Abstract
Proteins containing an Rho GTPase-activating protein (RhoGAP) domain work as molecular switches involved in the regulation of diverse cellular functions. The ability of these GTPases to regulate a wide number of cellular processes comes from their interactions with multiple effectors and inhibitors, including the RhoGAP family, which stimulates their intrinsic GTPase activity. Here, a phylogenetic approach was applied to study the evolutionary relationship among 59 RhoGAP domain-containing proteins. The sequences were aligned by their RhoGAP domains and the phylogenetic hypotheses were generated using Maximum Parsimony and Bayesian analyses. The character tracing of two traits, GTPase activity and presence of other domains, indicated a significant phylogenetic signal for both of them.
Key words: Bayesian analysis, character tracing, parsimony, phylogenomics, protein domain, RhoGAP
Introduction
The Rho GTPase-activating proteins (RhoGAPs) are defined by the presence of a 150-amino-acid homolog region that is designated as the RhoGAP domain. This domain is necessary and sufficient for GAP activity and shares at least 20% sequence identity among its family members 1., 2.. Proteins containing an RhoGAP domain act as molecular switches involved in the regulation of diverse cellular functions, including actin cytoskeleton rearrangements, regulation of gene transcriptions, cell cycle regulation, control of apoptosis, and membrane trafficking 2., 3., 4., 5.. Rho GTPases cycle between active and inactive GTP-bound states. The control of these states is regulated by three main classes of proteins: guanine nucleotide exchange factors, guanine nucleotide dissociation inhibitors, and GAPs. To date, at least 21 Rho GTPases have been defined, among which only three (RhoA, Cdc42, and Rac1) are well characterized. Therefore, most studies have been focusing on these three proteins.
The ability of Rho GTPases to regulate a wide number of cellular processes comes from their interactions with multiple effectors or inhibitors. One class of these inhibitors is the RhoGAP family, which stimulates GTPase activity by enhancing the intrinsic rate of GTP hydrolysis.
In the early analyses of the human genome sequence, 77 different genes containing the RhoGAP domain were found. Further studies have demonstrated that many of these genes are simple gene sequence variations or single nucleotide polymorphisms 6., 7.. The structural data available for RhoGAP domain-containing proteins showing their complexity with Rho GTPases (Cdc42 and RhoA) demonstrated the 3D workflow for RhoGAP-mediated GTP-hydrolysis, and highlighted the importance of a well-conserved arginine residue present in the active site that acts as a conformation stabilizer needed for hydrolysis 8., 9., 10..
Recently, a novel member of the RhoGAP family, ARHGAP21 (Rho GTPase-activating protein 21, alias ARHGAP10), was cloned and characterized in our laboratory (11). In addition to the RhoGAP domain, ARHGAP21 presents a PH domain and a P-loop-containing PDZ domain. This gene is widely expressed at high levels in muscle and brain, and is up-regulated during myeloid and erythroid maturation, suggesting a potential role for this RhoGAP in regulating cell differentiation (11).
The aim of this study is to infer the evolution of the RhoGAP superfamily using a phylogenetic approach, to determine the roles of other domains and their main GTPase activities in this evolutionary history, and to provide a tool that could render some insights regarding subfamily protein functions using RhoGAP-containing proteins as a model.
