Abstract
A novel high-throughput system, called the stacked slice-gel system for separation and reactions (4SR), was developed for the analysis of DNA/RNA and protein/peptide. The system provides a novel three-dimensional gel electrophoresis approach that exploits the property of stacked slice gels. It allows multiple samples simultaneously to react as well as to be separated, offering a two-dimensional (m × n) sample loading system. For this purpose, high-throughput multi-micro vessels (MMVs) containing variable numbers of wells (100 wells in this paper) have been used, which are made of 25 mm square-size polyacrylamide gels. Furthermore, after electrophoretic separation, a slice gel containing a desired sample can be easily removed and proceeded to the next step. Different biological reactions as well as successive separation of products were effectively carried out dealing with DNA/RNA and protein/peptide. It shows that this system has a diversity of potentials to be developed.
Key words: three-dimensional polyacrylamide gel electrophoresis (3D-PAGE), high-throughput screening, three-dimensional separation, microarray technology
Introduction
The completion of the Human Genome Project has triggered large-scale screening of genomes (1) and proteomes (2) in aims to find out candidate genes related to diseases (3), perform expression analyses at the mRNA level (4) or at the protein level (5), discover new drugs (6), and analyze molecular interactions (7). For such purposes, technologies handling a tiny amount of samples should be developed, of which the importance has already been described as the ambient analyte theory (8). To get a move on screening analysis, microarray technologies such as DNA and protein chips have been developed and used in the recent years 9., 10., 11., 12.. However, the microarray high-throughput systems developed thus far are limited to be useful in their purposes of development with a highly specific nature. Therefore, a novel high-throughput system is required to meet a diversity of needs.
Techniques based on gel electrophoresis have a smart nature of allowing scientists to add devices and have brought about great successes in separating macromolecules at a high resolution (13). Here we introduce a novel technology that has the potential to widen (by introducing various reaction-and/or-separation analysis systems) and deepen (by introducing different array systems in micro-gel) the conventional gel electrophoresis and array technologies. This system, termed as stacked slice-gel system for separation and reactions (4SR), is a high-throughput three-dimensional (3D) gel electrophoresis technology, which can allow many samples simultaneously to react as well as to be separated. It consists of a primordial 3D electrophoresis apparatus including slice gels and know-how operations. The breakthrough of 4SR is in the introduction of slice gels for 3D electrophoresis and reactions. Acrylamide gel is used to form a high-throughput reaction container (that is, the gel containing m×n wells) for different reactions and is also used for the successive separation of biomolecules. The high-throughput gels, termed as multi-micro vessels (MMVs), which contain different numbers of wells (100 wells in this paper), have been devised in-house in 25 mm square-size gels for different reactions of biomolecules. The reaction products in MMVs could be simultaneously separated using a stack of slice gels, allowing easy detection and recovery of separated samples. These advantageous properties of 4SR are expected to develop the formerly unattainable analytical technologies such as micro-amount, multi-step, parallel, or huge-number analyses.
