Abstract
GINS, a heterotetramer of SLD5, PSF1, PSF2, and PSF3 proteins, is an emerging chromatin factor recognized to be involved in the initiation and elongation step of DNA replication. Although the yeast and Xenopus GINS genes are well documented, their orthologous genes in higher eukaryotes are not fully characterized. In this study, we report the genomic structure and transcriptional regulation of mammalian GINS genes. Serum stimulation increased the GINS mRNA levels in human cells. Reporter gene assay using putative GINS promoter sequences revealed that the expression of mammalian GINS is regulated by 17β-Estradiol-stimulated estrogen receptor α, and human PSF3 acts as a gene responsive to transcription factor E2F1. The goal of this study is to present the current data so as to encourage further work in the field of GINS gene regulation and functions in mammalian cells.
Key words: DNA replication initiation, bioinformatics, gene structure, gene regulation, 17β-Estradiol, transcription factor E2F1
Introduction
In eukaryotic cells, chromosomal DNA replication requires the formation of a prereplicative complex (pre-RC) on origins, and assembly of other replication proteins in the S phase of the cell cycle to load the DNA polymerases to initiate DNA synthesis (1). The Cdc45 protein is crucial for the latter step of DNA replication initiation, as it activates the pre-RC that contains the MCM helicase. In addition to these proteins, GINS, a heterotetramer consisting of SLD5, PSF1 (partner of Sld five 1), PSF2, and PSF3 proteins, was recently identified as a loading factor for DNA polymerases, and as being required to separate the DNA strands at the replication fork in yeast and Xenopus egg extracts 2., 3., 4., 5., 6.. Although GINS is supposed to play a significant role in eukaryotic cells, the biological functions of its orthologous genes in higher eukaryotes are not fully characterized. A comparative genomic approach for the study of the GINS orthologous genes in higher eukaryotes could help in the understanding of a common regulatory mechanism for mammalian GINS genes.
In this study, we characterized the human and mouse GINS orthologous genes by using bioinformatics techniques. The gene structures, chromosomal localization, protein homology, and expression profiles are discussed. In addition, transcriptional regulations of human and mouse GINS genes are characterized. This is the first report of the identification of GINS up-regulation by serum stimulation and 17β-Estradiol (E2)-stimulated estrogen receptor α (ERα), and also is the first report that the putative promoter region of human PSF3 is responsive to transcription factor E2F1.
Results
Characterization of the GINS orthologous genes in higher eukaryotes
To investigate the functional importance of the GINS proteins in higher eukaryotes, we initially compared the amino acid sequences among human, chimpanzee, orangutan, dog, cow, mouse, and rat GINS proteins. As summarized in Table 1, human PSF1, PSF2, PSF3, and SLD5 proteins showed 92%−94%, 86%−100%, 89%−99%, and 84%−88% total amino acid identity, respectively, with the mammalian orthologous genes. Next, we sought to determine the gene structures of the human and mouse GINS genes. For this purpose, we searched for a genome database using human and mouse GINS cDNAs as query sequences in the BLAST program. Consequently, human (mouse) PSF1, PSF2, PSF3, and SLD5 gene fragments were identified within the genome sequences NT_011387 (AL808125), NT_010498 (AC103360), NT_086851 (AC113951), and NT_086740 (AC126038), respectively. The gene structures, especially the exon length and location of the human and mouse GINS genes were well conserved. Human (mouse) PSF1, PSF2, PSF3, and SLD5 genes had 7 (7), 5 (5), 3 (3), and 8 (8) exons, respectively (Table 2, Table 3, Table 4, Table 5). The precise exon-intron boundaries of the human and mouse GINS genes were determined based on the consensus sequence (“gt…ag” rule of intronic sequence) of exon-intron junctions and the codon usage. One exception was found in intron2 (“gg…ag”) of the human PSF3 gene (Table 4). The human (mouse) PSF1, PSF2, PSF3, and SLD5 genes were mapped to 20p11.21 (2G3), 16q24.1 (8E1), 16q21 (8C5), and 8p11.21 (8A3), respectively (Table 6).
Table 1.
