Abstract
Proteomics is the study of proteins and their interactions in a cell. With the completion of the Human Genome Project, the emphasis is shifting to the protein compliment of the human organism. Because proteome reflects more accurately on the dynamic state of a cell, tissue, or organism, much is expected from proteomics to yield better disease markers for diagnosis and therapy monitoring. The advent of proteomics technologies for global detection and quantitation of proteins creates new opportunities and challenges for those seeking to gain greater understanding of diseases. High-throughput proteomics technologies combining with advanced bioinformatics are extensively used to identify molecular signatures of diseases based on protein pathways and signaling cascades. Mass spectrometry plays a vital role in proteomics and has become an indispensable tool for molecular and cellular biology. While the potential is great, many challenges and issues remain to be solved, such as mining low abundant proteins and integration of proteomics with genomics and metabolomics data. Nevertheless, proteomics is the foundation for constructing and extracting useful knowledge to biomedical research. In this review, a snapshot of contemporary issues in proteomics technologies is discussed.
Key words: proteomics, mass spectrometry, protein chip
Introduction
Proteomics is the protein equivalent of genomics and has captured the imagination of biomolecular scientists worldwide. It encompasses a broad range of technologies aimed at determining the identity and quantity of expressed proteins in cells, their three-dimensional structure and interaction partners. In the postgenomic era, proteomics is a rapidly growing field of research that is becoming increasingly important. For example, it is extensively applied to the study of proteins involved in carcinogenesis, as well as to discover biomarkers for clinical use (1). Disease markers are widely used for screening, diagnosis, staging, prognosis, monitoring response to treatment, and detection of recurrent diseases.
As protein-protein interactions are the key to signal transduction to many regulatory events, a systematic classification of protein-protein interfaces is a valuable resource for understanding the principles of molecular recognition and for modeling protein complexes (2). The question of how one protein regulates the activity of another by binding to it requires an integration of structural, functional, and dynamic information. In the last decade, there have been many improvements in protein separation techniques, including two-dimensional polyacrylamide gel electrophoresis (2-D PAGE), liquid chromatography (LC), isotope-coded affinity tag (ICAT) labeling, and so on. Much progress has also been recently achieved in the analysis of proteins from tryptic digests using mass spectrometry (MS) and database searching. The newest generation of MS, combined with good separation techniques, is capable of providing rapid and confident protein identifications at the low femtomole level 3., 4., 5., 6..
From Genomics to Proteomics
With the completion of the Human Genome Project, the emphasis is shifting to the protein compliment of the human organism. This has given rise to the science of proteomics, the study of all the proteins produced by cell and organism, which involves the identification of proteins in the body and the determination of their roles in physiological and pathophysiological functions. As sequencing of the entire genomes of many prokaryotes and eukaryotes has been completed, there is a revival of interest in proteomics.
The term “proteome” refers to all the proteins expressed by a genome. While a genome remains unchanged to a large extent, the proteins in any particular cell change dramatically as genes are turned on or off in response to the environment. Since it is proteins that are directly involved in both normal and disease-associated biochemical processes, a more complete understanding of disease may be gained by looking directly into the proteins within a diseased cell or tissue. As a reflection of the dynamic nature of the proteome, some researchers use the term “functional proteome” to describe all the proteins produced by a specific cell in a single time frame. Whilst humans are estimated to have 30,000 to 40,000 genes potentially encoding 40,000 different proteins, alternative RNA splicing and post-translational modification (PTM) may increase this number up to 2,000,000 proteins or protein fragments (7). As a consequence, the proteome is far more complex than the genome.
Genomics has provided a vast amount of information linking gene activity with disease, but it does not predict PTM that most proteins undergo. Although DNA/RNA is easier to work with, there are limitations to the information that can be derived from DNA/RNA analysis. There are a number of reasons why gene sequence information and the pattern of gene activity in a cell do not provide a complete and accurate profile of a protein’s abundance or its final structure and state of activity. After synthesis on ribosomes, proteins are cut to eliminate initiation, transit, and signal sequences, as well as simple chemical groups or complex molecules that are attached. The gene transcript can be spliced in different ways prior to translation into protein. Following translation, most proteins are chemically changed through PTM, mainly through the addition of carbohydrate and phosphate groups. Such modification plays a vital role in modulating the function of many proteins but is not directly coded by genes. The PTMs are numerous (more than 300 types have been documented), static, and dynamic, including phosphorylation, glycosylation, acetylation, deamidation, palmitoylation, sulfation, and so on. As a consequence, the information from a single gene can encode as many as 50 different protein species. Therefore, DNA sequence analysis does not predict the active form of a protein and RNA quantitation does not always reflect the corresponding protein levels. Multiple proteins can be obtained from each gene when PTM and mRNA splicing are taken into account. DNA/RNA analysis cannot predict the amount of a gene product, including the type and amount of PTM when a gene is translated, or the events involving multiple genes such as aging, stress responses, drug responses, and pathological transformations. Beyond the genomic level, it is clear to see why genomic information often does not provide an accurate profile of protein abundance, structure, and activity.