System
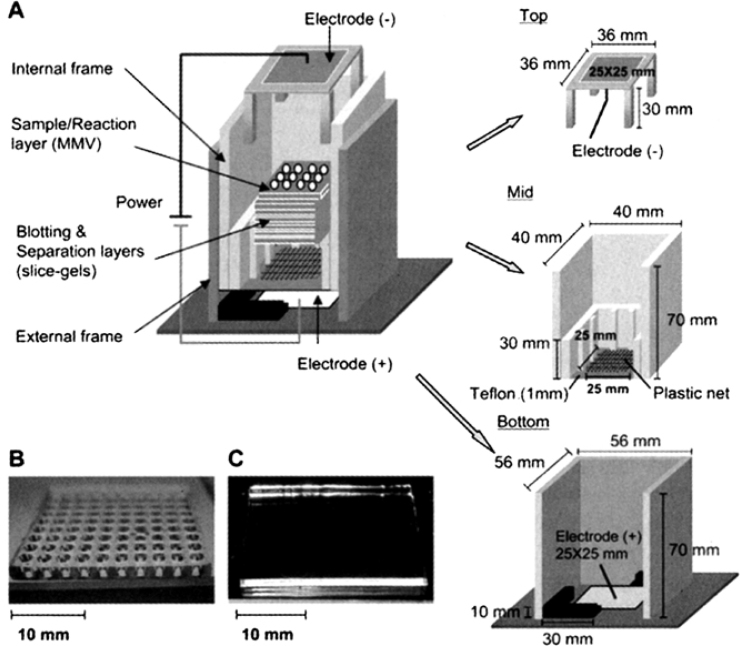
A schematic presentation of 4SR is shown in Figure 1. The system is made of acrylic plastics with three main parts: the external frame, the internal frame, and the upper electrode parts. The external frame contains an electrode and holds the overall assemblies. The internal frame serves as a container of slice gels and MMVs. At the bottom of the internal frame there is a teflon border, which holds the total frame and a plastic net of the slice-gel size (25 mm × 25 mm). The gels are put on the plastic net so that the current can pass without resistance. The innate advantage of this system is in accommodating stacked gel layers inside the internal frame where the separation of molecules occurs. The gels stacked are composed of three different types: (1) Reaction/Incubation layer (samples are loaded on this layer for reaction or separation); (2) Blotting/Trapping layer (sample stacking is done on this layer); and (3) Filtering/Separation layer (products are separated in different slice gels for consecutive analysis). The upper electrode part is placed over the sample layer to serve as an electrode as well as a part of the holder for gel layers.
Fig. 1.
Schematic presentation of 4SR. A. The 4SR apparatus consisting of top part (upper electrode), middle part (internal frame), and bottom part (external frame and lower electrode); B. A picture of a 100-well MMV made of polyacrylamide gels; C. A picture of a piece of slice gel. Both MMVs and slice gels are of the same size (25 mm × 25 mm) except their thicknesses (1 mm for slice gels and variable thicknesses for MMVs). The MMV shown in Panel B has a thickness of 3.5 mm (5 μL capacitor).
In this system, the top layer can be either a slab gel that contains the samples innerly separated by conventional 2D electrophoresis or an arrayed vessel. In the latter case, samples are loaded onto 2D-arrayed wells (Figure 1). In other words, gels containing m×n wells can be used for reactions prior to the migration of samples. For this purpose, the high-throughput MMVs with 100 (10×10) wells or more in 25 mm square size are devised using polyacrylamide gels (to be published elsewhere).
Results
Various applications of 4SR were successfully performed as listed in Table 1, including future implications. Throughout the experiments, the resolution for separation was proved to be much improved by implementing a blotting gel just beneath the MMV.
Table 1.