Conservation of Amino Acids in the GINS Proteins*
| Mammal | PSF1 |
PSF2 |
PSF3 |
SLD5 |
||||
|---|---|---|---|---|---|---|---|---|
| Identify (%) | ORF (aa) | Identify (%) | ORF (aa) | Identify (%) | ORF (aa) | Identify (%) | ORF (aa) | |
| Human | 100 | 196 | 100 | 185 | 100 | 216 | 100 | 223 |
| Chimpanzee | 92 | 268 | 100 | 185 | 99 | 216 | – | – |
| Orangutan | – | – | – | – | 99 | 216 | – | – |
| Dog | 94 | 196 | 94 | 185 | 94 | 216 | 88 | 292 |
| Cow | 92 | 196 | 86 | 340 | 93 | 216 | 84 | 489 |
| Mouse | 92 | 196 | 92 | 185 | 89 | 216 | 87 | 223 |
| Rat | 93 | 196 | 92 | 434 | 90 | 216 | 88 | 223 |
The percentage conservation in mammals, including chimpanzee, orangutan, dog, cow, mouse, and rat proteins relative to the sequence of the human protein (taken as 100%) is presented. The lengths of the amino acid (aa) sequences are indicated. “–” indicates that the protein is not deposited in the databases.
Table 2.
Exon-Intron Structures of the Human PSF1 and Mouse Psf1 Genes*
| Exon No. | Nucleotide sequence around exon-intron boundaries of human PSF1 gene | Nucleotide position of exon in human genome sequence | Exon (bp) | Intron (bp) |
|---|---|---|---|---|
| 1 | AGCGCG—TTCAAC gtgagg | 25328321–25328531 of NT_011387 | 211 | 5,896 |
| 2 | ttctag GAGGAT—TGATGT gtaagt | 25334426–25334490 of NT_011387 | 65 | 3,251 |
| 3 | tatcag GAATGA—ATACCT gtaagc | 25337740–25337838 of NT_011387 | 99 | 904 |
| 4 | tcctag GTATGA—GAAGAA gtgagt | 25338741–25338831 of NT_011387 | 91 | 7,017 |
| 5 | ttgcag ATGGAG—ATTGAA gtatgt | 25345847–25345963 of NT_011387 | 117 | 16,376 |
| 6 | tttcag GTCCGG—AGCCAG gtattt | 25362338–25362412 of NT_011387 | 75 | 4,148 |
| 7 | cctcag CACTTT—ATGGAG | 25366559–25367702 of NT_011387 | 1,144 | |
| Exon No. | Nucleotide sequence around exon-intron boundaries of mouse Psf1 gene | Nucleotide position of exon in mouse genome sequence | Exon (bp) | Intron (bp) |
| 1 | GGAGCT—TTTAAT gtgagg | 76468–76660 of AL808125 | 193 | 2,989 |
| 2 | tttcag GAGGAC—TGATGT gtaagt | 79648–79712 of AL808125 | 65 | 3,292 |
| 3 | tctcag GAATGA—ATACCT gtgagt | 83003–82101 of AL808125 | 99 | 1,636 |
| 4 | tcctag GTATGA—GAAGAA gtaagt | 84736–84826 of AL808125 | 91 | 7,924 |
| 5 | tcacag ACGGAG—ATTGAA gtatgt | 92749–92865 of AL808125 | 117 | 2,045 |
| 6 | tttcag GTGCGG—AGTCAG gtagtt | 94909–94983 of AL808125 | 75 | 2,726 |
| 7 | ttgtag CACTTT—TTTCAC | 97708–98145 of AL808125 | 438 | |
The nucleotide sequences around the exon-intron boundaries are shown in upper-case (exon) and lower-case letters (intron). The exon and intron lengths (bp) are shown.
Table 3.