Actually, genomics and proteomics are complementary fields, with proteomics extending the functional analysis. It is believed that through genomics and proteomics, new disease markers and drug targets can be identified, which will ultimately help design products to prevent, diagnose, and treat diseases. The future of medicine and biotechnology will be impacted greatly by genomics and proteomics, but there is much needed to be done to realize the potential benefits.
Proteomics Technologies
Two-dimensional polyacrylamide gel electrophoresis
Over three decades, 2-D PAGE has proven to be a reliable and efficient method for separation of proteins based on mass and charge. It can achieve the separation of several thousand different proteins in one gel. High-resolution 2-D PAGE can resolve up to 10,000 protein spots per gel. Stains such as Coomassie blue, silver, SYPRO Ruby and Deep Purple can be employed to visualize the proteins (8). The proteins are separated by charge (isoelectric point) in the first dimension and by mass in the second dimension. In isoelectric focusing, the proteins migrate in a pH gradient to the pH at which they have no net charge. The most common proteins are separated by isoelectric point in the horizontal direction and by size in the vertical direction.
Unfortunately, 2-D PAGE is a time-consuming and labor-intensive process. It implies some inherent limitations especially concerning hydrophobic and alkaline proteins, which are therefore often underrepresented in 2-D PAGE (9). Besides, this technique has low dynamic range and gel-to-gel variability. As an improvement, fluorescence-based difference gel electrophoresis overcomes the problems associated with traditional 2-D PAGE and allows more accurate and sensitive quantitative proteomics studies. Nowadays, 2-D PAGE analysis is often coupled with the MS technology. The 2-D PAGE with immobilized pH gradients, IPG-Dalt, has provided higher resolution, improved reproducibility, and higher loading capacity for preparative purposes with the intent of subsequent spot identification by MS.
Liquid chromatography
LC is a method of separating compounds within a sample so that they can be identified and quantified. There are increasing applications in proteomics research due to its ability to analyze large, fragile biomolecules. With advancements in ionization methods and instrumentation, LC combined with MS has become a powerful technology for the characterization and identification of peptides and proteins in a complex mixture (10). The bottom-up LC-MS approach to proteomics generally involves protease digestion followed by LC-MS with peptide mass fingerprinting or LC-MS/MS to derive sequence of individual peptides. Another significant improvement is the development of multidimensional protein identification technology, which separates peptides in 2-D LC. The separation can be interfaced directly with the ion source of a mass spectrometer (11).
Isotope-coded affinity tag labeling
Quantitative proteomics has recently emerged as a complementary technology to mRNA profiling with the ability to comprehensively characterize the structural and functional aspects of protein species. Because of the ionization variability for different peptides, the best internal standard for peptide quantitation is the same peptide labeled with stable isotopes. There are several protocols for the use of stable isotopes in proteomics, in which the ICAT labeling technique has received the most attention. The relative quantification is highly accurate because it is based on stable isotope dilution techniques (12). One of the major advantages of the ICAT labeling strategy is that the labeling reaction is compatible with commonly used reagents for 2-D PAGE. However, the ICAT labeling technique has some limitations in that it requires cystein-containing proteins and it provides only relative quantitation. A more recent development of ICAT is isobaric tags for relative and absolute quantification (iTRAQ), in which a set of isobaric reagents allows for the identification and quantitation of up to four different samples simultaneously. The multiplexing capacity of iTRAQ provides additional statistical validation within a given experiment (13). Moreover, the technique offers more extensive proteome coverage and enhanced sensitivity, making the tracking of PTMs feasible. The isobaric nature of the gross tag requires that the quantitation occurs in MS/MS rather than in MS mode, simplifying the analysis, but necessitating higher performance instrumentation. Anyway, the ICAT labeling strategy has been adopted into a number of proteomics studies 14., 15..
