Abstract
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-TOF/TOF-MS), incorporated with online database searching, were performed to investigate differential proteins of breast cancer and adjacent normal breast tissues. Considering that serum albumin is abundantly presented in normal control samples, 15 differential spots detected in 11 out of 12 (91.7%) breast cancer samples were identified by online SIENA-2DPAGE database searching and MALDI-TOF/TOF-MS analysis. The results indicate that pathological changes of breast cancer are concerned with augmentation of substance metabolism, promotion of proteolytic activity, decline of activity of some inhibitors of enzymes, and so on. Some important proteins involved in the pathological process of breast cancer with changed expression may be useful biomarkers, such as alpha-1-antitrypsin, EF-1-beta, cathepsin D, TCTP, SMT3A, RPS12, and PSMA1, among which SMT3A, RPS12, and PSMA1 were first reported for breast cancer in this study.
Key words: biomarker, breast cancer, 2D-PAGE, mass spectrometry, molecular mechanism
Introduction
Breast cancer is a much commonly diagnosed cancer that is a leading cause of death for women (1). The tumorigenesis and progression toward malignancy of breast cancer are an extremely complex event involved in multiple agents, including hormones, genetic mutations, and various physical, chemical, and organismic agents from outside environment 2., 3.. Although breast cancer has been studied extensively during the past several decades, the underlying molecular mechanism is currently not fully understood. Genes are the essential genetic materials to control cellular biological characteristics while proteins perform varied and vital cellular functions in movement, and all cellular behaviors in life can almost be interpreted in the protein level. In both category and abundance, the expressed proteins in breast cancer cells are different from those in normal breast cells. To elucidate the underlying molecular mechanism and search for new markers involved in clinical diagnosis, treatment, and prognosis, in recent years many efforts have been made to study the differentially expressed proteins in breast cancer cells compared with normal controls by proteomic methods 4., 5., 6.. The known genetic background and consistent cellular composition are the main characteristics of cell lines distinguished from tissues; therefore, breast cancer cell lines are