Exon-Intron Structures of the Human PSF2 and Mouse Psf2 Genes
| Exon No. | Nucleotide sequence around exon-intron boundaries of human PSF2 gene | Nucleotide position of exon in human genome sequence | Exon (bp) | Intron (bp) |
|---|---|---|---|---|
| 1 | GCGGCC—ATCGGG gtgagg | 39336779–39336614 of NT_010498 | 116 | 1,236 |
| 2 | tggcag GGGGAC—ATGTAG gtaagg | 39335379–39335265 of NT_010498 | 115 | 5,780 |
| 3 | ttttag AAAAGT—AAATCA gtaagt | 39329486–39329387 of NT_010498 | 100 | 2,917 |
| 4 | ctttag TGCTTC—GCCAAG gtaggt | 39326471–39326345 of NT_010498 | 127 | 204 |
| 5 | ctttag CTGGAT—AACTCA | 39326142–39325479 of NT_010498 | 664 | |
| Exon No. | Nucleotide sequence around exon-intron boundaries of mouse Psf2 gene | Nucleotide position of exon in mouse genome sequence | Exon (bp) | Intron (bp) |
| 1 | GGGAAA—ATCGGG gtgagc | 111624–111788 of AC103360 | 165 | 88 |
| 2 | ttgcag GGGGAC—ATGTGG gtgagc | 111875–111989 of AC103360 | 115 | 2,534 |
| 3 | tcagag AGAAAC—GAATCA gtgagt | 114522–114621 of AC103360 | 100 | 3,886 |
| 4 | cttcag TGCTTC—GCCAAG gtaggc | 118506–118632 of AC103360 | 127 | 272 |
| 5 | tttcag CTGGAC—TATACT | 118903–122081 of AC103360 | 3,179 | |
Table 4.
Exon-Intron Structures of the Human PSF3 and Mouse Psf3 Genes
| Exon No. | Nucleotide sequence around exon-intron boundaries of human PSF3 gene | Nucleotide position of exon in human genome sequence | Exon (bp) | Intron (bp) |
|---|---|---|---|---|
| 1 | CCGCTT—CCACAG gtgagc | 2010301–2010569 of NT_086851 | 269 | 9,759 |
| 2 | ctgcag GGTTCC—CTGCAG ggcaag | 2020327–2020560 of NT_086851 | 234 | 1,169 |
| 3 | tcccag ACTTTT—TTAGCA | 2021728–2023378 of NT_086851 | 1,651 | |
| Exon No. | Nucleotide sequence around exon-intron boundaries of mouse Psf3 gene | Nucleotide position of exon in mouse genome sequence | Exon (bp) | Intron (bp) |
| 1 | GAGTTT—CCTCAG gtgagg | 126499–126244 of AC113951 | 256 | 4,028 |
| 2 | acgcag GGTACA—CTGAAG gtaagt | 122217–121984 of AC113951 | 234 | 5,061 |
| 3 | cctcag ACTTTT—GCCTGC | 116924–115517 of AC113951 | 1,407 | |
Table 5.
Exon-Intron Structures of the Human SLD5 and Mouse Sld5 Genes
| Exon No. | Nucleotide sequence around exon-intron boundaries of human SLD5 gene | Nucleotide position of exon in human genome sequence | Exon (bp) | Intron (bp) |
|---|---|---|---|---|
| 1 | GCGACT—CCCCGA gtgagc | 28874824–28874993 of NT_086740 | 170 | 789 |
| 2 | cattag GTTCCT—GAGCAG gtaagc | 28875781–28875895 of NT_086740 | 115 | 6,063 |
| 3 | ttttag GCCTGG—CACATG gtaaga | 28881957–28882043 of NT_086740 | 87 | 735 |
| 4 | taatag GAAGAA—ATGAAG gtttga | 28882777–28882890 of NT_086740 | 114 | 2,436 |
| 5 | tcacag ATAGAG—CAGAGA gtgagt | 28885325–28885422 of NT_086740 | 98 | 142 |
| 6 | tttcag GTTCAT—GGGCAG gtaaac | 28885563–28885651 of NT_086740 | 89 | 1,851 |
| 7 | tggcag TTCCCA—GCAGAG gtgagt | 28887501–28887591 of NT_086740 | 91 | 93 |
| 8 | tttcag GGACTA—AAAAAA | 28887683–28888085 of NT_086740 | 403 | |
| Exon No. | Nucleotide sequence around exon-intron boundaries of mouse Sld5 gene | Nucleotide position of exon in mouse genome sequence | Exon (bp) | Intron (bp) |
| 1 | GTGTTT—TGCGAG gtgagc | 79323–79134 of AC126038 | 190 | 372 |
| 2 | tttcag GTGTAG—GAGCAG gtaagc | 78763–78644 of AC126038 | 120 | 2,141 |
| 3 | ttttag GCCTGG—CACATG gtaaga | 76504–76418 of AC126038 | 87 | 2,037 |
| 4 | taatag GAAGAA—ATGAAG gtttga | 74382–74269 of AC126038 | 114 | 2,903 |
| 5 | tcacag ATAGAG—CAAAGA gtaagt | 71367–71270 of AC126038 | 98 | 103 |
| 6 | ttccag GTATAT—GGGCAG gtaaga | 71168–71080 of AC126038 | 89 | 2,078 |
| 7 | tggcag TTCCCA—GCAGAG gtgagt | 69003–68913 of AC126038 | 91 | 75 |
| 8 | tttcag AGACTA—AAGCTT | 68839–68289 of AC126038 | 501 | |
Table 6.