Mass spectrometry
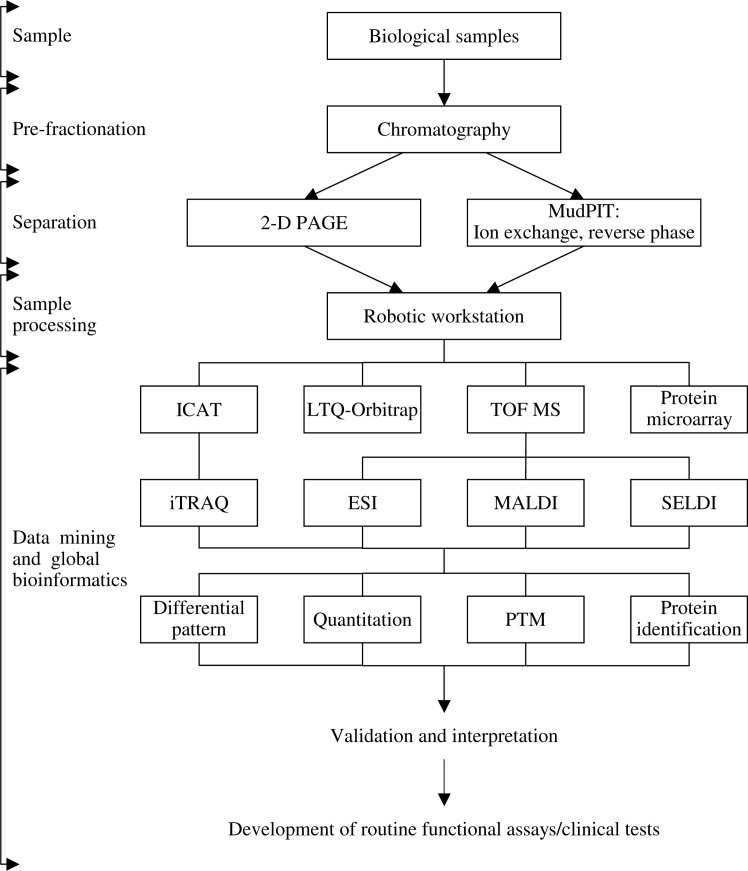
In order to exploit the protein profiles expressed under different physiological and pathophysiological conditions, protein analysis by MS is usually used as a powerful platform in proteomics. The technology is applied for mass determination and can be adapted for protein identification. A mass spectrometer separates proteins and other analytes according to their mass-to-charge (m/z) ratio. The molecule is ionized and the ion is propelled into a mass analyzer by an electric field that resolves each ion according to its m/z ratio (ionization of proteins allows them to be propelled towards the analyzer by virtue of charge repulsion). Then the detector passes the information to the computer for analysis. Frequently used ionization methods include electrospray ionization (ESI), matrix-assisted laser desorption/ionization (MALDI), and surface-enhanced laser desorption/ionization (SELDI), because they cause little or no fragmentation of the molecule during the ionization and desorption process (Figure 1).
Fig. 1.
The workflow of proteomics technologies. 2-D PAGE: two-dimensional polyacrylamide gel electrophoresis; ESI: electrospray ionization; ICAT: isotope-coded affinity tag; iTRAQ: isobaric tags for relative and absolute quantification; LTQ: linear ion trap quadrupole; MALDI: matrix-assisted laser desorption/ionization; MudPIT: multidimensional protein identification technology; PTM: post-translational modification; SELDI: surface-enhanced laser desorption/ionization; TOF MS: time-of-flight mass spectrometry.
Electrospray ionization
This technology involves the production of gaseous ions by application of a potential to a flowing liquid, resulting in the formation of a spray of small droplets with solvent-containing analyte. Solvent is removed from the droplet by heat or another form of energy such as collision with a gas, and multiply charged ions are formed. As the droplet size further decreases, they reach a point at which they become unstable and explode into even finer droplets. Finally, electrostatic repulsion is sufficiently high to cause desorption of the analyte ions, which are then passed to the mass spectrometer. Ions generated by ESI usually bear multiple charges (M+nH)n+. This can be mathematically transformed into a simple mass spectrum that reveals the molecular weights of the fragments. A quadrupole analyzer is frequently used with ESI. Only ions of a certain m/z will have the correct oscillation path that enables it to reach the detector in the presence of an electric field. One benefit of this method for analysis is that an LC column may be used as the source for the proteins entering the ESI mass spectrometer, which facilitates automation. A novel linear ion trap (LIT) mass spectrometer with MALDI and ESI ionization sources has been built in the MALDI-LIT-ESI configuration. The design features two independent ion source/ion optical channels connected to opposite ends of a single mass analyzer (16).
