. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp. Cell Res. 2001;268:189–200. doi: 10.1006/excr.2001.5278. [DOI] [PubMed] [Google Scholar]
- 23.Silva W.A., Jr. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells. 2003;21:661–669. doi: 10.1634/stemcells.21-6-661. [DOI] [PubMed] [Google Scholar]
- 24.Panepucci R.A. Comparison of gene expression of umbilical cord vein and bone marrow-derived mesenchymal stem cells. Stem Cells. 2004;22:1263–1278. doi: 10.1634/stemcells.2004-0024. [DOI] [PubMed] [Google Scholar]
- 25.Hishikawa K. Gene expression profile of human mesenchymal stem cells during osteogenesis in three-dimensional thermoreversible gelation polymer. Biochem. Biophys. Res. Commun. 2004;317:1103–1107. doi: 10.1016/j.bbrc.2004.03.165. [DOI] [PubMed] [Google Scholar]
- 26.Wang D. Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor betal stimulation. J. Biol. Chem. 2004;279:43725–43734. doi: 10.1074/jbc.M407368200. [DOI] [PubMed] [Google Scholar]
- 27.Kratchmarova I. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 28.Liang P., MacRae T.H. Molecular chaperones and the cytoskeleton. J. Cell Sci. 1997;110:1431–1440. doi: 10.1242/jcs.110.13.1431. [DOI] [PubMed] [Google Scholar]
- 29.Guay J. Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci. 1997;110:357–368. doi: 10.1242/jcs.110.3.357. [DOI] [PubMed] [Google Scholar]
- 30.Mehlen P. HSP27 as a switch between differentiation and apoptosis in murine embryonic stem cells. J. Biol. Chem. 1997;272:31657–31665. doi: 10.1074/jbc.272.50.31657. [DOI] [PubMed] [Google Scholar]
- 31.Gilmore W.S. Annexin A2 expression during cellular differentiation in myeloid cell lines. Biochem. Soc. Trans. 2004;32:1122–1123. doi: 10.1042/BST0321122. [DOI] [PubMed] [Google Scholar]
- 32.Prosperi M.T. A human cDNA corresponding to a gene overexpressed during cell proliferation encodes a product sharing homology with amoebic and bacterial proteins. J. Biol. Chem. 1993;268:11050–11056. [PubMed] [Google Scholar]
- 33.Lim M.J. The type II peroxiredoxin gene family of the mouse: molecular structure, expression and evolution. Gene. 1998;216:197–205. doi: 10.1016/s0378-1119(98)00290-x. [DOI] [PubMed] [Google Scholar]
- 34.Kim W., Kang K. Recent developments of cathepsin inhibitors and their selectivity. Expert Opin. Ther. Patents. 2002;12:419–432. [Google Scholar]
- 35.Kaplan D.D. Identification of a role for beta-catenin in the establishment of a bipolar mitotic spindle. J. Biol. Chem. 2004;279:10829–10832. doi: 10.1074/jbc.C400035200. [DOI] [PubMed] [Google Scholar]
- 36.Mbalaviele G. Beta-catenin and BMP-2 synergize to promote osteoblast differentiation and new bone formation. J. Cell. Biochem. 2005;94:403–418. doi: 10.1002/jcb.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurlstone A.F. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T. A Wnt- and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornet F. Effect of dexamethasone on moesin gene expression in rabbit bone marrow stromal cells. Mol. Cell. Biochem. 2004;265:79–83. doi: 10.1023/b:mcbi.0000044309.43612.3b. [DOI] [PubMed] [Google Scholar]
- 40.Melhem R.F. Involvement of OP18 in cell proliferation. Biochem. Biophys. Res. Commun. 1991;179:1649–1655. doi: 10.1016/0006-291x(91)91764-4. [DOI] [PubMed] [Google Scholar]
- 41.Di Paolo G. The phosphoprotein stathmin is essential for nerve growth factor-stimulated differentiation. J. Cell Biol. 1996;133:1383–1390. doi: 10.1083/jcb.133.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C.L. Oncoprotein 18 levels and phosphorylation mediate megakaryocyte polyploidization in human erythroleukemia cells. Proteomics. 2001;1:1415–1423. doi: 10.1002/1615-9861(200111)1:11<1415::AID-PROT1415>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Rubin C.I. Stathmin expression and megakaryocyte differentiation: a potential role in polyploidy. Exp. Hematol. 2003;31:389–397. doi: 10.1016/s0301-472x(03)00043-2. [DOI] [PubMed] [Google Scholar]