Chromosomal Localization of the Human and Mouse GINS Genes
| PSF1 | PSF2 | PSF3 | SLD5 | |
|---|---|---|---|---|
| Human | 20p11.21 | 16q24.1 | 16q21 | 8p11.21 |
| Mouse | 2G3 | 8E1 | 8C5 | 8A3 |
Expression profiles of human and mouse GINS mRNAs
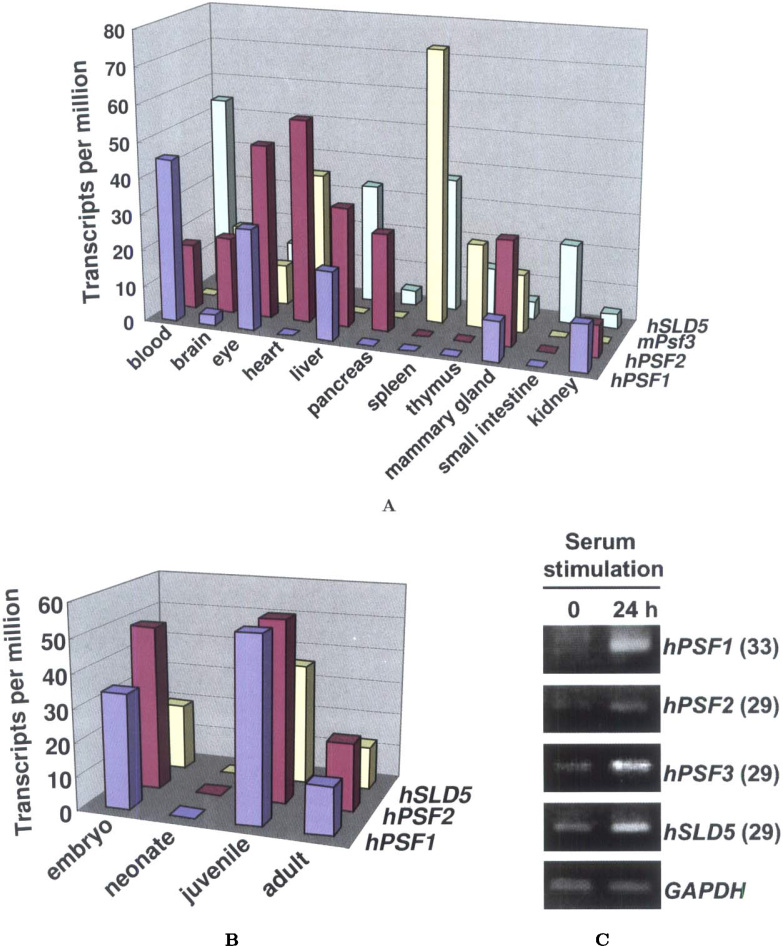
In silico analysis revealed that the expression patterns of the GINS mRNA in human/mouse tissues and organs were quite different (Figure 1A). On the other hand, during the developmental stages, the GINS mRNA expression patterns were quite similar: highest in the embryo and juvenile stages, lower in adults, and no expression in the neonatal stage (Figure 1B). Next, we attempted a search for GINS mRNA in public databases. The Gene Expression Omnibus (GEO) database search revealed that the SLD5 gene is down-regulated under two growth arrest conditions, namely, serum deprivation and contact inhibition, in T98G cancer cells (Accession No. GDS911), and PSF1 is an estrogen target in human breast carcinoma MCF7 cells (Accession No. GDS118). It is of interest that GINS is regulated by serum stimulation, because the mRNA level of a known DNA replication initiator is reported to be induced when cells enter the G1/S phase of the cell cycle 7., 8.. To substantiate this evidence, growth arrest of Saos-2 cells was induced by incubation in serum-free medium for three days, then the cells were re-entered into the cell cycle with the re-addition of 20% fetal bovine serum (FBS). Twenty-four hours after the serum re-addition, the GINS mRNA level was found to be up-regulated, while the GAPDH mRNA level remained unchanged (Figure 1C). At this time point, cyclin A protein accumulation was clearly observed as the cells re-entered the cell cycle (data not shown).