Results
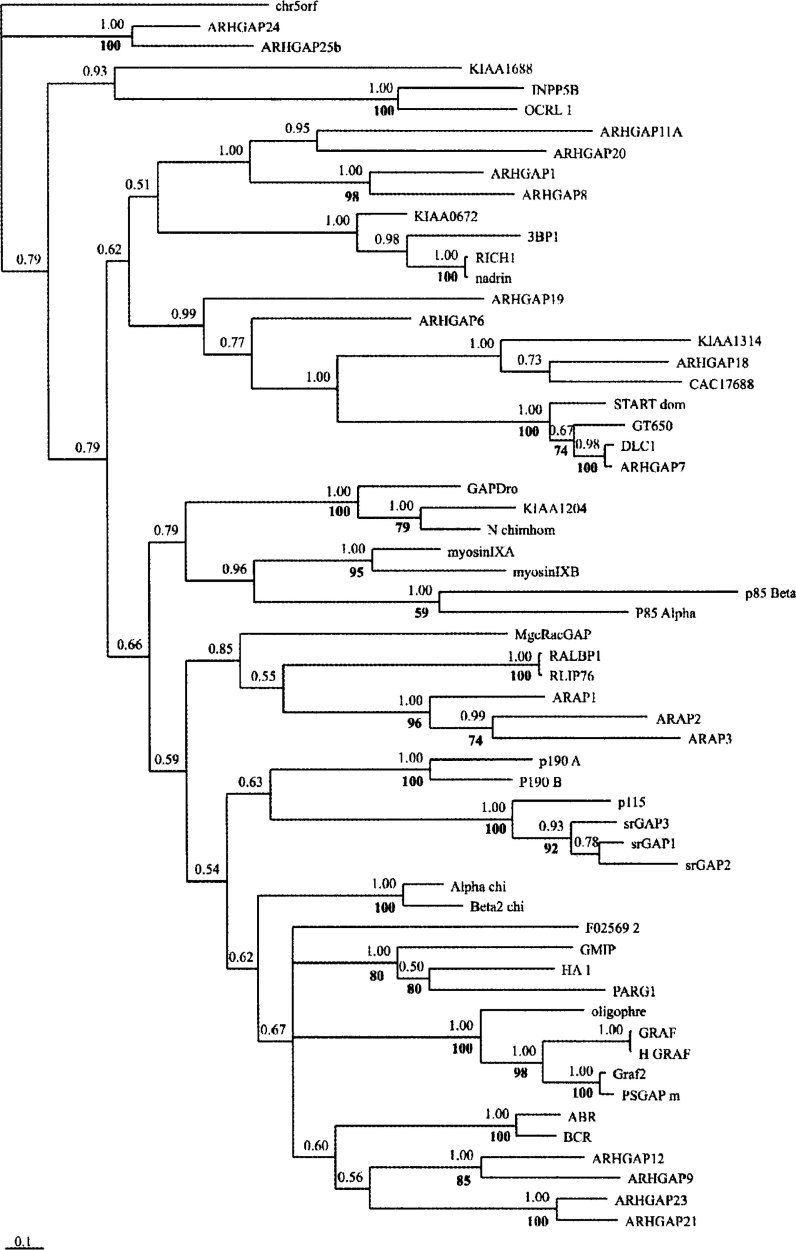
The full dataset contains 267 amino acids, of which 161 are parsimony-informative. Parsimony searches of the equally weighed dataset resulted in 12 equally parsimonious trees with 3,213 steps (CI=0.449; RI=0.525). The strict consensus tree was mostly not resolved at internal nodes (data not shown), but almost all terminal branches showed strong bootstrap support (Figure 1).
Fig. 1.
The Bayesian tree based on amino acid sequences of the RhoGAP domain from RhoGAP domain-containing proteins. Values above the branches indicate the Posterior Probability (PP) and the bold numbers indicate the bootstrap values from 500 replications (where it exceeds 50%).
A Bayesian tree recovered mostly the same terminal relationships by Maximum Parsimony (MP) analyses with strong values of Posterior Probability (PP). However, internal branches have from low to moderate PP values (Figure 1).
