Abstract
This study describes methods for predicting and classifying voltage-gated ion channels. Firstly, a standard support vector machine (SVM) method was developed for predicting ion channels by using amino acid composition and dipeptide composition, with an accuracy of 82.89% and 85.56%, respectively. The accuracy of this SVM method was improved from 85.56% to 89.11% when combined with PSI-BLAST similarity search. Then we developed an SVM method for classifying ion channels (potassium, sodium, calcium, and chloride) by using dipeptide composition and achieved an overall accuracy of 96.89%. We further achieved a classification accuracy of 97.78% by using a hybrid method that combines dipeptide-based SVM and hidden Markov model methods. A web server VGIchan has been developed for predicting and classifying voltage-gated ion channels using the above approaches. VGIchan is freely available at www.imtech.res.in/raghava/vgichan/.
Key words: ion channels, prediction, VGIchan, SVM, HMM
Introduction
Voltage-gated ion channels are integral membrane proteins that enable the passage of selected inorganic ions across cell membranes. They open and close in response to changes in transmembrane voltage, and play a key role in electric signaling by excitable cells such as neurons (1). They also have a critical role in the function of the nervous system, where they instigate and conduct nerve impulses by asserting control over the voltage potential across the plasma membrane. These ion channels are important for physiological functions and are critical in producing hyperexcitability. Many drugs that are routinely used in clinical setting, as well as several novel experimental drugs, have shown interactions with voltage-gated ion channels (2). Ion channels are valuable targets for antiepileptic drug design (3), antihypertensives (4), anesthetics (5), and antipsychotics against diseases such as schizophrenia, the main phase of manicdepressive illness, and other acute idiopathic psychotic illness (6). Ion channels are also helpful in understanding the mechanism of various activities in the cell, and each ion channel has its own specific importance.
To our knowledge, currently there is no server available to classify ion channels into subclasses like potassium, sodium, calcium, and chloride ion channels from protein sequences. Keeping this in mind, we compiled all the annotated ion channels from the Swiss-Prot database, developed prediction methods for voltage-gated ion channels, and further classified them into potassium, sodium, calcium, and chloride ion channels.
Results and Discussion
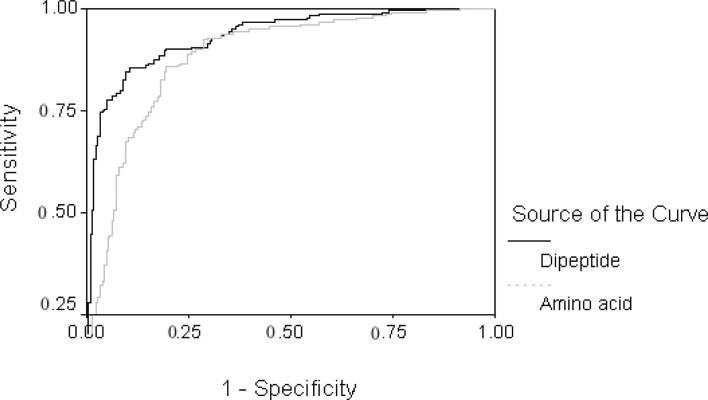
Firstly, we developed methods to discriminate ion channels and non-ion channels from a given protein sequence. The performance of various methods for discriminating ion channels from non-ion channels is shown in Table 1. The support vector machine (SVM) module achieved an accuracy of 82.89% and 85.56% by using amino acid composition and dipeptide composition, respectively, while an accuracy of 89.11% was achieved by using a hybrid approach that combines dipeptide-based SVM and PSI-BLAST similarity search (7). In the prediction of voltage-gated ion channels, we did not use hidden Markov model (HMM) since it was difficult to align all the different ion channels by using ClustalW (8) in one group. The receiver operating characteristic (ROC) plot of the SVM module based on amino acid composition and dipeptide composition is shown in Figure 1. We also developed modules for classifying voltage-gated ion channels based on their types. The performance of various methods used for classification of voltage-gated ion channels is shown in Table 2. The SVM module information regarding the kernel is available in the supplementary data (www.imtech.res.in/raghava/vgichan/supplemantary.html). The results indicate that the accuracy of the dipeptide-based SVM method (96.89%) is comparable with that of HMM (96.86%) in classifying voltage-gated ion channels. The overall classfication accuracy achieved by PSI-BLAST was 69.33%. We combined the best two methods, namely the dipeptide-based SVM and HMM, and obtained an overall accuracy of 97.78%. The reliability index (RI) was assigned based on the dipeptide-based SVM module to know the prediction reliability. The calculation showed that nearly 77.78% of the sequences have RI ≥ 3, and the expected accuracy of these sequences is 100.00%. The prediction accuracy with RI equal to a given value is shown in the supplementary data (www.imtech.res.in/raghava/vgichan/supplemantary.html). In contrast, there is a database of voltage-gated potassium channel that only allows BLASTP to match for the query sequence (9). The accuracy levels of the classification for potassium (60%) and chloride (~66%) ion channels in PSI-BLAST search were low as compared with those of the dipeptide-based SVM (100% for potassium and ~87% for chloride ion channels) and HMM (98% for potassium and ~86% for chloride ion channels).