the most common samples used to make proteomic analysis of human breast cancer in the mechanism of tumorigenesis, the effect and resistance of drugs, and prognosis 7., 8., 9.. However, the growth and progression of breast tumor cells not only depend on their malignant potential but also depend on agents presented in the tumor microenvironment. There are interactions among cells and between cells and agents in their surroundings. Proteins extracted from cell lines are only a part of the total proteins correlated to breast cancer. To make the best of obtaining all the information of proteins correlated to breast cancer, some other samples have been used for breast cancer proteomic analysis in recent several years, including tissue, serum, nipple aspirate fluid, interstitial cells, and so on 10., 11., 12., 13..
In this study, two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-TOF/TOF-MS), incorporated with online database searching, were performed to investigate differential proteins of breast cancer and adjacent normal breast tissues. The majority of pI values of proteins are within 5.0–8.0, therefore, the linear immobilized pH gradient (IPG) strips of pH 5.0–8.0 were used.
Results
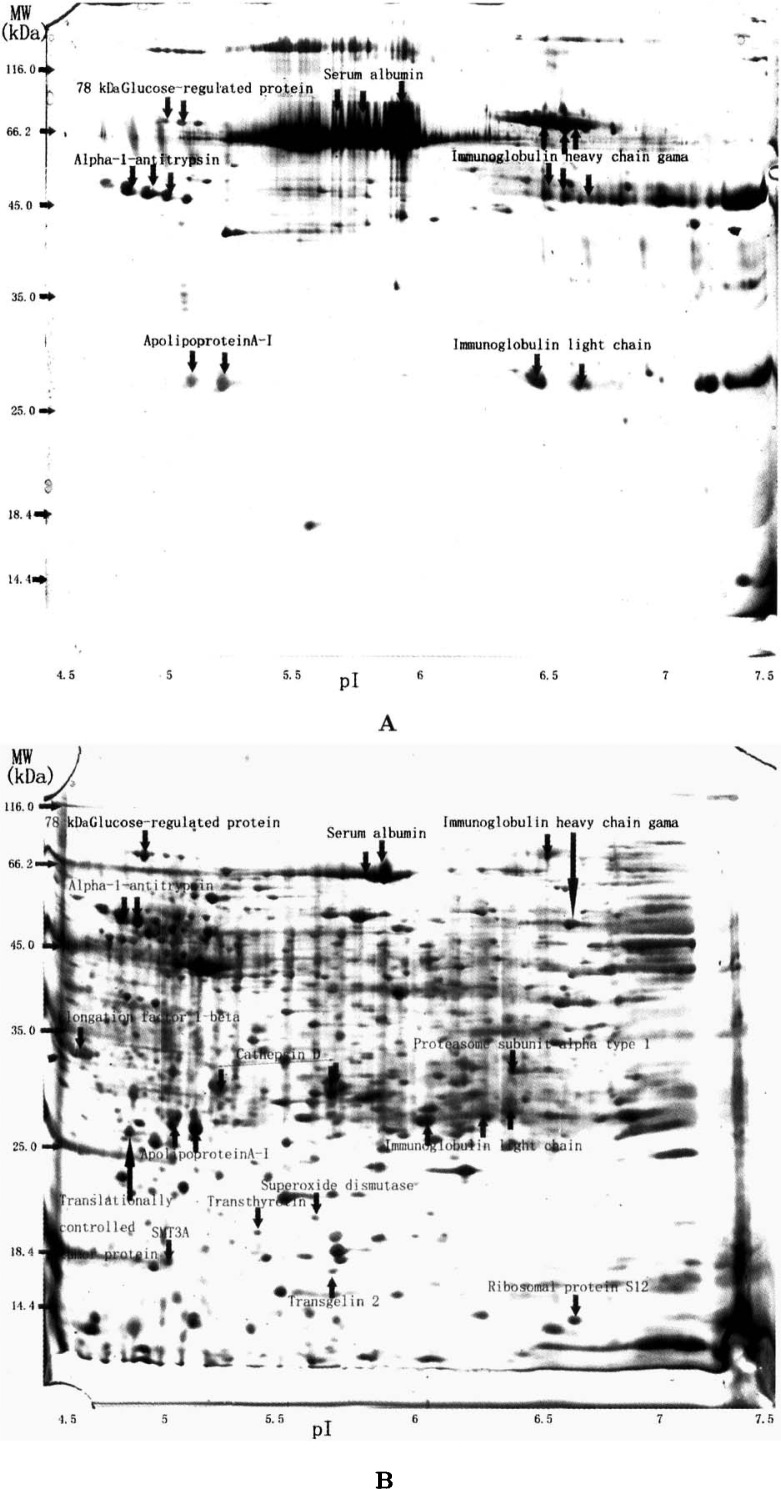
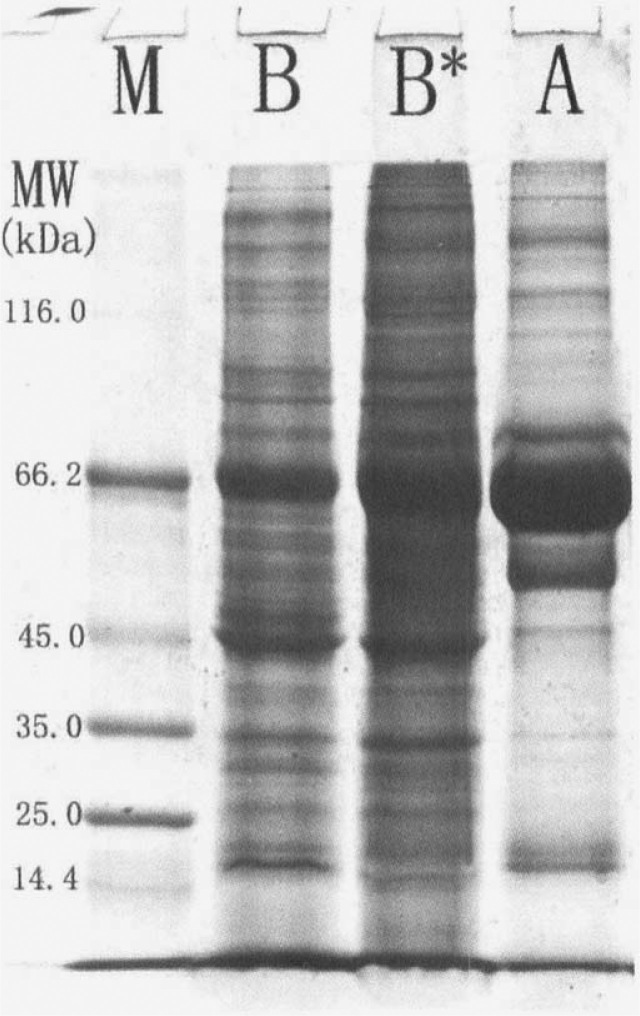
More than 85% of the 2D gel images of all samples from both breast cancer and normal breast tissues were analyzed. As a result, about 1,000 spots from breast cancer tissue and 150 spots from normal breast tissue were detected (Figure 1). According to the image of sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE; Figure 2), we found that the protein content of the main strip (66.2 kDa) in normal control was two folds more than that in breast cancer sample. The majority of the proteins in normal control was identified as serum albumin (10). The proportion of serum albumin to the total protein in normal breast tissue was reckoned to be approximately 80%. Furthermore, only about 400 spots were detected in breast cancer tissue when 50% loading content of samples was used in isoelectronic focusing (IEF). Therefore, proteins were thought to be up-regulated in breast cancer tissue when their abundance was four folds more than that in normal control. Proteins that increased less than four folds were thought to be possibly up-regulated. The other proteins with unchanged or decreased abundance were thought to be down-regulated in breast cancer tissue.