Matrix-assisted laser desorption/ionization
Ionization by MALDI involves a protein suspended or dissolved in a crystalline structure (the matrix) of small, organic, and UV-absorbing molecules. The crystal absorbs energy at the same wavelength of the laser that is used to ionize the protein. The laser energy strikes the matrix to cause rapid excitation of the matrix, and then the matrix and analyte ions are subsequently passed into gaseous phase. The principal ion detected using threshold laser intensity for MALDI is (M+H+), although signals for multiply charged ions and oligomeric forms of the analyte may be seen, especially for large proteins. The ionized protein is accelerated by an electrostatic field and expelled into a flight tube. As it exits the flight tube, the mass analyzer is encountered. The analyzer is often a time-of-flight (TOF) analyzer. This is based on the principle that when accelerated by application of a constant voltage, the velocity with which an ion reaches the detector is determined by its mass. MALDI is able to analyze proteins down to femtomole quantities. It can tolerate small amounts of contaminants. The information obtained from MALDI analysis can be automatically submitted to a database search for further examination (17). Currently, there is a development of direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded (FFPE) tissue sections using the strategy based on in situ enzymatic digestion of the tissue section after paraffin removal. The investigators therefore pinpointed the exact locations of proteins in an FFPE sample and anticipated that adding protein profiles to histological data would help physicians with difficult diagnoses (18).
Surface-enhanced laser desorption/ionization
SELDI is designed to perform mass spectrometric analysis of protein mixtures retained on chromatographic chip surfaces. Differentially expressed proteins may be determined from these protein profiles by comparing peak intensity. It can be used for protein purification, expression profiling, or protein interaction profiling. With only one microliter of sample, a high resolution mass spectrum following a complete chromatographic separation can be performed. There are many types of substances bound to the protein arrays, including antibodies, receptors, ligands, nucleic acids, carbohydrates, or chromatographic surfaces (that is, cationic, anionic, hydrophobic, or hydrophilic). Some surfaces have broad specificity that bind the whole classes of proteins, while others are highly specific that only a few proteins from a complex sample are bound. After the capture step, the array is washed to reduce nonspecific binding. When subjected to short bursts of laser light, the retained proteins are uncoupled from the array surface and are analyzed by laser desorption/ionization TOF MS. Some protein arrays contain antibodies covalently immobilized onto the array surface that capture corresponding antigens from a complex mixture. Many analyses can be followed, for example, analysis of proteolytic digests of the proteins bound to the array can disclose the antigenic determinant, other proteins of interest can be immobilized on the array, bound receptors can reveal ligands, and binding domains for protein-protein interactions can be detected. The power of the technology is the integration of on-chip selective capture, relative or absolute quantitation, and partial characterization of proteins and peptides from small samples, which allows rapid protein discovery and differential expression analysis. However, a difficulty of SELDI is that proteins must often remain folded in the correct conformation during the preparation and incubation with the array for protein-protein interactions to occur (19).
Linear ion trap quadrupole-Orbitrap mass spectrometry
Linear ion trap quadrupole (LTQ)-Orbitrap MS couples an LIT mass spectrometer to an Orbitrap mass analyzer via a radio frequency-only trapping quadrupole with a curved axis. The latter injects pulsed ion beams into a rapidly changing electric field in the Orbitrap wherein they are trapped at high kinetic energies around an inner electrode. Image current detection is subsequently performed after a stable electrostatic field is achieved. Fourier transformation of the acquired transient allows wide mass range detection with high resolving power, mass accuracy, and dynamic range. The entire instrument operates in LC/MS mode (1 spectrum/s) with nominal mass resolving power of 60,000 and uses automatic gain control to provide high accuracy mass measurements, within 2 ppm using internal standards and within 5 ppm with external calibration. The maximum resolving power exceeds 100,000 full width at half maximum. Rapid, automated data-dependent capabilities enable real-time acquisition of up to 3 high mass accuracy MS/MS spectra per second (20). LTQ-Orbitrap MS is often used as a powerful tool in top-down proteomics with its high performance for mass spectrometric protein characterization 21., 22..