Table 1.
Performance of Various Methods on Prediction of Voltage-Gated Ion Channels
| Method | ACC (%) | MCC | ROC |
|---|---|---|---|
| Amino acid-based SVM (A)*1 | 82.89 | 0.66 | 0.89 |
| Dipeptide-based SVM (B)*2 | 85.56 | 0.71 | 0.93 |
| PSI-BLAST (C)*3 | 84.22 | – | – |
| Hybrid (B+C) | 89.11 | 0.78 | – |
RBF kernel, ᵞ=60; C=100; j=0.1; threshold value=0.3.
RBF kernel, ᵞ=40; C=10; j=1; threshold value=0.4.
E-value=0.01. ACC, Accuracy; MCC, Matthew’s correlation coefficient; ROC, receiver operating characteristic.
Fig. 1.
The overall performance of the SVM module using amino acid composition and dipeptide composition in predicting voltage-gated ion channels. The ROC plot was obtained between sensitivity (Y-axis) and 1—specificity (X-axis) at different thresholds.
Table 2.
Performance of Various Methods on Classification of Voltage-Gated Ion Channels
| Method | Potassium |
Sodium |
Calcium |
Chloride |
Overall ACC (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | MCC | ACC (%) | MCC | ACC (%) | MCC | ACC (%) | MCC | ||
| Amino acid-based | 100 | 0.86 | 80.00 | 0.88 | 80.00 | 0.86 | 73.33 | 0.84 | 93.78 |
| SVM (A)*1 | |||||||||
| Dipeptide-based | 100 | 0.95 | 88.00 | 0.91 | 92.00 | 0.93 | 86.67 | 0.91 | 96.89 |
| SVM (B)*2 | |||||||||
| PSI-BLAST*3 | 65.62 | – | 92.00 | – | 76.00 | – | 60.00 | – | 69.33 |
| HMM*4 | 98.12 | – | 96.00 | – | 96.00 | – | 86.17 | – | 96.86 |
| SVM (B) + HMM | 99.38 | 0.96 | 96.00 | 0.93 | 96.00 | 0.98 | 86.67 | 0.92 | 97.78 |
Amino acid composition as input vector; RBF kernel, ᵞ=500; C=10; j=0.1.
Dipeptide composition as input vector; RBF kernel, ᵞ=50; C=10; j=1.
E-value=0.01.
E-value=1. ACC, Accuracy; MCC, Matthew’s correlation coefficient.