Fig. 1.
The silver-staining 2D gels of proteins from normal breast tissue (A) and breast infiltrating ductal carcinoma tissue (B).
Fig. 2.
The SDS-PAGE gel (coomassie brilliant blue R-250-staining). M: Protein marker; A: Normal breast tissue; Β: Breast infiltrating ductal carcinoma tissue; Β*: Breast infiltrating ductal carcinoma tissue with two-fold loading content more than that in A or Β.
Consequently, 15 differential spots were detected in 11 out of 12 (91.7%) breast cancer samples. These spots were then identified by searching in the online SIENA-2DPAGE database and MALDI-TOF/TOF-MS analysis. Among the 15 spots, 6 were up-regulated, 5 were down-regulated, and 4 (<4 folds) were possible to be up-regulated in the infiltrating ductal carcinoma samples (Table 1).
Table 1.
Protein Annotations from Breast Cancer and Normal Breast Tissues Identified by 2-DE*
| Group | Spot ID | MW | pI | Accession No. | Spots identified |
|---|---|---|---|---|---|
| Down-regulation |
Pd1 | 67,502 | 5.98 | P02768 | Serum albumin |
| Pd2 | 88,029 | 7.54 | P99006 | Immunoglobulin heavy chain gama | |
| Pd3 | 58,614 | 4.91 | P01009 | Alpha-1-antitrypsin | |
| Pd4 | 23,989 | 5.12 | P02647 | ApolipoproteinA-I (apoA-I) | |
| Pd5 | 25,705 | 6.41 | P99007 | Immunoglobulin light chain | |
| Probably Up-regulation |
Pd6 | 73,578 | 4.99 | P11021 | 78 kDa Glucose-regulated protein (GRP78) (immunoglobin heavy chain binding protein) (BIP) |
| Pd7 | 16,086 | 5.41 | P00441 | Superoxide dismutase (SOD) | |
| Pd8 | 14,000 | 5.05 | P02766 | Transthyretin (TBPA) | |
| Pms1 | 21,072.5 | 7.63 | P37802 | Transgelin 2 | |
| Up-regulation | Pd9 | 29,514 | 4.57 | P24534 | Elongation factor 1-beta (EF-1-beta) |
| Pd10 | 28,206 | 5.27 | P07339 | Cathepsin D | |
| Pd11 | 21,788 | 4.87 | P13693 | Translationally controlled tumor protein (TCTP) | |
| Pms2 | 11,704 | 5.89 | P55854 | Small ubiquitin-related modifier 3 precursor (SMT3A) (SUM03) | |
| Pms3 | 14,905 | 6.81 | P63324 | Ribosomal protein S12 (RPS12) | |
| Pms4 | 29,578.9 | 6.15 | P25786 | Proteasome subunit alpha type 1 (PSMA1) | |
Pd: Proteins identified by searching in the SIENA-2DPAGE database; Pms: Proteins identified by MALDI-TOF-TOF-MS; MW: molecular weight.