Protein identification
MS can also be used to identify proteins. Proteolytic digestion of gel-separated proteins into peptides and mass analysis of the peptides provide a peptide mass fingerprint, which can be searched against theoretical fingerprints of sequences in the databases. Alternatively, the peptides can be sprayed into a tandem mass spectrometer that has the ability to resolve peptides, isolate one peptide, and dissociate it into amino or carboxy terminal containing fragments. Although it is more complex than the peptide mass approach, the sequence information is more specific for the identification of a protein than a list of peptide masses. The data can be used to search protein sequence and nucleotide databases. Protein identification using ESI-MS/MS can be fully automated. This involves using three quadrupole mass analyzers in series. Peptides resulting from chemical or enzymatic cleavage are separated by high performance LC and sent to the first analyzer. One peptide is chosen by the first quadrupole, passed to the second quadrupole where it is fragmented, and the fragmented ions are analyzed by the third quadrupole. The difference between neighboring ions in the product ion spectrum represents the amino acid at that position of the sequence and its mass can be determined by subtraction. The data may also be analyzed by application of algorithms and comparison with theoretical production spectra of proteins in the databases (23). On the other hand, the shotgun proteomics strategy, based on digesting proteins into peptides and sequencing them using tandem MS, has become widely adopted (24). There is also a new fast method for identification and characterization of proteolytic digests of proteins by monolithic LC coupled with MS. The advantages of the monolithic columns are a high-pressure stability and low back pressure, resulting in higher flow rates for capillary or nanosize columns simplifying the system handling. It is successfully applied for the detection of PTMs and the analysis of membrane proteins (25).
Post-translational modification
Proteomics has the significant advantage of being able to discern not only changes in expression levels but also in PTMs. Proteomics-based profiling uniquely allows delineation of global changes in expression patterns resulting from transcriptional and post-transcriptional control, PTMs, and shifts in proteins between cellular compartments (26). Analysis of a protein for PTM such as phosphorylation or glycosylation is very important for understanding issues such as activity, stability, and turnover. Peptides that do not have the expected molecular mass are further analyzed. Post-translational regulation of proteins via protein phosphorylation is one of the major means of protein regulation. Phosphorylation is a very rapid and reversible method of changing the function of proteins (27). Reversible protein phosphorylation is crucially involved in all aspects of cell physiology. The highly challenging task of revealing and characterizing the dynamic protein phosphorylation networks has only recently begun to become feasible. MS is especially useful for the analysis of expressed proteins with PTM, such as alkylation, glycosylation, and phosphorlylation, which may be the most important regulatory proteins in a biological cell. To identify the specific sites of modification in the analysis of enzymatically digested protein, tandem MS with collision-induced dissociation of peptides inside MS can be used (28). Immobilized metal affinity chromatography is most useful for phosphopeptide enrichment after methylation of the peptides in the complex protein digests. The subsequent tandem MS of the isolated phosphopeptides results in both the identification of phosphorylated proteins and the mapping of the in vivo phosphorylation sites (29). Phosphoproteins can be revealed by comparing the map of a sample digested with a proteolytic enzyme to the same sample digested with the proteolytic enzyme and a phosphatase. Similarly, n-linked sugars can be detected by use of glycosylases. Antibodies recognizing phosphotyrosines or phosphoserines confirm the presence of phosphate groups. These phosphoantibodies can be employed to precipitate phosphorylated proteins before MS. In addition to MS, stains are also used to detect phosphoproteins in polyacrylamide gels (30). Proteomics approaches have been used for a number of PTM studies. For example, Odhiambo et al. have used MALDI-TOF MS and LC-MS/MS to identify oxidative PTMs of serum albumin in patients with idiopathic pulmonary arterial hypertension and pulmonary hypertension of sickle cell anemia (31).