Various Types of Reaction and Separation Experiments to Be Performed Using 4SR
| Type | Experiment | ||
|---|---|---|---|
| Separation only |
Pre-separated | Separation (2D/3D) | |
| Direct loading | Blotting experiment* | ||
| Long-distance separation of ssDNAs using blotting gel* | |||
| Reaction and separation | Solution type | Isothermal | RNA-Z and separation* |
| Cathepsin E inhibition and analysis* | |||
| Temperatrue programmed | Random/Specific PCR and analysis | ||
| Single molecule PCR and analysis | |||
| Gel buried type | Restriction digestion and separation* | ||
| 2D analysis of reaction products | |||
Long distance separation
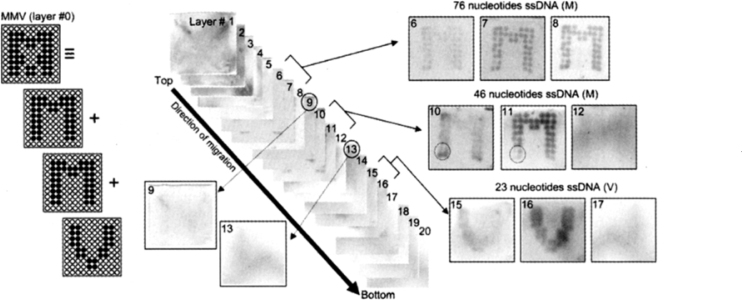
In order to test the effect of long distance migration performed with 4SR, three single-stranded DNA (ssDNA) fragments (76, 46, and 23 nucleotides) were electrophored using a 100-well MMV as shown in Figure 2. After electrophoresis, the ssDNAs were made detectable by silver-staining of individual slice gels, showing reasonable resolutions by separation (gel layers 6–8, 10–12, and 15–17 for 76-, 46-, and 23-nucleotide ssDNAs, respectively). Since most of the spots in a slice gel were stained similarly, the so-called “smiling effect” caused by the thermally or electrically inhomogeneous field was sufficiently small. In other words, ssDNAs of the same size in different wells migrated in the same mobility, reaching the same slice gel during electrophoresis. Although there were some dissimilarities in mobility as shown in gel layers 10 and 11 with a dotted circle, they can be improved by introducing blotting gels as shown in Figure 3. Therefore, this micro-gel-based system can be competently used for long distance separation of ssDNAs.
Fig. 2.
Long-distance separation of ssDNAs with 4SR. The separation was done using a 100-well MMV loaded with three ssDNAs of 76, 46, and 23 nucleotides (0.1 pmol/well for each letter), forming an overlapped shape of alphabetical letters “M”, “M”, and “V”, respectively. Electrophoresis was carried out using 3D gels (composed of slice gels of 25 mm square size) at 50 V/cm for 15 min and then the detached gel sheets were silver-stained. Gel layer numbers are labeled ascending from top to bottom, which is the direction of electrophoretic migration, throughout these experiments.
Fig. 3.
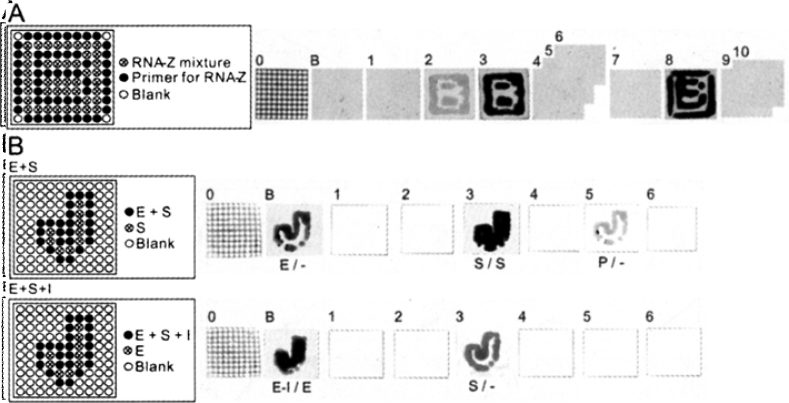
Reaction and separation with 4SR. A. Isothermal RNA-Z reaction in a 100-well MMV and successive separation. The separation was carried out using 3D gels composed of one sheet of blotting gel (4% and 20% skin gels) and 10 sheets of slice gels (10%). RNA-Z reaction was carried out at 50°C in the wells (inside and outside of the line of the letter B, shown by ⊗) of MMV charged with a full RNA-Z reaction mixture. The primer used for this reaction was FITC-labeled. Electrophoresis was performed at 25 V/cm for 60 min. DNAs were viewed by fluorescence. B. Cathepsin E reaction inhibition and separation. In the upper row, the wells of MMV tracing the letter J (shown by ⊗) were filled with fluorescent substrate (S) while those surrounding wells around the letter J (•) were filled with enzyme (Cathepsin E) and substrate (E+S). After the enzymatic reaction, the cleaved peptides and the enzyme were separated by 3D electrophoresis (50 V/cm, 60 min) of the system equipped with a blotting gel (4% and 40% skin gels). The stacked slice gels used were made of 40% acrylamide (6 sheets), detached and sliver-stained after electrophoresis. The outlines of J appeared in gel layers B (blotting gel), 3, and 5, corresponding to the enzyme, the uncleaved substrate peptide (1,375 Da), and the cleaved peptides (674 Da and 719 Da), respectively. For the lower row, the enzyme reaction was carried out in J-outlined wells (•) in vain due to the addition of the inhibitor of pepstatin A, thus failing to generate cleaved peptides. The interpretation of J-outlined (out) and J-figured (in) molecules are shown below in the manner of (out/in) using the symbols, E (enzyme), S (substrate), P (products), and I (inhibitor).