Discussion
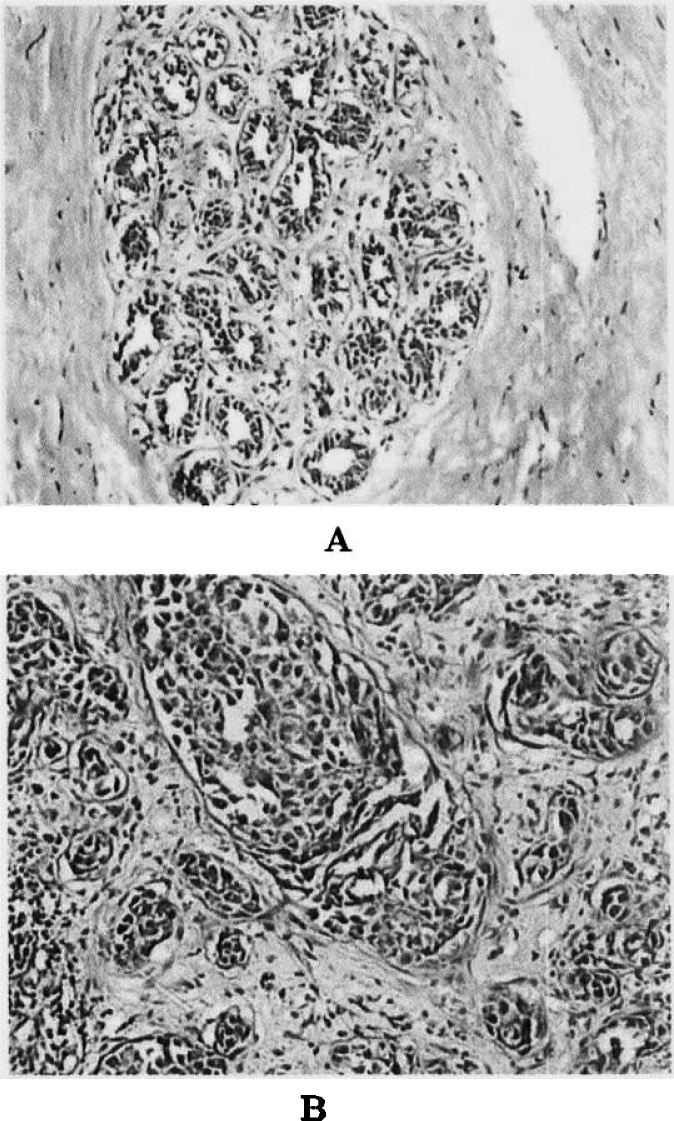
Tumorigenesis of breast cancer is a much more complicated event correlated with multiple agents in vivo and in vitro 14., 15., 16.. Carcinomatous changes of cells are tightly concerned with agents in microenvironment 17., 18.. Changes of protein expressions in breast cancer tissues may play an important role in the tumorigenesis of breast cancer. Proteins from tissues are better to show cellular biological characteristics than those from cell lines (19). The structure and function of cancer cells are different from those of normal cells, thus proteins expressed by cancer cells are different from those by normal cells. Proteins from tissues comprise not only intracellular proteins but also proteins secreted to the outside of cells, and extracellular proteins may be secreted from either non-breast cells or non-breast cancer cells. Furthermore, in this study the normal breast tissues contained rich connective tissues but few breast ductal and lobular cells that presented non-active function in the non-lactation period of volunteers with the age between 32–60 years (Figure 3). Therefore, many proteins with low abundance from normal breast cells were not detected. The results in both single SDS-PAGE gel (Figure 2) and 2D-PAGE gel (Figure 1) showed that proteins expressed in cancer tissue were extremely different from those in normal tissue. Since serum albumin is the majority of the total protein content in normal tissue sample, the loading content of each protein in normal control was quite different from that in cancer tissue sample. Consequently, many proteins were not detected by 2D-PAGE, and differential protein spots in 2D-PAGE gels were not completely differential proteins in expression but maybe the same abundant protein that was possibly different in expression. On the basis of serum albumin being abundantly presented in normal tissue, proteins were thought to be up-regulated in breast cancer tissue when their abundance was four folds more than that in normal control. Proteins with unchanged or decreased abundance were thought to be down-regulated in breast cancer tissue. Proteins that increased less than four folds in 2D-PAGE were thought to be possibly up-regulated because the difference may be not resulted from breast cancer.
Fig. 3.
The hematoxylin and eosin (H&E)-staining images (100 folds) of normal breast tissue (A) and breast infiltrating ductal carcinoma tissue (B).