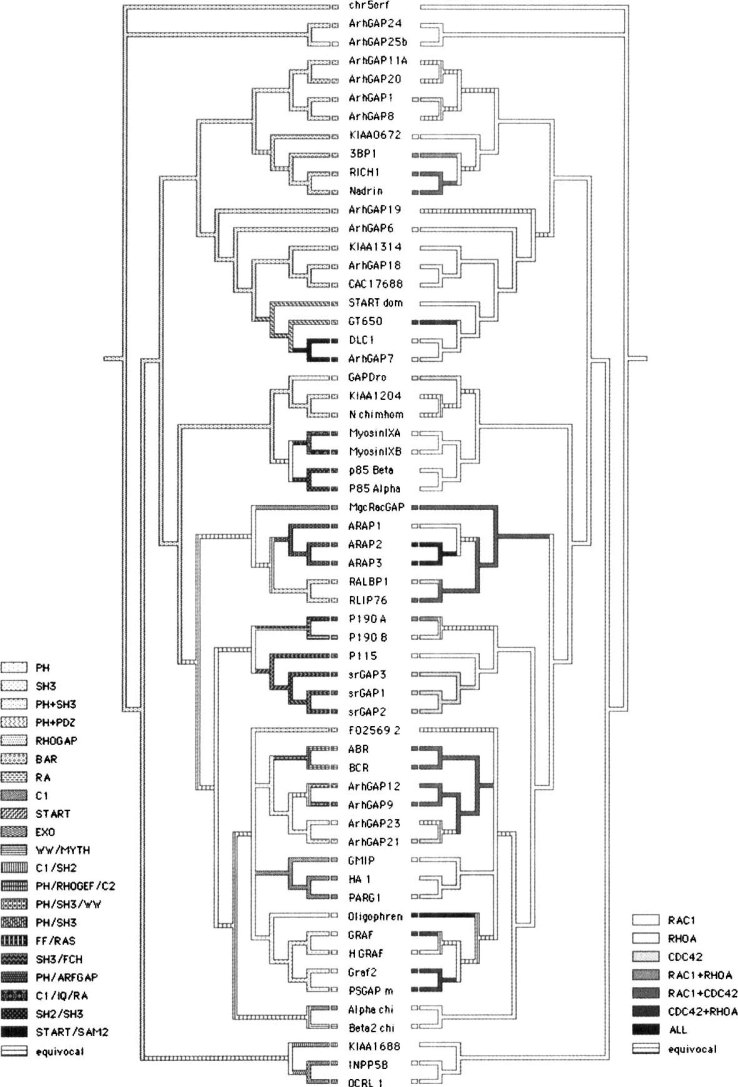
The two characters investigated (domains and GTPase activity) showed a significant phylogenetic signal (P=0.003), which suggests that the distribution of these traits among the proteins can be explained by their phylogenetic relationships (Figure 2). The optimization of the other domains over our phylogenetic hypothesis suggests that the ancestral state of these proteins involves solely the presence of the RhoGAP domain and the activity toward Rac1.
Fig. 2.
Domain presence (left side) and GTPase activity (right side) in the RhoGAP domain-containing proteins traced along the proposed phylogeny.
The character tracing of these traits suggests an overall pattern on which proteins sharing equal domains also share equal GTPase activity. The clade joining KIAA0672 + 3BP1 + RICH1 + Nadrin was recovered both by MP and Bayesian analyses with strong support. All proteins in this clade share the presence of the BAR (Bin-Amphiphysin-Rvs) domain and GTPase activity toward Rac1, solely or in addition to other GTPases. The same rule can be applied to the clade joining STARTdom + GT650 + DLC1 + AHRGAP7. All of them share the presence of the START (steroidogenic acute regulatory proteinrelated lipid transfer) domain and all, but STARTdom, have GTPase activity over RhoA. GTPase activity is unknown for STARTdom. The clade joining GMIP + HA1 + PARG is composed by proteins containing the C1 domain in addition to their GTPase activity toward RhoA.
The clade joining AHRGAP11A + AHRGAP20 + AHRGAP1 + AHRGAP8 has in common the absence of other domains that are not RhoGAP. The GTPase activity of this group is known only for AHRGAP1, which is active toward Cdc42. Considering other terminal clades, we can presume that other AHRGAPs within this group can show the same GTPase activity toward Cdc42.
On the other hand, the clade joining P115 + srGAP3 + srGAP1 + srGAP2 shares the presence of the FCH (FER/CIP4-homology) domain, however, P115 has GTPase activity over Rac1, while the other three proteins have activity toward Cdc42.
Discussion
Phylogenetic reconstruction and bioinformatics analyses that integrate evolutionary considerations are becoming increasingly important tools for applied fields. Numerous gene sequences were generated in the genomics age with little or no accompanying experimental determination of functional information or evolutionary relationships. Previous works from Peck et al. (12) and Moon and Zheng (2) also present a phylogenetic approach on the RhoGAP family; however, the authors did not indicate the methodology applied neither did they present any support analyses for their cladograms.