Fig. 1.
Expression profiles of GINS genes. A search was conducted based on the EST counts in human/mouse tissues (A) and during the developmental stages (B) in the UniGene database (EST Profile Viewer, NCBI). The datasets used for PSF1, PSF2, Psf3, and SLD5 genes were Hs. 360033, Hs. 433180, Mm. 35546, and Hs. 521557, respectively. The number of transcripts per million was calculated from the gene EST/total EST in the pool. C. Semi-quantitative RT-PCR analysis of the transcripts of GINS genes in Saos-2 cells after serum stimulation for 24 h. The number of PCR cycles is shown in parentheses. The housekeeping gene, GAPDH, was used as the control.
Transcriptional regulation of the human and mouse GINS genes
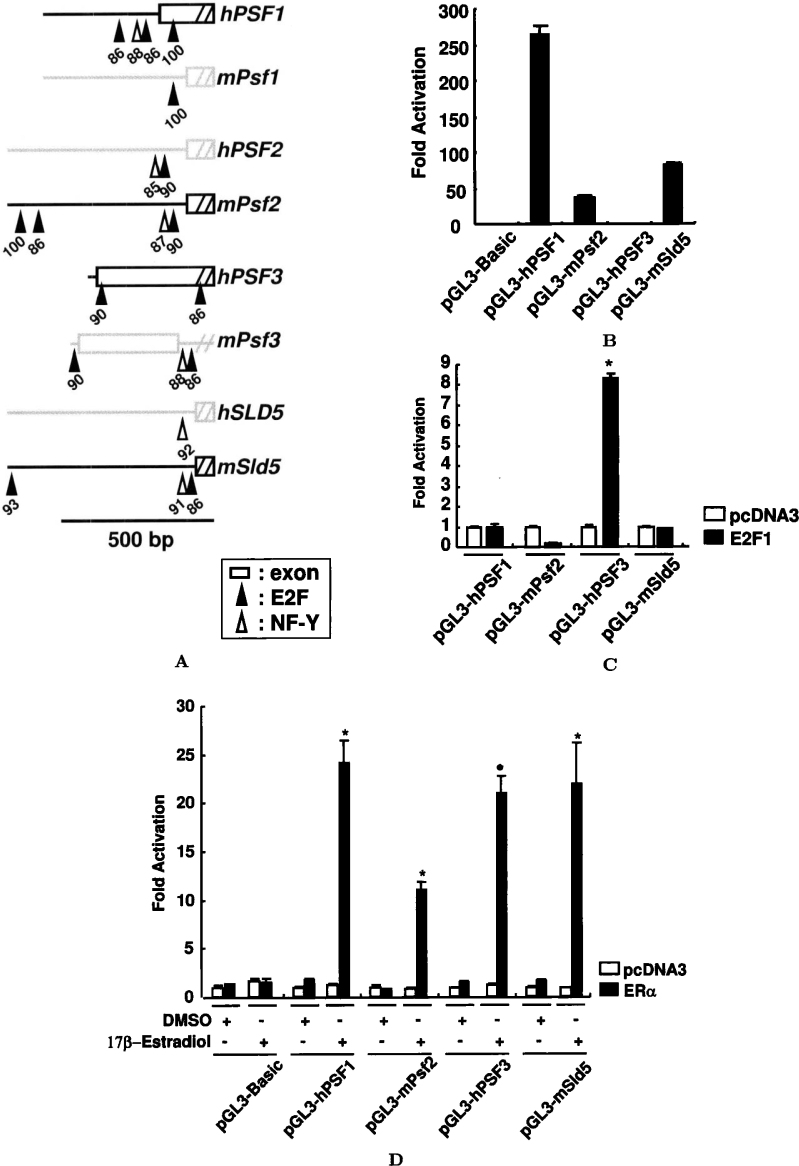
To gain more insights into the functions of the human and mouse GINS genes, we searched the proximal region of the transcription start site for potential cis-elements using the Transfac software (ver. 4.0, cut-off 85). The E2F- and/or NF-Y-binding consensus sequences were identified to be surrounded by GC-rich sequences in the putative promoter region of the human and mouse GINS genes (Figure 2A). In addition to that by serum stimulation, many genes involved in DNA replication initiation were found to be regulated by transcription factor E2F1 7., 8.. To demonstrate the importance of the putative E2F-binding element in the basal promoter activity, we generated human PSF1, mouse Psf2, human PSF3, and mouse Sld5 promoter-luciferase constructs. As shown in Figure 2B, human PSF1, mouse Psf2, human PSF3, and mouse Sld5 promoter constructs showed approximately 265-, 37-, 0.6-, and 82-fold increases in activity, respectively, as determined by measuring the relative luciferase activities, taking the activity in the control luciferase vector pGL3-Basic as 1. Exogenous coexpression of E2F1 caused up to approximately 1-, 0.2-, 8.3-, and 0.9-fold increases in the human PSF1, mouse Psf2, human PSF3, and mouse Sld5 promoter activities, respectively, as compared to that of the pcDNA3 control vector (Figure 2C). These results suggest that the E2F-binding motif plays critical roles in the E2F1-mediated human PSF3 promoter activity. This is the first report of the E2F1-dependent regulation of the PSF3 gene.