In this study, five proteins were down-regulated in breast cancer tissue, including serum albumin, immunoglobulin heavy chain gama, immunoglobulin light chain, apolipoproteinA-I (apoA-I), and alpha-1-antitrypsin. Serum albumin, whose main function is the regulation of the colloidal osmotic pressure of blood, is the main protein of plasma. Both immunoglobulin heavy chain gama and immunoglobulin light chain are components of seroglobulin. ApoA-I, which is the major protein of plasma high density lipoprotein (HDL), participates in the reverse transport of cholesterol from tissues to the liver for excretion by promoting cholesterol efflux from tissues and by acting as a cofactor for the lecithin cholesterol acyltransferase. The above four proteins are the exactly components of plasma proteins, and down-regulation of those proteins suggests that blood supply for cancer tissue is less than that for normal breast tissue.
The concentrations of key proteins in diverse regulatory pathways are controlled by post-translational ubiquitination and degradation by the 26S proteasome. Therefore, alterations in this proteolytic system are associated with pathologies of breast cancer 20., 21., 22.. The up-regulation of small ubiquitin-related modifier 3 precursor (SMT3A) and proteasome subunit alpha type 1 (PSMA1) indicate that the action of ubiquitin-proteasome system in breast cancer is strengthened. In addition, cathepsin D, which possesses acid proteases active in intracellular protein breakdown, is currently thought to correlate to the pathogenesis of breast cancer 23., 24., 25.. The over-expressin of cathepsin D in breast cancer may be involved in tissue infiltration. Proteolytic enzymes could be very important in the spread of cancer, but the role of the body’s natural inhibitors of these enzymes in this process is rarely reported. Alpha-1-antitrypsin is an inhibitor of serine proteases. Its primary target is elastase, but it also has a moderate affinity for plasmin and thrombin. It was reported that the activity of alpha-1-antitrypsin was significantly lower in patients with breast cancer before surgery than that in the controls, while after six months an increase in alpha-1-antitrypsin activity was observed (24). Goodarzi and Turner (26) reported that patients were unresponsive to treatment when high amounts of alpha-1-antitrypsin were extracted, and the carbohydrate structure of alpha-1-antitrypsin was also altered in cancer. Therefore, the down-regulation of alpha-1-antitrypsin suggests that it might play an important role in pathological progression in breast cancer.
Malignant cells proliferate actively and substance metabolism is concomitantly enhanced. The up-regulation of elongation factor 1-beta (EF-1-beta) and ribosomal protein S12 (RPS12) may be related to the promotion of protein synthesis. The elongation step of protein synthesis involves binding of aminoacyl-tRNA to the ribosomal A site, formation of a peptide bond, and translocation of the newly formed peptidyl-tRNA to the Ρ site. The nucleotide exchange factor EF-1-beta plays a major role in the regulation of this process by regenerating a GTP-bound EF-1-alpha that is necessary for each elongation cycle (27). However, the function of RPS12 is currently unknown. Cheng et al. 28., 29. found that the gene expression of RPS12 altered in the adjacent histopathologically “normal” cervical squamous epithelial tissue from cervical cancer patients, and thought that RPS12 may be potentially useful as an early pre-transformation diagnostic marker for human cervical cancer.
Translationally controlled tumor protein (TCTP) is involved in calcium binding and microtubule stabilization, and may be concerned with cell migration. Some antihistaminic compounds and other pharmacological compounds with a related structure could kill tumor cells and significantly decrease the gene level of TCTP (30). Vercoutter-Edouart et al. (31) also found that the up-regulation of TCTP occurred in MCF-7 human breast cancer cells induced by fibroblast growth factor-2 (FGF-2) stimulation. The up-regulation of TCTP in breast cancer indicates that TCTP might be a useful biomarker for breast cancer.
In 2D-PAGE gels, there were many low abundant proteins, the majority of which were not detected when the loading content of samples in IEF was decreased at 50%. Besides, serum albumin is abundantly presented in normal control sample, which may impact on experimental results. Therefore, only four proteins, namely 78 kDa glucose-regulated protein (GRP78), superoxide dismutase (SOD), transthyretin (TBPA), and transgelin 2, were identified and analyzed. These four proteins increased in breast cancer but were less than four folds, thus they were possible to be up-regulated in breast cancer. The alteration of transthyretin concentrations might compensate for changes in free thyroxine and thus the change of transthyretin may be involved in the substance metabolism in breast cancer (32). Over-expression of the GRP78 gene in malignant but not benign human breast lesions (33) with the changes of SOD and GRP78 might be correlated with stress reaction in cancer. Transgelin 2 was identified by MALDI-TOF/TOF-MS but its function has not been understood. Shi et al. (34) found that transgelin 2 mRNA was over-expressed in hepatocellular carcinoma (HCC) patients and thought that transgelin 2 mRNA may be a diagnostic marker for HCC. The report related to the role of transgelin 2 in breast cancer has not been seen, and its function needs to be further verified.