In this work, bioinformatics and phylogenomics tools were used to present a phylogenetic relationship of 59 members of the RhoGAP superfamily. All amino acid alignments and subsequent phylogenetic tree constructions were based on the RhoGAP domain sequence. We demonstrated that these RhoGAP domain-containing proteins, with the conservative argenine residue, form a monophyletic group, that is, all of them share a common protein ancestor in their evolutionary history.
The tracing for GAP activity toward the most studied RhoGTPases (RhoA, Rac1, and Cdc42) (Figure 2) indicates that this trait presents a strong phylogenetic signal (P=0.003), contrasting with previous findings of Peck et al. (12).
The analysis of the resulting phylogenetic tree has suggested that the ancestral state for GTPase activity is the affinity to Rac1. It is still difficult to determine the gap activity by only analyzing the protein sequence; the GAP assay 13., 14. is the most reliable way to determine activity. The phylogenetic approach may give a clue, once it is capable of clustering together different proteins that share common substrates as can be seen on the clades of KIAA0672 + 3BP1 + RICH1 + Nadrin and srGAP3 + srGAP1 + srGAP2. Speculations regarding protein specific functions (only using the GTPase activity character) may be avoided for now, because the affinity for the same GTPase does not imply in the same function, since each GTPase may present contrasting functions in different pathways (15).
Furthermore, structural and molecular biology studies are needed to elucidate the exact amino acid composition involved in determining specificity and how the differences in this composition can affect the 3D protein structure and its interaction with Rho GTPases.
In addition to the RhoGAP domain, the members of this superfamily usually contain other functional motifs. Therefore, RhoGAPs might catalyze or participate in enzymatic reactions other than the enhancement of the intrinsic GTP hydrolysis of Rho GTPases, and sometimes apparently aiding the Rho protein to signaling (2).
Somehow the presence of additional domains was linked to the RhoGAP domain structure because, even focusing the alignment exclusively on the RhoGAP domain sequence, the phylogeny joined in clades of different proteins sharing the same additional domains with strong bootstrap and PP, that is, the ARHGAPs were divided into two groups. One is composed of a clade including the ARHGAPs 9, 12, 21, and 23, presenting an RhoGAP domain and a pH domain accompanied or not by additional domains (Figure 2). The other is composed of the terminals ARHGAPs 1, 8, 11A, and 20, presenting only the RhoGAP domain, except the ARHGAP20 that present an RA domain.
Interactions among genomics, evolution, and bioinformatics go further than sequence alignment and relationship elucidation among species. Evolutionary analysis may help researchers design new strategies to understand protein or gene interactions and their functionalities and might provide an insight for new experiments. In conclusion, a phylogenetic study of the RhoGAP domain-containing proteins has demonstrated that there is a strong evolutionary relationship among the RhoGAP superfamily members, especially when they share common motifs or GAP activity.
Materials and Methods
Materials
All protein sequences used here were obtained from the GenBank database (http://ncbi.nlm.nih.gov/Genbank/) at the National Center for Biotechnology Information (NCBI), as well as from the Swiss-Prot/TrEMBL database (http://expasy.org/sprot/) at the Swiss Institute for Bioinformatics and at the European Bioinformatics Institute (Table 1).
Table 1.