Amplification and separation
Amplification of DNA and RNA is an indispensable process in genetic engineering and other analyses such as gene expression analysis (14), random amplified polymorphic DNA (15), and genome profiling (16). Therefore, RNA-Z reaction (17), which is a derived reaction from the self-sustained sequence replication (3SR), a kind of isothermal amplification of DNA and RNA molecules in vitro (18), was tentatively carried out in a 100-well MMV (Figure 3A). Samples were loaded in the 100-well MMV forming the shape of the alphabet “B”. The RNA-Z reaction was performed at 50°C for 2 h and the products obtained were separated with 4SR (Figure 3A). The images in gel layers 2-3 showed that the reaction was almost equally propagated and the products thus obtained had the same mobility judging from the equality of the fluorescence intensity. By introducing the blotting gel layer that works for stacking samples, the fine resolution of DNAs was ensured reproducibly (six trials) as can be seen here. Evidently, the electrophoresis performed at a lower voltage (less than 25 V/cm) was effective in this experiment in increasing the resolution and preventing from the uneven migration, which generates the so-called smiling pattern of DNAs probably due to the reduction of Joule heat.
Inhibition and separation
To detect an enzyme-inhibition reaction, a cathepsin E inhibition system was chosen. Cathepsin E is an intracellular aspartic proteinase belonging to the pepsin family, mainly expressed in the cells for the immune system, and is believed to contribute to homeostasis by participating in host defense mechanisms (19). The enzyme activity and its inhibition by an inhibitor, pepstatin A, could be clearly monitored by 4SR (Figure 3B). A slice gel used for the detection of small peptides was prepared according to the protocol described by West et al. (20) and could effectively work in measuring the enzyme activity by discriminating enzyme-cleaved and non-cleaved peptides, as shown in Figure 3B.
3D separation
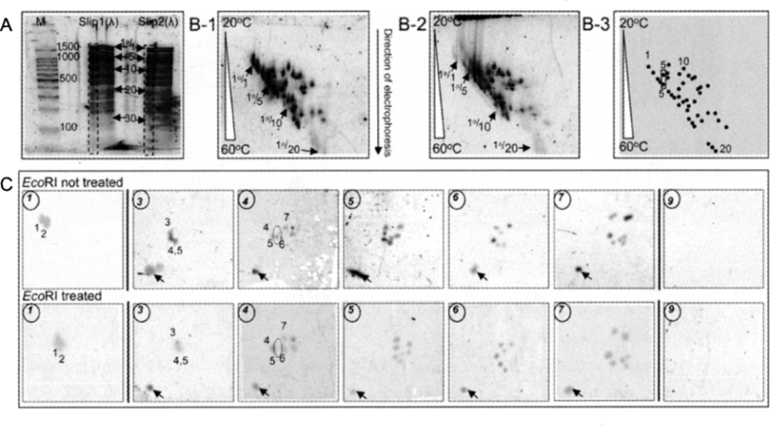
We tried a novel challenge of 3D separation of DNAs with 4SR (Figure 4). To date, 2D gel electrophoresis has been performed on DNA separation, often being accompanied with mass spectrometric (MS) identification (21). This system is well established but is rather costly, time-consuming, and thus less high-throughput. In our experiment, for the first-dimensional (1D) separation, DNA fragments (totally 116) obtained by digesting λ-DNA with Sau3AI were applied to polyacrylamide gel electrophoresis (PAGE) using multi-slot μ-gel, identifying a total of 34 bands (Figure 4A). A slip of μ-gel thus obtained was applied to micro-temperature gradient gel electrophoresis (μ-TGGE) for 2D separation, generating a total of 66 bands (Figure 4B). The Band 5, which was assigned for 1D separation (Figure 4A) containing five DNA fragments theoretically, was separated into two spots upon the 2D separation using μ-TGGE. 3D separation was tried with 4SR after either being treated with restriction enzyme EcoRI overnight or not (Figure 4C). Upon the 3D separation with 4SR, a spot derived from Band 5 (shown with a dotted circle), known to be