Conclusion
On the whole, breast cancer has a very complicated pathological process. In this study, we verified that pathological changes of breast cancer are concerned with augmentation of substance metabolism, promotion of proteolytic activity, decline of activity of some inhibitors of enzymes, and so on. Some important proteins with changes in expression are involved in the pathological process of breast cancer and may be useful biomarkers correlated to diagnosis, treatment effects, and prognosis for patients with breast cancer, such as alpha-1-antitrypsin, EF-1-beta, cathepsin D, TCTP, SMT3A, RPS12, and PSMA1. Among these proteins, SMT3A, RPS12, and PSMA1 were first reported for breast cancer in this study.
Materials and Methods
Tissue specimens
The fresh breast cancer tissues and adjacent normal breast tissues were obtained at excision of surgical operation from 12 volunteers between 32–60 years old with breast cancer at different clinical stages [T1N1M0 (n=1), T2N1M0 (n=4), T2N2M0 (n=1), T2N3M0 (n=2), T3N2M0 (n=3), and T3N3M1 (n=1)] at the Affiliated Hospital of North Sichuan Medial College, Nanchong, China. The tissues were frozen in −150°C super low temperature icebox (Revco, Asheville, USA) as soon as mastectomy was done. All samples were examined pathologically to obtain representative, viable, and non-necrotic tissues. The breast cancer was pathologically identified as infiltrating ductal carcinoma (Figure 3).
Protein sample preparation
Tissue protein extraction was accomplished by the ReadyPre™ Protein Extraction Kit (Total Protein) provided by Bio-Rad (Hercules, USA). The protein samples were then stored in aliquots of 200 μL at −80°C. To measure the concentration of the total protein, the Bio-Rad RC DC Protein Assay was used. Since the protein samples contained substances like salts, nucleic acids, and lipids, all of which are known to interfere with IEF, the ReadyPre™ 2-D Cleanup Kit (Bio-Rad) was used to wash them away and to concentrate proteins in samples at the same time. Before IEF, the concentration of protein samples was measured by using the Bio-Rad RC DC Protein Assay once more.
2D-PAGE and gel staining
Before 2D-PAGE, SDS-PAGE was firstly performed to preliminarily analyze the differences of proteins expressed in breast cancer and normal tissues (Figure 2), respectively. The loading content of protein samples was 35 μg. The first-dimension IEF was performed with precast IPG strips (pH 5.0–8.0, linear pH IPG, 178×3.3×0.5 mm; Bio-Rad) using the focusing tray (Bio-Rad). The loading volume of protein samples was 320 μL (The loading content of protein samples was 500 μg stained with silver nitrate and 2 mg stained with coomassie brilliant blue R-250 in each gel). IEF was carried out at 20°C with 50 μA/strip under the following conditions: Rehydration, 50 V, 12 h, passive; S1., 250 V, 25 min, slow; S2, 1,000 V, 2 h, rapid; S3, 8,000 V, 5 h, linear; S4, 8,000 V, 60,000 Vh, rapid; S5, 500 V, arbitrary. After IEF, it was necessary to equilibrate the IPG strips in a buffer containing SDS. The strips were first equilibrated in a buffer containing 6 M urea, 0.375 M Tris-HCl (pH 8.8), 2% SDS, 20% glycerol, and 2% (w/v) dithiothreitol for 15 min, and then in a buffer containing 6 M urea, 0.375 M Tris-HCl (pH 8.8), 2% SDS, 20% glycerol, and 2.5% (w/v) iodoacetamide for another 15 min. The equilibrated strips were placed on top of the prepared 12% SDS polyacrylamide gels and sealed in place with 0.5% low-melt agarose (Sigma, St. Louis, USA). The SDS-PAGE was performed at a current of 16 mA/gel for 30 min, then at another current of 24 mA/gel at 4°C for about 5 h using PROTEANII® Xi Cell (Bio-Rad).