Selected Rho GTPase-Activating Proteins
| No. | Protein | GenBank | SwissProt |
|---|---|---|---|
| 1 | 3BP-1 | Q9Y3L3 | |
| 2 | ABR | NP_068781.2 | |
| 3 | α-Chimaerin | CAA35769.1 | |
| 4 | ARAP1 | NP_056057.1 | |
| 5 | ARAP2 | BAA25506.1 | |
| 6 | ARAP3 | CAC83946.1 | |
| 7 | ARHGAP1 | NP_004299 | |
| 8 | ARHGAP6 | NP_038286.1 | |
| 9 | ARHGAP7 | Q63744 | |
| 10 | ARHGAP8 | CAB90248.1 | |
| 11 | ARHGAP9 | BAB56159.1 | |
| 12 | ARHGAP11A | NP_055598.1 | |
| 13 | ARHGAP12 | NP_060757.4 | |
| 14 | ARHGAP18 MacGAP | NP_277050 | |
| 15 | ARHGAP19 | NP_116289.4 | |
| 16 | ARHGAP20 | AAS45466.1 | |
| 17 | ARHGAP21 | AF480466.1 | |
| 18 | ARHGAP23 | BAA96025.1 | |
| 19 | ARHGAP24 | NP_112595.1 | |
| 20 | ARHGAP25b | NP_055697.1 | |
| 21 | β-Chimaerin | AAA16836.1 | |
| 22 | BCR | NP_004318.2 | |
| 23 | CAC17688.2 | CAC17688.2 | |
| 24 | CHR50RF | NP_057687.1 | |
| 25 | DLC-1 | NP_006085.2 | |
| 26 | GAPDro | AAF44627.1 | |
| 27 | GMIP | NP_057657.1 | |
| 28 | GRAF | NP_055886.1 | |
| 29 | GRAF-2 | BAB61771 | |
| 30 | GT650 (DLC2) | NP_443083.1 | |
| 31 | HA-1 (KIAA0233) | BAA13212.1 | |
| 32 | H-Graf | CAA71414.2 | |
| 33 | INPP5B | AAA79207.1 | |
| 34 | KIAA0672 | BAA31647 | |
| 35 | KIAA1204 | BAA86518.1 | |
| 36 | KIAA1314 | BAA92552.1 | |
| 37 | KIAA1688 | BAB21779.1 | |
| 38 | MgcRacGAP | NP_037409.2 | |
| 39 | Myosin_IXA | NP_008832.1 | |
| 40 | Myosin_IXB | NP_004136.2 | |
| 41 | Nadrin | NP_060524.4 | |
| 42 | N-Chimaerin homolog | AAB81198.1 | |
| 43 | OCRL-1 | NP_001578.2 | |
| 44 | Oligophrenin-1 | NP_002538.1 | |
| 45 | p190-A | AAF80386.1 | |
| 46 | p190-B | NP_001164.2 | |
| 47 | P85-alpha | P27986 | |
| 48 | P85-beta | NP_005018.1 | |
| 49 | PARG1 | NP_004806.2 | |
| 50 | PSGAP | AAK18175.1 | |
| 51 | RALBP1 | NP_006779.1 | |
| 52 | RHG4 (p115) | CAA55394.1 | |
| 53 | RICH-1 | CAC37948.1 | |
| 54 | RLIP76 | AAB00103.1 | |
| 55 | srGAP1 | BAA92542.1 | |
| 56 | srGAP2 | BAA32301.1 | |
| 57 | srGAP3 | CAC22407.1 | |
| 58 | START domain | NP_055540.2 | |
| containing 8 (KIAA0189) | CAC22407.1 | ||
| 59 | HA-1 (KIAA0233) | BAA13212.1 |
The sequences were aligned by their RhoGAP domains with additional 100 N-terminal residues, primarily using ClustalW version 1.83 (16) under default settings, followed by adjustment by eye using the BioEdit version 6.0.7 (Ibis Therapeutics, Carlsbad, USA). All alignment files, the protein sequences in the FASTA format, and other related colored materials are available for download at http://www.hemocentro.unicamp.br/submission/.
Phylogenetic analyses
The Bayesian analysis was carried out by using MrBayes version 3.1.2 17., 18. with the mixed model of amino acid substitution provided in the package. Six simultaneous chains were conducted for 1.0×106 generations, sampling trees every 100 cycles. The first 1,000 trees were discarded as “burn in”. For all analyses, chr5orf was used as an outgroup to root the tree, based on the absence of the conservative argentine residue.
The MP analyses were performed with PAUP* 4.0b10 (19) on the entire dataset using a heuristic search with 500 random taxon addition replicates, TBR branch-swapping, gaps scored as missing data, and all characters equally weighted. A strict consensus tree was computed whenever multiple equally parsimonious trees were obtained. The robustness of each branch was determined using the nonparametric bootstrap test (20) with 500 replicates and 10 random taxon additions.