susceptible to EcoRI digestion, became faint in EcoRI-treated gel in comparison with non-EcoRI-treated one, indicating that the enzyme reaction occurred with EcoRI-susceptible DNA fragment contained in gel. Obviously, the two independent experiments performed here with 4SR are showing the reproducible results with a sufficiently high resolution [though a light degree of spot swellings was observed for the relevant spots due to the treatment of the shrink-and-swell procedure (see Materials and Methods)]. In this experiment, prior to 3D separation, a slab gel (or 2D gel) was clearly allowed to be treated with an additional reaction (which was a restriction enzyme reaction in this case). The reaction can be versatile, such as DNA-to-DNA, protein-to-protein, protein-to-DNA interactions, or various enzyme reactions. This is another reason of the high performance of 4SR in addition to the ability to deal with a large number of samples at a time provided by MMV just before the first separation.
Fig. 4.
3D separation of DNAs with 4SR. A. 1D electrophoretic separation of bacteriophage λ-DNAs in μ-gel at 100 V/cm for 8 min. In Panel A, 100 bp ladder DNA marker and Sau3AI-digested λ-DNAs were migrated. The DNA bands assigned are 1°/1 (2,225 bp), 1°/5 (1,094, 1,082, 1,064, 1,057, and 1,049 bp), 1°/10 (720 and 707 bp), 1°/20 (349, 348, and 346 bp), and 1°/30 (169, 165, 159, and 153 bp), which are composed of individual DNAs indicated as in each parenthesis. The gels of slips 1 and 2 shown with dotted boxes were used for 2D electrophoresis B-1 and B-2, respectively. B. Panel B-1 and B-2: 2D separation by parallel μ-TGGE, that is, both the direction of electrophoresis and temperature gradient are the same. Those spots evidently derived from the 4 bands assigned in 1D gel are indicated. All detectable spots are illustratively shown in Panel B-3. C. 3D separation by 4SR. 3D electrophoresis was performed at 50 V/cm for 30 min. The spots expected to be diminished after the digestion with EcoRI (digested from 1,082 bp to 1,068 bp DNA fragment) are marked with a dotted circle. The arrows indicate the position of the internal marker used (100 bp DNA ladder spots).
Discussion
In the post genomic era, the attention of scientists in the field of analytical sciences is gradually shifting from deciphering the entire genomes and proteomes of particular organisms to elucidating structural and functional variations among various populations of organisms. For such purposes, it is of great importance to establish high-throughput analytical systems. The analytical system developed here, termed as 4SR, was aimed for such purposes and was first realized as a miniaturized apparatus to separate DNAs and proteins in 3D mode with a high-throughput property (Figure 1). Evidently, this owes to the 2D loading of samples (Figure 1B). The MMV of 100 wells in 25 mm square-size gel is already a high-throughput system equivalent to those systems that are currently used (for example, 1,536 wells in a microplate of 8 cm × 12 cm) (10). It will not be so difficult to increase the number of wells per plate once the method is established to handle such a small amount of samples, which is currently being pursued.