After 2D-PAGE, the gels were stained with silver nitrate for analysis or with coomassie brilliant blue R-250 for preparative purposes.
Scanning and image analysis
The stained gels were scanned by UMAXpowerlook 1120 (UMAX, Taiwan, China) and analyzed by using the PDQuest 7.0 software (Bio-Rad), and the results in differential protein spots in gels from breast cancer and adjacent normal tissues were determined.
Identification of protein spots in gels
The 2D-PAGE images were compared with the online counterparts of the SIENA-2DPAGE database (http://www.bio-mol.unisi.it/cgi-bin/map1), and then the correlative information of differential protein spots was acquired by clicking on a spot located in the same point in the corresponding 2D-PAGE maps of the SIENA-2DPAGE database. Some differential spots whose correlative information was not obtained from the SIENA-2DPAGE database was identified by MALDI-TOF/TOF-MS (35).
Authors’ contributions
SSD carried out the protein extraction, 2-DE separation and identification, participated in the PDQuest analysis of 2-DE gel images, and drafted the manuscript. TYX carried out the collection of tissue samples. HYZ participated in the design of the study. RHX participated in the protein extraction and 2-DE separation. YGL participated in the PDQuest analysis of 2-DE gel images. BW carried out the pathological identification of tissue samples. SQL participated in the 2-DE separation. HJY conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank Dr. Wei-Min Hu and Prof. Zheng-Wei Yang for technical assistance.
References
- 1.Chen K. Incidence, mortality and survival rates of female breast cancer in Tianjin, China. Chin. J. Oncol. 2002;24:573–575. [PubMed] [Google Scholar]
- 2.Axelson H. Hypoxia-induced dedifferentiation of tumor cells—a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin. Cell Dev. Biol. 2005;16:554–563. doi: 10.1016/j.semcdb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Singh R.R., Kumar R. Steroid hormone receptor signaling in tumorigenesis. J. Cell Biochem. 2005;96:490–505. doi: 10.1002/jcb.20566. [DOI] [PubMed] [Google Scholar]
- 4.Pucci-Minafra I. Proteomic patterns of cultured breast cancer cells and epithelial mammary cells. Ann. N. Y. Acad. Sci. 2002;963:122–139. doi: 10.1111/j.1749-6632.2002.tb04103.x. [DOI] [PubMed] [Google Scholar]
- 5.Benvenuti S. Differential proteome analysis of replicative senescence in rat embryo fibroblasts. Mol. Cell. Proteomics. 2002;1:280–292. doi: 10.1074/mcp.m100028-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Harris R.A. Cluster analysis of an extensive human breast cancer cell line protein expression map database. Proteomics. 2002;2:212–223. doi: 10.1002/1615-9861(200202)2:2<212::aid-prot212>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 7.Somiari R.I. High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast. Proteomics. 2003;3:1863–1873. doi: 10.1002/pmic.200300560. [DOI] [PubMed] [Google Scholar]
- 8.Huber M. Comparison of proteomic and genomic analyses of the human breast cancer cell line T47D and the antiestrogen-resistant derivative T47D-r. Mol. Cell. Proteomics. 2004;13:43–55. doi: 10.1074/mcp.M300047-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Adam P.J. Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol. Cancer. Res. 2003;1:826–835. [PubMed] [Google Scholar]
- 10.Luo Y. Comparative proteome analysis of breast cancer and normal breast. Mol. Biotechnol. 2005;29:233–244. doi: 10.1385/MB:29:3:233. [DOI] [PubMed] [Google Scholar]
- 11.Vlahou A. A novel approach toward development of a rapid blood test for breast cancer. Clin. Breast Cancer. 2003;4:203–209. doi: 10.3816/cbc.2003.n.026. [DOI] [PubMed] [Google Scholar]
- 12.Alexander H. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin. Cancer Res. 2004;10:7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- 13.Celis J.E. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics. 2004;3:327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Fu Y.P. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. [PubMed] [Google Scholar]
- 15.Liu L. Passive smoking and other factors at different periods of life and breast cancer risk in Chinese women who have never smoked—a case-control study in Chongqing, People’s Republic of China. Asian Pac. J. Cancer Prev. 2000;1:131–137. [PubMed] [Google Scholar]