VGIchan
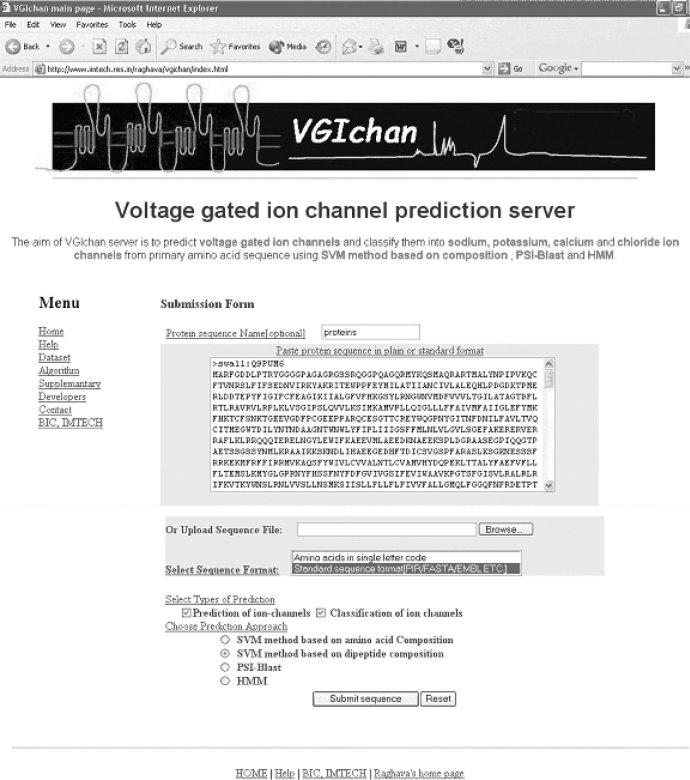
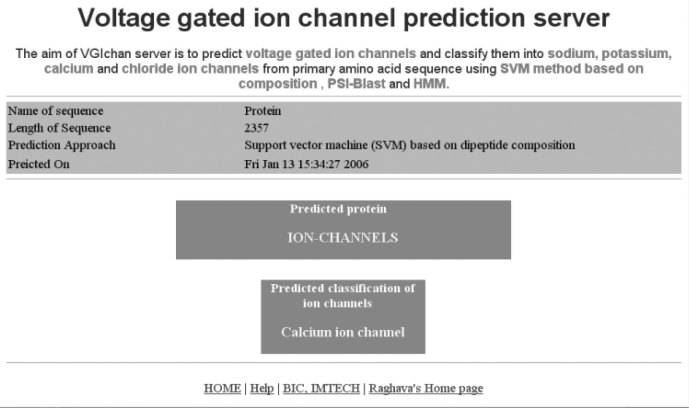
A web server VGIchan has been developed for predicting and classifying voltage-gated ion channels using the above approaches. VGIchan is freely available at http://www.imtech.res.in/raghava/vgichan/. The common gateway interface script of VGIchan is written by using the PERL language (version 5.03). The VGIchan server is installed on a Sun Server (420E) under UNIX (Solaris 7) environment. Users can provide the input sequence by cut-paste or directly uploading sequence file from disk. The server accepts the sequence in raw format as well as in standard formats, such as EMBL, FASTA, and GCG acceptable to ReadSeq (developed by Dr. Don Gilbert). A snapshot of the sequence submission page of the server is shown in Figure 2. Users can predict the type of voltage-gated ion channels by choosing SVM, PSI-BLAST, or HMM methods, where the SVM method is based on either amino acid composition or dipeptide composition. On submission the server will give results in a user-friendly interface (Figure 3). This method can be used for automated annotation of genomic data and will assist the preliminary analysis of possible types of new ion channels.
Fig. 2.
Snapshot of the input page of VGIchan server.
Fig. 3.
Snapshot of the results obtained after the analysis of submission.
Materials and Methods
Collection and compilation of ion channels
We searched ion channels in the Swiss-Prot database using keyword ion channels in the Swiss-Prot full text (http://au.expasy.org/sprot/). We examined each protein obtained from our query search manually in order to eliminate non-ion channels. Finally we obtained 473 proteins, including 307 potassium, 66 sodium, 61 calcium, and 39 chloride ion channels. These protein sequences were retrieved from Swiss-Prot. The non-ion channel protein sequences were obtained from Swiss-Prot by using SRS (http://au.expasy.org/srs5bin/cgi-bin/wgetz). We carried out combined searches in the query form by using two information fields: (1) comment with the query word “function” and (2) comment with the query word “ion channels” with “BUTNOT” option. We examined all the retrieved protein sequences and checked their functions in order to eliminate ion channel proteins. A final dataset of 236 non-redundant proteins was created using the PROSET software (10), where sequences with more than 90% sequence identity were removed. This is a fast procedure to create non-redundant sets of protein sequences. The final dataset is available online at http://www.imtech.res.in/raghava/vgichan/dataset.html. We further classified these 236 non-redundant ion channels into potassium (164), sodium (27), calcium (27), and chloride (18) ion channels.
Support vector machine
SVM was implemented using the freely downloadable software package SVM_light (11). The amino acid composition (20 vectors) and dipeptide composition (400 vectors) of each protein sequence were used as input vectors.
Amino acid composition
Amino acid composition is the fraction of each amino acid in a protein. The fraction of each of the 20 natural amino acids was calculated using the following equation:
where i can be any one of the 20 amino acids.
Dipeptide composition
Dipeptide composition is used to encapsulate the global information about each protein sequence, which gives a fixed pattern length of 400 (20×20). This representation encompasses the information about amino acid composition along the local order of amino acids. The fraction of each dipeptide was calculated using the following equation:
where dipep (i) is one out of 400 dipeptides.