Fig. 2.
Transcriptional regulation of the human and mouse GINS genes. A. Schematic representation of the vicinity of the transcription start sites of the human and mouse GINS genes. The putative transcription factor E2F- (closed isosceles triangles) and NF-Y-binding sites (open isosceles triangles) are indicated with their scores (maximum score 100) calculated by the Transfac program. Exon 1 (open boxes) is indicated. Scale bar equals 500 bp. B. Human and mouse GINS promoter activities in asynchronously growing human cells. HeLa cells were transfected with 200 ng of reporter constructs and 400 ng of the expression vector for E2F1, together with 0.6 ng of pRL-TK. The pcDNA3 vector was used as the negative control. At 48 h after the transfection, the cells were harvested, and extracts were prepared to measure the firefly and Renilla luciferase activities. Values are represented as relative luciferase activities, with that of pGL3-Basic being taken as 1. C. The E2F-binding motif of the PSF3 promoter is sufficient to confer responses to ectopic E2F expression. The experiment was performed as described in panel B. Values are represented as relative luciferase activities, with that of the control vector pcDNA3 being taken as 1. Statistically significant (P < 0.05) induction is indicated by an asterisk. D. E2-induced activation of GINS promoter constructs in HeLa cells. Cells were transfected with the indicated reporter plasmids and pcDNA3 or pSG5-ERα expression vector, and the effects of E2 or DMSO on the luciferase activities were determined. Results are expressed as mean±S.D. for at least three triplicate determinations for each treatment group. Statistically significant (P < 0.05) induction is indicated by an asterisk.
To determine whether E2 also regulates GINS promoter activity, HeLa cells were cultured with E2, and the effect of cotransfection with ERα was measured. Interestingly, the initial transient transfection studies with pGL3-hPSF1, -mPsf2, -hPSF3, and -mSld5 reporter vector constructs showed that treatment with 100 nM of E2 resulted in 1.2-, 0.8-, 1.3-, and 0.9-fold increases in the reporter gene activities, respectively, as compared to that after DMSO treatment (Figure 2D). However, following cotransfection with ERα, there was an enhanced (11~24-fold) induction of reporter gene activity after treatment with 100 nM of E2 as compared to that in the pcDNA3 control vector (Figure 2D). These results suggest that the GINS genes are regulated by E2 via ERα in cultured cells.
Discussion
In the present study, we attempted to characterize the gene structure and transcriptional regulation of the human and mouse GINS genes. Studies using bioinformatics techniques revealed that the GINS proteins are well conserved among several mammals. In addition, conservation of the gene structure, including the putative promoter region between human and mouse GINS genes, suggests that their transcriptional regulation might also be similar. We succeeded in identifying PSF3 as an E2F1-regulated gene, and that the human and mouse GINS genes are regulated by growth stimuli, including serum addition and E2 treatment. In contrast to the rapid progress of research on the yeast and Xenopus GINS, only a few reports have been published regarding mammalian GINS. For example, mice homozygous for the PSF1 gene died in utero around the time of implantation, indicating that PSF1 protein may be required during early embryogenesis (9). cDNA microarray analyses revealed that the PSF2 gene is frequently up-regulated in cholangiocarcinoma, suggesting that PSF2 protein may play an important role in the development of this type of liver cancer (10).