Phage display
Another technique making an important contribution to proteomics is phage display. It involves the creation of peptide or protein libraries on viral surfaces that are screened for activity as a group. The peptides or proteins remain associated with their corresponding genes allowing identification. An alternative to phage display is cloning of ligand targets. It uses ligands, which are small peptide sequences that bind to larger domain units within proteins, to discover new domains and subsequently new proteins (32). Phage display has been used extensively in vitro and in animal models to generate ligands and to identify disease-relevant targets. A series of patients have been reported to receive phage library for serial panning of disease targeting ligands (33). For example, Geuijen et al. have obtained a panel of myeloid cell binding single chain variable fragments by applying phage display selection on myeloid cell lines, followed by the identification of several antibody target molecules on malignant cells (34).
Bioinformatics
Bioinformatics is an integration of mathematical, statistical, and computational methods to analyze biological, biochemical, and biophysical data. Sophisticated bioinformatics tools are being developed to handle the vast quantity of data resulting from genomics and proteomics studies. Collections of proteomics databases for organisms contain biological information on proteins that assists with annotating and interpreting experimental results from proteomics studies. Databases of computationally derived protein structures are available to aid in the design of molecules capable of interacting with a protein of interest (35). A wide range of research areas in bioinformatics, molecular biology, and medicinal chemistry require precise chemical structure information about molecules and reactions, such as drug design, ligand docking, metabolic network reconstruction, and systems biology. A database of metabolites called BioMeta that augments the existing pathway databases by explicitly assessing the validity, correctness, and completeness of chemical structure and reaction information has been recently presented (36). Bioinformatics tools are essential for converting raw proteomics data into knowledge and subsequently into useful applications. A good example would be the development of methodology for predicting protein-protein interactions by Shen et al. based on sequence information (37).
Challenges and Perspectives
Challenges
There are several difficulties in the study of proteins that are not inherent in the study of nucleic acids. Proteins are more difficult to work with than DNA and RNA. They have secondary and tertiary structure that must often be maintained during their analysis. Proteins can be denatured by the action of enzymes, heat, light, or by aggressive mixing as in beating egg whites. Some proteins are difficult to analyze due to their poor solubility.
On the other hand, proteins cannot be amplified like DNA, therefore less abundant species are more difficult to detect. Approximately half of the total protein content in plasma comes from albumin (∽55 mg/mL), and together with another 10 proteins, they make up 90% of the total content. Low abundant proteins such as cytokines are normally present at 1–5 pg/mL. Each proteomics technology can only analyze proteins within 3–4 orders of magnitude, and mostly at the higher concentration end of the spectrum. The removal of high abundant proteins from plasma or serum is thus a prerequisite for conducting more detailed proteomics studies on low abundant proteins. However, many potentially important biomarkers may be lost in this process due to non-specific binding or the co-removal of proteins/peptides intrinsically bound to the high abundant carrier proteins. In particular albumin, removal has been shown to result in significant loss of cytokines (38). Thulasiraman et al. developed the new deep proteome approach via ligand library beads. The use of equalizer beads coupled with a combinatorial library of ligands has been shown to allow access of many low abundant proteins or polypeptides undetectable by classical analytical methods. The population of beads has such diversity that a binding partner should exist for most proteins in a sample. Each bead has an equivalent binding capacity. High abundant proteins saturate their binding partners and the excess proteins are washed away, whereas trace proteins are concentrated on their specific affinity ligands. This panoply of methods could offer a strong step forward in “mining below the tip of the iceberg” for detecting the “unseen proteome” (39).
To tackle with the sensitivity issue, Nettikadan et al. constructed the ultramicroarrays, combining the advantages of microarray including multiplexing capabilities, higher throughput, and cost savings with the ability to screen very small sample volumes. These ultramicroarrays were found to have a high specificity and sensitivity with detection levels using purified proteins in the attomole range. This strategy enables the proteomics analysis of materials that are available in very limited quantities, such as those collected by laser capture microdissection, neonatal biopsy microspecimens, and forensic samples (40).
Apparently, a number of technical obstacles remain before routine proteomics analysis can be achieved in the clinic. However, the standardization of methodologies and dissemination of proteomics data into publicly available databases is starting to overcome these hurdles. The cost is also a precluding factor for the widespread use of proteomics in clinical laboratory. Most proteomics technologies use complex instrumentation, critical computing power, and expensive consumables. Another major challenge will be the integration of proteomics with genomics and metabolomics data, as well as their functional interpretation in conjunction with clinical results and epidemiology (41).