Another great advantage of this system resides in the fact that the gels are sliced in advance, which seems to have never been adopted up to now by any other system (Figure 1C). We confirmed through a number of experiments that a stack of slice gels can work as if they were a seamless gel. The convenience of slice gels is that, after the use for electrophoresis, a dump of gels can be separated into pieces and a particular slice gel containing a desired sample can be easily collected and proceeded to the next procedure without the difficulty of slicing gels. The unique merit of using slice gels is, of course, in the ability that they can be stacked in a various combination allowing to adopt affinity gels (which traps a particular kind of molecules) and others.
Since the MMV-facilitated 4SR method is so simple and easy to operate, it can be readily extended to a large-scale analysis of DNA/RNA and protein/peptide in various ways. This technology together with further improvements and developments can be expected to contribute greatly to future studies of genome and proteome analyses.
Conclusion
A novel high-throughput 3D gel electrophoresis method, 4SR, applied for separation and reactions of biopolymers, has been invented and shown to be practically usable. This method can be reinforced as a high-throughput system by introducing the MMV where different reactions can be carried out at the same time. Most significantly, 3D separation of DNAs could be realized for the first time utilizing this method, which enables us to develop further versatile high-throughput measures.
Materials and Methods
Multi-micro vessel
The MMVs used for different experiments were of 16% polyacrylamide gel. The polyacrylamide gel mixture (25 mL) contained 10 mL of 40% acrylamide:bis (9:1) solution, 1.25 mL of 20× TBE (130 mM Tris, 40 mM Boric acid, and 3 mM EDTA) buffer, 12.5 g sucrose, 20% ammonium persulfate (250 μL), 0.0025 g riboflavin, and 13.5 mL twice-distilled H2O.
Slice gel and blotting gel
Slice gels and blotting gels were prepared by the same procedure as described for MMVs (Figure 1). A blotting gel layer was composed of two skins of gels: an upper thin skin (4%) and a lower thick skin (20% or else). After polymerization of the thick skin, the thin skin was formed over it. Usually, slice gels were made of 10% polyacrylamide gel sized (25 mm × 25 mm) by cutting out from a large sheet of gel (12 cm × 12 cm × 1 mm).
Discrete separation of ssDNAs
Three ssDNA fragments of 76, 46, and 23 nucleotides were loaded into a 100-well MMV in the overlapped shape of alphabetical letters “M”, “M”, and “V” with a volume of 0.1 pmol/well. A total of 20 slice gels (10%) were stacked and used for the separation experiment, which tested the effect of long distance separation of 4SR. The slice gels were stacked with care not to allow any air bubble in between the gels. After electrophoresis of around 15 min at 50 V/cm, the slice gels were drawn out from the system, position-marked by carving and stained with silver.