- 16.Muti P. The role of endogenous hormones in the etiology and prevention of breast cancer: the epidemiological evidence. Recent Results Cancer Res. 2005;166:245–256. doi: 10.1007/3-540-26980-0_16. [DOI] [PubMed] [Google Scholar]
- 17.Morris D.R. Protease-activated receptor-2 is essential for factor VIIa and Xa-induced signaling, migration, and invasion of breast cancer cells. Cancer Res. 2006;66:307–314. doi: 10.1158/0008-5472.CAN-05-1735. [DOI] [PubMed] [Google Scholar]
- 18.Toi M. Vascular endothelial growth factor and its relationships with endogenous inhibitors in a breast cancer microenvironment manipulated by hormonal therapy: a hypothetical consideration. Biomed. Pharmacother. 2005;59:S344–S347. doi: 10.1016/s0753-3322(05)80071-x. [DOI] [PubMed] [Google Scholar]
- 19.Huang H.L. Biomarker discovery in breast cancer serum using 2-D differential gel electrophoresis/MALDI-TOF/TOF and data validation by routine clinical assays. Electrophoresis. 2006;27:1641–1650. doi: 10.1002/elps.200500857. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q. Inducible expression of a degradation-resistant form of p27Kipl causes growth arrest and apoptosis in breast cancer cells. FEBS Lett. 2005;579:3932–3940. doi: 10.1016/j.febslet.2005.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dornan D. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 22.Boyer L. CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1-/- cells. Mol. Biol. Cell. 2006;17:2489–2497. doi: 10.1091/mbc.E05-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vyas S. Insulin-like growth factor II mediates resveratrol stimulatory effect on cathepsin D in breast cancer cells. Growth Factors. 2006;24:79–87. doi: 10.1080/08977190500366068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak A. Activity of cathepsin D and alpha(1)-antitrypsin in the blood serum of patients with mammary carcinoma. Exp. Oncol. 2005;27:233–237. [PubMed] [Google Scholar]
- 25.Chahed K. Expression of fibrinogen E-fragment and fibrin E-fragment is inhibited in the human infiltrating ductal carcinoma of the breast: the two-dimensional electrophoresis and MALDI-TOF-mass spectrometry analyses. Int. J. Oncol. 2005;27:1425–1431. [PubMed] [Google Scholar]
- 26.Goodarzi M.T., Turner G.A. Decreased branching, increased fucosylation and changed sialylation of alpha-1-proteinase inhibitor in breast and ovarian cancer. Clin. Chim. Acta. 1995;236:161–171. doi: 10.1016/0009-8981(95)06049-j. [DOI] [PubMed] [Google Scholar]
- 27.Mamoun C.B., Goldberg D.E. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor lbeta and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 2001;39:973–981. doi: 10.1046/j.1365-2958.2001.02289.x. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Q. Identification and characterization of genes involved in the carcinogenesis of human squamous cell cervical carcinoma. Int. J. Cancer. 2002;98:419–426. doi: 10.1002/ijc.10177. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Q. Identification of molecular markers for the early detection of human squamous cell carcinoma of the uterine cervix. Br. J. Cancer. 2002;86:274–281. doi: 10.1038/sj.bjc.6600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuynder M. Translationally controlled tumor protein is a target of tumor reversion. Proc. Natl. Acad. Sci. USA. 2004;101:15364–15369. doi: 10.1073/pnas.0406776101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vercoutter-Edouart A.S. Proteomic detection of changes in protein synthesis induced by fibroblast growth factor-2 in MCF-7 human breast cancer cells. Exp. Cell Res. 2001;262:59–68. doi: 10.1006/excr.2000.5066. [DOI] [PubMed] [Google Scholar]
- 32.Ramaker J., Wood W.G. Transthyretin—an explanation of “anomalous” serum thyroid hormone values in severe illness? J. Clin. Chem. Clin. Biochem. 1990;28:155–161. doi: 10.1515/cclm.1990.28.3.155. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez P.M. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res. Treat. 2000;59:15–26. doi: 10.1023/a:1006332011207. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y.Y. Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br. J. Cancer. 2005;92:929–934. doi: 10.1038/sj.bjc.6602460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qualtieri A. Proteomics of bovine myelin sheath: characterization of a truncated form of P0 by MALDI-TOF/TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:117–123. doi: 10.1016/j.jasms.2005.09.011. [DOI] [PubMed] [Google Scholar]