Perspectives
With the availability of DNA microarray analysis permitting the expression of thousands of genes to be monitored simultaneously, it may be asked why proteomics is so important. The importance of the proteome cannot be overstated as it is the proteins within the cell that provide structure, produce energy, as well as allow communication, movement, and reproduction. Basically, proteins provide structural and functional framework for cellular life. Genetic information is static while the protein complement of a cell is dynamic.
Differential proteomics is a scientific discipline that detects the proteins associated with a disease by means of their altered levels of expression between the control and disease states. It enables correlations to be drawn between the range of proteins produced by a cell or tissue and the initiation or progression of a disease state. Proteomics research permits the discovery of new protein markers for diagnostic purposes and the study of novel molecular targets for drug discovery. The protein markers identified have a broad range of potential applications. They may be used for clinical diagnostic or prognostic purposes. Biomarkers may also be used to help devising an optimal therapeutic treatment plan for different patient subsets and to monitor the effect of treatment. In this way, protein markers may be used to accelerate the speed and efficacy of clinical trials. If further biochemical research reveals that proteins have a causal role in disease pathology, they may have the utility as molecular targets for therapeutic intervention in the disease.
Proteomics has much promise in novel drug discovery via the analysis of clinically relevant molecular events. New types of proteomics technologies combined with advanced bioinformatics are currently being used to identify molecular signatures of diseases based on protein pathways and signaling cascades. Applying their findings will improve our understanding of the roles of individual proteins or the entire cellular pathways in the initiation and development of disease. It is envisaged that analyzing the cellular circuitry of ongoing molecular networks will become a powerful clinical tool in individualized patient management (42).
The abundance of information provided by proteomics research is entirely complementary with the genetic information being generated by genomics research. Proteomics makes a key contribution to the development of functional genomics. The combination of genomics and proteomics will play a major role in biomedical research, and it will have a significant impact on the development of diagnostic and therapeutic products in the future.
References
- 1.Cho W.C. Contribution of oncoproteomics to cancer biomarker discovery. Mol. Cancer. 2007;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim W.K. The many faces of protein-protein interactions: a compendium of interface geometry. PLoS Comput. Biol. 2006;2 doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormack A.L. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 4.Bogan M.J., Agnes G.R. Wall-less sample preparation of microm-sized sample spots for femtomole detection limits of proteins from liquid based UV-MALDI matrices. J. Am. Soc. Mass Spectrom. 2004;15:486–495. doi: 10.1016/j.jasms.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Cho W.C., Cheng C.H. Oncoproteomics: current trends and future perspectives. Expert Rev. Proteomics. 2007;4:401–410. doi: 10.1586/14789450.4.3.401. [DOI] [PubMed] [Google Scholar]
- 6.Cho W.C. Application of proteomics in Chinese medicine research. Am. J. Chin. Med. 2007 doi: 10.1142/S0192415X07005375. (In press) [DOI] [PubMed] [Google Scholar]
- 7.Kosak S.T., Groudine M. Gene order and dynamic domains. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- 8.Lauber W.M. Mass spectrometry compatibility of two-dimensional gel protein stains. Electrophoresis. 2001;22:906–918. doi: 10.1002/1522-2683()22:5<906::AID-ELPS906>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson C.L. Identification of protein vaccine candidates from Helicobacter pylori using a preparative two-dimensional electrophoretic procedure and mass spectrometry. Anal. Chem. 2000;72:2148–2153. doi: 10.1021/ac9912754. [DOI] [PubMed] [Google Scholar]
- 10.Chen G. Applications of LC/MS in structure identifications of small molecules and proteins in drug discovery. J. Mass Spectrom. 2007;42:279–287. doi: 10.1002/jms.1184. [DOI] [PubMed] [Google Scholar]
- 11.Florens L., Washburn M.P. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 12.Gygi S.P. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 13.Zieske L.R. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 14.Dean R.A., Overall C.M. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Sethuraman M. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic. Biol. Med. 2007;42:823–829. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.A. Dual-source mass spectrometer with MALDI-LIT-ESI configuration. J. Proteome Res. 2007;6:837–845. doi: 10.1021/pr060514i. [DOI] [PubMed] [Google Scholar]