Character optimization
MacClade 4.08 (21) was used to perform character optimization analyses. We investigated the evolution of two characters that were superimposed onto the Bayesian tree proposed for the RhoGAP-containing proteins: the presence of different domains in addition to RhoGAP, and the GTPase enhancing activity toward the most studied Rho GTPases (Racl, RhoA, and Cdc42). For domain identification, a search of the PFAM database version 19.0 (22) was performed using the HMMPFAM tool from the HMMER suite version 2.3.2 (23) with the Ε-value cutoff for the persequence set to 1.0E-10. This character had 20 unordered character states plus a 21st character state corresponding to the absence of additional domains other than RhoGAP. The GTPase character had eight character states representing the affinity toward one or two Rho GTPases; these data was mined by searching PubMed (http://www.ncbi.nlm.nih.gov/entrez) at NCBI.
To test whether there was a phylogenetic signal in the characters traced, we used the methodology proposed by Wahlberg (24) that was modified from the PTP test described by Faith and Cranston (25). The test compared the number of steps of the tree constructed with the actual data, with the number of steps in the trees obtained for each random reshuffling of the separated character states. We performed 300 random reshufflings of character states among the fixed terminal proteins by using Mesquite version 1.06 (http://www.mesquiteproject.org). The probability (P) that the observed pattern does not differ from random is given as (n + 1)/300, where n is the number of replications no bigger than that of the actual steps. A significant phylogenetic signal was observed when Ρ is less than 0.05 (25).
Authors’ contributions
MMB participated in the design of the study, sequence alignment, bioinformatics analyses, and drafted the manuscript. KLSB participated in all automatic alignment and eye refinements, interpretations of the bioinformatics results, and drafted the manuscript. FFC and STOS conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; Grant 03/06621-5 to MMB) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; fellowship to KLSB). We thank Ms. Raquel Foglio for English revision.
References
- 1.Aitsebaomo J. p68RacGAP is a novel GTPase-activating protein that interacts with vascular endothelial zinc finger-1 and modulates endothelial cell capillary formation. J. Biol. Chem. 2004;279:17963–17972. doi: 10.1074/jbc.M311721200. [DOI] [PubMed] [Google Scholar]
- 2.Moon S.Y., Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 3.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 4.Lamarche N., Hall A. GAPs for rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 5.Van Aelst L., D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 6.Sachidanandam R. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Nassar N. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 9.Rittinger K. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 10.Rittinger K. Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 11.Besseres D.S. ARHGAPIO, a novel human gene coding for a potentially cytoskeletal Rho-GTPase activating protein. Biochem. Biophys. Res. Commun. 2002;294:579–585. doi: 10.1016/S0006-291X(02)00514-4. [DOI] [PubMed] [Google Scholar]
- 12.Peck J. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 2002;528:27–34. doi: 10.1016/s0014-5793(02)03331-8. [DOI] [PubMed] [Google Scholar]
- 13.Manser E. Identification of GTPase-activating proteins by nitrocellulose overlay assay. Methods Enzymol. 1995;256:130–139. doi: 10.1016/0076-6879(95)56018-1. [DOI] [PubMed] [Google Scholar]
- 14.Manser E. Diversity and versatility of GTPase activating proteins for the p21rho subfamily of ras G proteins detected by a novel overlay assay. J. Biol. Chem. 1992;267:16025–16028. [PubMed] [Google Scholar]
- 15.Bishop A.L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson J.D. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 18.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 19.Swofford D.L. PAUP*: Phylogenetic Analysis using Parsimony (and Other Methods), version 4.0b10. CD-ROM. Sinauer Associates; Sunderland, USA: 2002. [Google Scholar]
- 20.Felsenstein J. Confidence limits on phytogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Maddison W.P., Maddison D.R. MacClade: Analysis of Phylogeny and Character evolution, version 3.08. CD-ROM. Sinauer Associates; Sunderland, USA: 1999. [Google Scholar]
- 22.Sonnhammer E.L. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 23.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 24.Wahlberg N. The phylogenetics and biochemistry of host-plant specialization in Melitaeine butterflies (Lepidoptera: Nymphalidae) Evolution. 2001;55:522–537. doi: 10.1554/0014-3820(2001)055[0522:tpaboh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Faith D.P., Cranston P.S. Could a cladogram this short have arisen by chance alone? On permutation tests for cladistic structure. Cladistics. 1991;7:1–28. [Google Scholar]