RNA-Z reaction
The isothermal RNA-Z reaction (17) was accomplished in a 100-well MMV and then the long-distance separation of the reaction products was done with 4SR. Prior to the addition of the reaction mixtures, the MMV was soaked twice (1 h for each) in 10 mL of a solution composed of 1 mL of 10× 3SR buffer (600 mM Tris-HCl of pH 8.1, 100 mM KCl, and 100 mM magnesium acetate) (18), 480 μL of 50 mM dithiothreitol (DTT; Wako Pure Chemical Industries Ltd., Osaka, Japan), 200 μL of 100 mM spermidine (Sigma Chemical Co., St. Louis, USA), 400 μL of 25 mM dNTP (N = G, A, T, C), 800 μL of 25 mM NTP (N = G, A, T, C) and 7.12 mL of twice-distilled H2O. The solutions in MMV wells were wiped out cleanly after shaking with the MMV soaked in the permeation solution mentioned above. The MMV was then kept on ice and the RNA-Z reaction mixture was loaded into the individual well according to the order. The RNA-Z reaction mixture (100 μL) contained 10 μL of 10× 3SR buffer, 5 μL of 50 mM DTT, 2 μL of 100 mM spermidine, 4 μL of 25 mM dNTP, 8 μL of 25 mM NTP, 57 μL of 2.2 M trehalose (heated at 90°C before use; Wako Pure), 2 μL of 40 pmol/μL 5′-FITC-labeled primer (5′-CTTTACAAATTTTCCC-3′), 8 μL of 1 pmol template DNA (79 bp), 2 μL HIV-1 reverse transcriptase (Worthington Biochemical Co., Lakewood, USA), and 2 μL TT-7 RNA polymerase (Toyobo Co. Ltd., Osaka, Japan). The ingredients of RNA-Z reaction were mixed at 0°C in order to hold down the reaction. Reaction mixtures (3 μL) were poured into respective wells and the MMV was quickly covered by a bedding gel over it and then sealed in an air-tight plastic bag. The reaction was carried out in an incubator at 50°C for 2 h. The MMV was taken out of the plastic container after the completion of reaction and placed on the blotting (made of 4% and 20% skin gels) and separating (10%; 10 sheets) slice gels located at the inside of the 4SR instrument. Electrophoresis was performed at a low voltage of 25 V/cm for 60 min in order to reduce the uneven migration of DNAs. DNAs were detected by the fluorescence of FITC using a Molecular Imager FX (Bio-Rad Laboratories, Hercules, USA).
Cathepsin E inhibition reaction
The cathepsin E inhibition reaction mixture was prepared following the method reported by Yasuda et al. (22). Cathepsin E (42.8 kDa) and a substrate (peptide of 1.4 kDa), together with pepstatin (0.6 kDa), which is a standard inhibitor of cathepsin E, were kindly provided by Prof. K. Yamamoto of Kyushu University, Japan. The reaction was carried out in a 100-well (16% of acrylamide) MMV at 40°C for 10 min. For the electrophoretic separation of the products, the blotting gels (4% and 40% skin gels) and separating slice gels (40%; 6 sheets) were prepared according to the procedures described by West et al. (20) for the separation of small peptides in polyacrylamide gels. After the electrophoresis of 1 h at 50 V/cm, the gels were replaced from the system, position-marked by carving and stained with silver.
3D separation of DNAs
The bacteriophage λ-DNA (circular DNA of 48,502 bp; GenBank Accession No. J02459; Takara Bio Inc., Shiga, Japan) was digested with Sau3AI (Takara). The digestion reaction mixture contained 1 U of Sau3AI, 10× H buffer, 1 μg of λ-DNA, and sterilized water up to 20 μL. The 1D separation was carried out by applying 100 ng of DNAs for PAGE in a 6% multi-slot μ-gel (23). A portion (1/3) of the gel was cut off and placed in a long-slot μ-gel (24). The long-slot μ-gel was applied to μ-TGGE using a μ-TG apparatus (TAITEC, Saitama, Japan). A temperature gradient of 20°C–60°C was set parallel to the direction of migration of DNAs for the 2D separation. The gel was further subjected to a shrink-and-swell procedure: shrinking by drying at room temperature (to 90% volume of its original) and then swelling by soaking it in a 3 mL of EcoRI reaction mixture containing 2,250 U of EcoRI (Takara), 300 μL of 10× H buffer and twice-distilled H2O up to 3 mL, and then incubated at 37°C for 12 h. After incubation, the gel was set in the 4SR apparatus for the 3D separation of DNAs. The separated DNAs were then stained with SYBR® Green I (Cambrex Bio Science Rockland Inc., Rockland, USA) and detected by the fluorescence using a Molecular Imager FX (Bio-Rad).
Acknowledgements
We thank T. Tayama for providing us with MMVs. This work was supported by the Rational Evolutionary Design of Advanced Biomolecules (REDS) Project, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence sponsored by the Japan Science and Technology Agency (JST).