- 17.Andersson T. Automating MALDI sample plate loading. J. Proteome Res. 2007;6:894–896. doi: 10.1021/pr0603607. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire R. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 19.Cho W.C. Research progress in SELDI-TOF MS and its clinical applications. Chin. J. Biotech. 2006;22:871–876. doi: 10.1016/S1872-2075(06)60061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarov A. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2220. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 21.Adachi J. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi R. Analysis of the mouse liver proteome using advanced mass spectrometry. J. Proteome Res. 2007 doi: 10.1021/pr0605668. (In press) [DOI] [PubMed] [Google Scholar]
- 23.Sadygov R. Central limit theorem as an approximation for intensity-based scoring function. Anal. Chem. 2006;78:89–95. doi: 10.1021/ac051206r. [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii A.I. Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol. Biol. 2006;367:87–120. doi: 10.1385/1-59745-275-0:87. [DOI] [PubMed] [Google Scholar]
- 25.Marcus K. A new fast method for nanoLC-MALDI-TOF/TOF-MS analysis using monolithic columns for peptide preconcentration and separation in proteomic studies. J. Proteome Res. 2007;6:636–643. doi: 10.1021/pr060406w. [DOI] [PubMed] [Google Scholar]
- 26.Chignard N., Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127:S120–S125. doi: 10.1053/j.gastro.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Gafken P.R., Lampe P.D. Methodologies for characterizing phosphoproteins by mass spectrometry. Cell Commun. Adhes. 2006;13:249–262. doi: 10.1080/15419060601077917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J. Relative information content and top-down proteomics by mass spectrometry: utility of ion/ion proton-transfer reactions in electrospray-based approaches. Anal. Chem. 2007;79:1073–1081. doi: 10.1021/ac061798t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turkina M.V., Vener A.V. Identification of phosphorylated proteins. Methods Mol. Biol. 2007;355:305–316. doi: 10.1385/1-59745-227-0:305. [DOI] [PubMed] [Google Scholar]
- 30.Yanagida M. Matrix assisted laser desorption/ionization-time of flight-mass spectrometry analysis of proteins detected by antiphosphotyrosine antibody on two-dimensional-gels of fibrolast cell lysates after tumor necrosis factor-alpha stimulation. Electrophoresis. 2000;21:1890–1898. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1890::AID-ELPS1890>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Odhiambo A. Identification of oxidative post-translational modification of serum albumin in patients with idiopathic pulmonary arterial hypertension and pulmonary hypertension of sickle cell anemia. Rapid Commun. Mass Spectrom. 2007;21:2195–2203. doi: 10.1002/rcm.3074. [DOI] [PubMed] [Google Scholar]
- 32.Stratmann T., Kang A.S. Cognate peptidereceptor ligand mapping by directed phage display. Proteome Sci. 2005;3:7. doi: 10.1186/1477-5956-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krag D.N. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 34.Geuijen C.A. A proteomic approach to tumour target identification using phage display, affinity purification and mass spectrometry. Eur. J. Cancer. 2005;41:178–187. doi: 10.1016/j.ejca.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Evanko D. Systems biology for beginners. Nat. Methods. 2006;3:964–965. doi: 10.1038/nmeth1206-964b. [DOI] [PubMed] [Google Scholar]
- 36.Ott M.A., Vriend G. Correcting ligands, metabolites, and pathways. BMC Bioinformatics. 2006;7:517. doi: 10.1186/1471-2105-7-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen J. Predicting protein-protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Granger J. Albumin depletion of human plasma also removes low abundance proteins including the cytokines. Proteomics. 2005;5:4713–4718. doi: 10.1002/pmic.200401331. [DOI] [PubMed] [Google Scholar]
- 39.Thulasiraman V. Reduction of the concentration difference of proteins in biological liquids using a library of combinatorial ligands. Electrophoresis. 2005;26:3561–3571. doi: 10.1002/elps.200500147. [DOI] [PubMed] [Google Scholar]
- 40.Nettikadan S. Detection and quantification of protein biomarkers from fewer than 10 cells. Mol. Cell. Proteomics. 2006;5:895–901. doi: 10.1074/mcp.M500350-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Kolch W. The molecular make-up of a tumour: proteomics in cancer research. Clin. Sci. 2005;108:369–383. doi: 10.1042/CS20050006. (Lond.) [DOI] [PubMed] [Google Scholar]
- 42.Gulmann C. Array-based proteomics: mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J. Pathol. 2006;208:595–606. doi: 10.1002/path.1958. [DOI] [PubMed] [Google Scholar]