Hidden Markov model
HMM profiles of the four types of voltage-gated ion channels were constructed using the HMMER software package (12). Each protein sequence was aligned in a multiple sequence alignment using ClustalW. An HMM profile was built with the hmmbuild program for each class, and later each profile was calibrated with the hmmcalibrate program. We created our own HMM database by concatenation of each single HMM profile. The hmmpfam program was used for searching a query sequence against the created profile in the HMM database. We set an E-value threshold (E-value<0.01) while predicting the quality by a five-fold cross validation.
PSI-BLAST
A module was designed in which query sequences in testing datasets were searched against proteins in training datasets using PSI-BLAST (7). Three iterations of PSI-BLAST were carried out at a cut-off E-value of 0.01. The module could predict voltage-gated ion channels and their types (potassium, sodium, calcium, and chloride) depending upon the similarity of the query protein to the protein in the dataset.
Hybrid approach
In the hybrid approach of SVM and PSI-BLAST, we combined their outputs by giving weightage to PSI-BLAST results when there were hits in the database, and considered SVM results only when there was no hits found by PSI-BLAST search. Similarly, in the hybrid approach of SVM and HMM, weightage was given to HMM search, and SVM results were considered only when there was no hits obtained in the database.
Performance measures
Five-fold cross validation
The performance modules constructed in this study for discriminating voltage-gated ion channels and their types were evaluated using a five-fold cross validation technique. In the five-fold cross validation, the relevant dataset was randomly divided into five sets. The training and testing was carried out for five times, each time using one distinct set for testing and the remaining four sets for training. Five threshold-dependent parameters (13), namely sensitivity, specificity, accuracy, positive predictive value (PPV), ROC, and Mathew’s correlation coefficient (MCC) were used for predicting and classifying the ion channels.
Reliability index
RI is a commonly used measure of prediction that provides confidence about a prediction to the users. In this study, RI was assigned according to the difference (δ) between the highest and the second highest SVM output scores. We computed the RI score of the classification method of ion channels based on dipeptide composition using the following equation:
Authors’ contributions
SS developed SVM models and the VGIchan web server. JZ collected and complied voltage-gated ion channels from literature and databases. BS guided JZ in the annotation of voltage-gated ion channel proteins and refined the manuscript drafted by SS and JZ. GPSR conceived the idea and supervised the work. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by the Council of Scientific and Industrial Research (CSIR) and the Department of Biotechnology, Government of India (Grant No. CMM-17).
Supporting Online Material
References
- 1.Sands Z. Voltage-gated ion channels. Curr. Biol. 2005;15:R44–R47. doi: 10.1016/j.cub.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 2.Errington A.C. Voltage gated ion channels: targets for anticonvulsant drugs. Curr. Top. Med. Chem. 2005;5:15–30. doi: 10.2174/1568026053386872. [DOI] [PubMed] [Google Scholar]
- 3.Yogeeswari P. Ion channels as important targets for antiepileptic drug design. Curr. Drug Targets. 2004;5:589–602. doi: 10.2174/1389450043345227. [DOI] [PubMed] [Google Scholar]
- 4.Abernethy D.R., Schwartz J.B. Calciumantagonist drugs. N. Engl. J. Med. 1999;341:1447–1457. doi: 10.1056/NEJM199911043411907. [DOI] [PubMed] [Google Scholar]
- 5.Sirois J.E. The TASK-1 two-pore domain K+ channel is a molecular substrate for neuronal effects of inhalation anesthetics. J. Neurosci. 2000;20:6347–6354. doi: 10.1523/JNEUROSCI.20-17-06347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldessarini R.J. Drugs and the treatment of psychiatric disorders: antipsychotic and antianxiety agents. In: Hardman J.G., editor. Goodman and Gilman’s The Pharmacological Basis of Therapeutics. ninth edition. McGraw-Hill Press; New York, USA: 1996. pp. 399–430. [Google Scholar]
- 7.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson J.D. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., Gallin W.J. VKCDB: voltage-gated potassium channel database. BMC. Bioinformatics. 2004;5:3. doi: 10.1186/1471-2105-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel V. PROSET—a fast procedure to create non-redundant sets of protein sequences. Math. Comput. Model. 1992;16:37–43. [Google Scholar]
- 11.Joachims T. Making large-scale SVM learning particle. In: Scholkopf B., editor. Advances in Kernal Methods: Support Vector Learning. MIT Press; Cambridge, USA: 1999. pp. 42–56. [Google Scholar]
- 12.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 13.Baldi P. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]