References
- 1.Frith M.C. The amazing complexity of the human transcriptome. Eur. J. Hum. Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 2.Plebani M. Proteomics: the next revolution in laboratory medicine? Clin. Chim. Acta. 2005;357:113–122. doi: 10.1016/j.cccn.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Napoli C. New advances in microarrays: finding the genes causally involved in disease. Methods Mol. Med. 2005;108:215–234. doi: 10.1385/1-59259-850-1:215. [DOI] [PubMed] [Google Scholar]
- 4.Harbers M., Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat. Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 5.Bodovitz S. Protein biochips: the calm before the storm. Drug. Discov. Today. 2005;10:283–287. doi: 10.1016/S1359-6446(05)03373-8. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y.F. From traditional biomarkers to transcriptome analysis in drug development. Curr. Mol. Med. 2005;1:29–38. doi: 10.2174/1566524053152915. [DOI] [PubMed] [Google Scholar]
- 7.Ge H. UPA, a universal protein array system for quantitative detection of protein-protein, protein-DNA, protein-RNA and protein-ligand interactions. Nucleic Acids Res. 2000;28:e3. doi: 10.1093/nar/28.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekins R.P. Multi-analyte immunoassay. J. Pharm. Biomed. Anal. 1989;7:155–168. doi: 10.1016/0731-7085(89)80079-2. [DOI] [PubMed] [Google Scholar]
- 9.Schena M. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 10.Battersby B.J., Trau M. Novel miniaturized systems in high-throughput screening. Trends Biotechnol. 2002;20:167–173. doi: 10.1016/s0167-7799(01)01898-4. [DOI] [PubMed] [Google Scholar]
- 11.Hong B.J. DNA microarrays on nanoscale-controlled surface. Nucleic Acids. Res. 2005;33:e106. doi: 10.1093/nar/gni109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoll D. Protein microarrays: applications and future challenges. Curr. Opin. Drug Discov. Devel. 2005;8:239–252. [PubMed] [Google Scholar]
- 13.Rosenthal A. Solid-phase methods for sequencing of oligodeoxyribonucleotides and DNA. Methods Enzymol. 1987;155:301–331. doi: 10.1016/0076-6879(87)55022-4. [DOI] [PubMed] [Google Scholar]
- 14.Eisen M.B., Brown P.O. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 15.Williams J.G. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naimuddin M., Nishigaki K. Genome Analysis technologies: towards species identification by genotype. Brief. Funct. Genomic. Proteomic. 2003;1:356–371. doi: 10.1093/bfgp/1.4.356. [DOI] [PubMed] [Google Scholar]
- 17.Breaker R.R., Joyce G.F. Emergence of a replicating species from an in vitro RNA evolution reaction. Proc. Natl. Acad. Sci. USA. 1994;91:6093–6097. doi: 10.1073/pnas.91.13.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guatelli J.C. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl. Acad. Sci. USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasuda Y. A new selective substrate for cathepsin E based on the cleavage site sequence of alpha2-macroglobulin. Biol. Chem. 2005;386:299–305. doi: 10.1515/BC.2005.036. [DOI] [PubMed] [Google Scholar]
- 20.West M.H.P. Polyacrylamide gel electrophoresis of small peptides. Electrophoresis. 1984;5:133–138. [Google Scholar]
- 21.Gygi S.P. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda Y. Characterization of new fluorogenic substrates for the rapid and sensitive assay of cathepsin E and cathepsin D. J. Biochem. (Tokyo) 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- 23.Salimullah M. Efficient SNP analysis enabled by joint application of the μTGGE and heteroduplex methods. Cell. Mol. Biol. Lett. 2005;10:237–245. [PubMed] [Google Scholar]
- 24.Biyani M., Nishigaki K. Hundredfold productivity of genome analysis by introduction of microtemperature-gradient gel electrophoresis. Electrophoresis. 2001;22:23–28. doi: 10.1002/1522-2683(200101)22:1<23::AID-ELPS23>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]