The action of E2 on its target cells is mediated via ERα or ERβ, which function as ligand-activated transcription factors regulating gene expression. E2 has been shown to play a central role in breast cancer development; therefore it is important to find out the critical downstream targets of E2/ERa action. The majority of E2 target genes are characterized by imperfect estrogen-responsive element (ERE: AGGTCANNNTGACCT) sequences or complex organization with the presence of half an ERE site. Interestingly, many E2-responsive genes, which do not contain ERE but do contain other cis-elements, such as Spl, have also been described. ERs can regulate transcription by direct protein-protein interaction with Spl (11). Indeed, the GINS promoter constructs used in this study contained GC-rich sequences, but no ERE-like sequences. The mitogenic effect of ERα is mediated at the regulation level of kinases that govern the transition from the G0/G1 to S phase of the cell cycle, such as under the influence of cyclin D-Cdk4/6 and cyclin E-Cdk2, to increase phosphorylation of the retinoblastoma protein (pRb) (12). pRb physically interacts with E2F1 to yield a transcriptionally repressed complex. Phosphorylated pRb results in dissociation of the pRb-E2F1 complex and subsequent up-regulation of E2F1-dependent genes 13., 14., 15.. Although the regulation of E2F1-dependent transactivation is closely linked to pRb, E2 is also known to induce E2F1 mRNA and protein in human cultured cells (16). This could mean that the E2F1-dependent regulation of PSF3 expression may be partially mediated by E2. More detailed studies are needed to clearly elucidate the mechanism by which GINS affects the development of breast cancer.
In conclusion, our study clearly demonstrated that a comparative genomic approach for the GINS orthologous genes in higher eukaryotes could help in the understanding of common or unique regulatory mechanisms for important genes. We identified the conserved gene structure and for the first time characterized the promoter region of the human and mouse GINS genes. This is the first report of the identification of the induction of GINS by serum stimulation and E2/ERα-induced genes, and is also the first report that human PSF3 gene is regulated by transcription factor E2F1.
Materials and Methods
Bioinformatics
Bioinformatics techniques were employed as previously described 17., 18.. A search for GINS orthologous genes was conducted using the HomoloGene or BLAST program at NCBI (http://www.ncbi.nlm.nih.gov/). The Transfac software (http://motif.genome.jp/) was used to determine the transcription factor binding elements. The search was conducted based on the expression profile suggested by analysis of the EST (expressed sequence tag) counts in human/mouse tissues and organs in the UniGene database (EST Profile Viewer, NCBI). The datasets used for PSF1, PSF2, Psf3, and SLD5 genes were Hs. 360033, Hs. 433180, Mm. 35546, and Hs. 521557, respectively. The number of transcripts per million was calculated based on the gene EST/total EST in the pool, and this value was exported to an Excel file. The expression profile suggested by analysis of Affymetrix GeneChip or SAGE (serial analysis of gene expression) was confirmed by using the GEO database search at NCBI.
Plasmids
The human and mouse GINS promoter fragments were generated by polymerase chain reaction (PCR) from genomic DNA and ligated into the pGL3-Basic vector (Promega, Madison, USA). Initial PCR primers were designed to amplify 548 bp (−427/+121), 638 bp (−613/+25), 315 bp (−14/+301), and 499 bp (−409/+90) of human PSF1 (GenBank Accession No. AL353812), mouse Psf2 (GenBank Accession No. AC103360), human PSF3 (GenBank Accession No. AC009118), and mouse Sld5 (GenBank Accession No. AC147247) promoter sequences, respectively, which are numbered relative to the transcription initiation site at +1 described in the NCBI UniGene database. The forward (F) and reverse (R) PCR primers for the promoter constructs were: 5′-gcagtccctaccagcactag-3′ (−427F), 5′-cggccttgccaaccacca-3′ (+121R), 5′-acccggagctgactttga-3′ (—613F), 5′-gcagcgcctcaggcactt-3′ (+25R), 5′-tcctgtctgatgcgtccc-3′ (−14F), 5′-tgcgctccgctccaggaa-3′ (+301R), 5′-tagagaacaactctgcgc-3′ (−409F), and 5′-agcggtgggcgccggaga-3′ (+90R), respectively. The forward/reverse PCR primers added BglII/HindIII (for PSF1 and Sld5) or KpnI/BglII (for Psf2 and PSF3) sites to facilitate subcloning. The pcDNA3-HA-E2F1 expression plasmid was a generous gift from Dr. Joseph R. Nevins (Duke University, Durham, USA). The pSG5-ERα expression plasmid was kindly provided by Dr. Gary H. Perdew (The Pennsylvania State University, Univeristy Park, USA).
Cell culture and reporter assay
HeLa and Saos-2 cells were cultured in Earle’s modified Eagle’s medium (MEM) supplemented with 10% FBS and penicillin/streptomycin. To measure the growth-dependent induction of the human GINS gene expression, growth of the Saos-2 cells was arrested in the G0 phase by incubation in the absence of FBS for 3 d, and the cells were reintroduced into the cell cycle by culturing with 20% FBS. Total RNA and cell lysates were recovered at 0 and 24 h after the serum stimulation. To measure promoter activity, the cells were transfected with FuGENE6 (Roche, Indianapolis, USA) according to the manufacturer’s instruction. Briefly, 400 ng of expression plasmid, 200 ng of firefly luciferase reporter plasmid (pGL3, Promega), and 0.6 ng of Renilla luciferase reporter plasmid (pRL-TK, Promega) per 24-well dish were used in each transfection. After 24 h, cells were treated with 100 nM of E2 (Sigma, St. Louis, USA) or DMSO for 34-40 h. The cells were harvested, and luciferase assay was performed using the Dual-Luciferase Reporter Assay System following the manufacturer’s protocol (Promega). Experiments were done at least in triplicate, and the relative activities and S.D. values were determined. To control for transfection efficiency, firefly luciferase values were normalized to the values for Renilla luciferase.
RT-PCR
Total cellular RNA was extracted from Saos-2 cells using Trizol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instruction. RT-PCR was performed with the SuperScript RT-PCR system according to the manufacturer’s instruction (Invitrogen). PCR was performed as follows: denaturation at 94°C for 2 min, followed by multiple cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 30 s. The following primers were used: PSF1 (GenBank Accession No. NM_021067), 5′-tgagatcacaggcgtgac-3′ and 5′-ctgtccccttccaaagtg-3′; PSF2 (GenBank Accession No. NM_016095), 5′-cagcctctggagagtact-3′ and 5′-cacctctgtgagagagtc-3′; PSF3 (GenBank Accession No. NM_022770), 5′-ctgcctgcagggagttac-3′ and 5′-agagggcccttcatagtc-3′; SLD5 (GenBank Accession No. NM_032336), 5′-gcctctcgccggaagagt-3′ and 5′-cctgacctcatgatccgc-3′. As a control, a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primer set was used (R&D Systems, Minneapolis, USA). The amplified products were separated on 1% agarose gels and visualized under ultraviolet transillumination.
Authors’ contributions
RH, TA, MT, and YG carried out the reporter assays and performed the statistical analysis. KY conceived of the study, participated in its design, and completed the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Sato Memorial Foundation for Cancer Research.
References
- 1.Blow J.J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takayama Y. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambus A. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 4.Pacek M. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Kanemaki M., Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki T. GINS is a DNA polymerase epsilon accessory factor during chromosomal DNA replication in budding yeast. J. Biol. Chem. 2006;281:21422–21432. doi: 10.1074/jbc.M603482200. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida K., Inoue I. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida K., Inoue I. Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene. 2004;23:6250–6260. doi: 10.1038/sj.onc.1207829. [DOI] [PubMed] [Google Scholar]
- 9.Ueno M. PSF1 is essential for early embryogenesis in mice. Mol. Cell. Biol. 2005;25:10528–10532. doi: 10.1128/MCB.25.23.10528-10532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obama K. Up-regulation of PSF2, a member of the GINS multiprotein complex, in intrahepatic cholangiocarcinoma. Oncol. Rep. 2005;14:701–706. [PubMed] [Google Scholar]
- 11.Saville B. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Spl) promoter elements. J. Biol. Chem. 2000;275:5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- 12.Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 13.Nevins J.R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 14.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.Harbour J.W., Dean D.C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 16.Wang W. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Spl/estrogen receptor interactions. Mol. Endocrinol. 1999;13:1373–1387. doi: 10.1210/mend.13.8.0323. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi R. Comparative genomics on MCM8 orthologous genes reveals the transcriptional regulation by transcription factor E2F. Gene. 2006;367:126–134. doi: 10.1016/j.gene.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem. Biophys. Res. Commun. 2005;331:669–674. doi: 10.1016/j.bbrc.2005.03.222. [DOI] [PubMed] [Google Scholar]