Abstract
We propose a new method for tumor classification from gene expression data, which mainly contains three steps. Firstly, the original DNA microarray gene expression data are modeled by independent component analysis (ICA). Secondly, the most discriminant eigenassays extracted by ICA are selected by the sequential floating forward selection technique. Finally, support vector machine is used to classify the modeling data. To show the validity of the proposed method, we applied it to classify three DNA microarray datasets involving various human normal and tumor tissue samples. The experimental results show that the method is efficient and feasible.
Key words: independent component analysis, feature selection, support vector machine, gene expression data
Introduction
With the advent of DNA microarrays, it is now possible to simultaneously monitor the expression of all genes in the genome. Increasingly, the challenge is to interpret such data to gain insight into biological processes and the mechanisms of human disease. To deal with such challenge, new statistical methods must be introduced to analyze those large amounts of data yielded from microarray experiments.
One of the central goals in microarray expression data analysis is to identify the tumor category. A reliable and precise classification of tumors is essential for successful diagnosis and treatment of cancer. However, traditional methods for classifying human malignancies mostly rely on a variety of morphological, clinical, and molecular variables. Despite recent progress, there are still many uncertainties in diagnosis. Furthermore, it is likely that the existing classes of tumors are heterogeneous diseases that are molecularly distant. Recently, with the development of large-scale high-throughput gene expression technology, it has become possible for researchers to directly diagnose and classify diseases, particularly cancers 1., 2.. By monitoring the expression levels in cells for thousands of genes simultaneously, microarray experiments may lead to a more complete understanding of the molecular variations among tumors, and hence to a finer and more reliable classification.
With the wealth of gene expression data from microarrays being produced, more and more new prediction, classification, and clustering techniques are being used for analysis of the data. Up to now, several studies have been reported on the application of microarray gene expression data analysis for molecular classification of cancer 3., 4., 5.. The analysis of differential gene expression data has been used to distinguish between different subtypes of lung adenocarcinoma (6) and colorectal neoplasm (7). The method that predicted clinical outcomes in breast cancer 8., 9. and lymphoma (10) from gene expression data has been proven to be successful. Golub et al. (2) utilized a nearest-neighbor classifier method for the classification of acute myeloid lymphoma and acute leukemia lymphoma in children. Furey et al. (5) proposed to use SVM as the classifier. Nguyen and Rocke (11) used partial least squares to reduce the dimension of gene expression data and then used logistic discrimination and quadratic discriminant analysis to classify them. Pochet et al. (12) performed a systematic benchmarking study of microarray data classification, and gave some useful conclusions about how to use different classification methods. Dudoit et al. (13) performed a systematic comparison of several discrimination methods for classification of tumors based on microarray experiments. While linear discriminant analysis was found to perform the best, in order to utilize the method, the number of genes selected had to be drastically reduced from thousands to tens using a univariate filtering criterion.
In spite of the harvests achieved till now, one of the challenges of bioinformatics is to develop new efficient ways to analyze global gene expression data. A rigorous approach to gene expression data analysis must involve an up-front characterization of the structure of the data. In addition to a broader utility in analysis method, principal component analysis (PCA) (14) can be a valuable tool for obtaining such a characterization. In gene expression data analysis applications, PCA is a popular unsupervised statistical method for finding useful eigenassay or eigengene (14). One goal for the PCA technique is to find a “better” set of eigenassay so that in this new basis the snapshot coordinates (the PCA coefficients) are uncorrelated, that is, they cannot be linearly predicted from each other. One characteristic of the PCA technique is that only second-order statistical information is used. However, in the task such as classification, much of the important information may be contained in the high-order relationships among samples. Therefore, it is important to investigate whether the generalizations of PCA are sensitive to high-order relationships, not just second-order relationships. Generally, independent component analysis (ICA) (15) is one of such generalizations. A number of algorithms for performing ICA have been proposed (16). Here, we employ FastICA, which was proposed by Hyvärinen (17) and has been proven successful in many applications.
In this paper, we propose a new method for tumor classification from gene expression data. Firstly, the original DNA microarray gene expression data are modeled by ICA. Secondly, the most discriminant eigenassays extracted by ICA are selected by the sequential floating forward selection (SFFS) technique 18., 19.. Finally, support vector machine (SVM) is used to classify the modeling data. To validate the efficiency, the proposed method was applied to classify three different DNA microarray datasets of colon cancer (3), acute leukemia (2), and high-grade glioma (20). The prediction results show that our method is efficient and feasible.
Model
ICA
ICA is a useful extension of PCA that has been developed in context with blind separation of independent sources from their linear mixtures (15). Such blind separation techniques have been used in various applications such as auditory signal separating and medical signal processing (16). In a sense, the starting point of ICA is the uncorrelatedness property of the standard PCA. Roughly speaking, rather than requiring that the coefficients of a linear expansion of the data vectors be uncorrelated, in ICA they must be mutually independent (or as independent as possible). This implies that higher-order statistics are needed in determining the ICA expansion.
Considering an n × p data matrix X, whose rows ri (i = 1, · · ·, n) correspond to observational variables and whose columns Cj (j = 1, · · ·, p) are the individuals of the corresponding variables, the ICA model of X can be written as:
| (1) |
Without loss of generality, A is an n × n mixing matrix, and S is an n × p source matrix subject to the condition that the rows of S are as statistically independent as possible. Those new variables contained in the rows of S are called “independent components”, that is, the observational variables are linear mixtures of independent components. The statistical independence between variables can be quantified by mutual information , where is the marginal entropy of the variable sk, p(sk) is the probabilistic density function, and H(S) is the joint entropy (17). Estimating the independent components can be accomplished by finding the right linear combinations of the observational variables, since we can invert the mixing as:
| (2) |
So far there have been a number of algorithms for performing ICA 16., 21., 22.. In this paper, we employ the FastICA algorithm (17) to address the problems of tumor classification. In this algorithm, the mutual information is approximated by a “contrast function”:
| (3) |
where G is any nonquadratic function and v is a normally distributed variable. For more details please see the literature (17).
Like PCA, ICA can remove all linear correlations and only take into account higher-order dependencies in the data. Yet, ICA is superior to PCA since PCA is just sensitive to second-order relationships of the data. In addition, the ICA model usually leaves some freedom of scaling and sorting by convention; the independent components are generally scaled to unit deviation, while their signs and orders can be chosen arbitrarily. In general, the number of independent components equals to the number of the observational variables.
It should be noted that ICA is a very general technique. When super-Gaussian sources are used, ICA can be seen as doing something akin to nonorthogonal PCA and to cluster analysis. In particular, we have empirically observed that many gene expression data are “sparse” or “super-Gaussian” signals (all the three datasets used in this paper are “super-Gaussian” signals). When sparse source models are appropriate, ICA has the following potential advantages over PCA: (1) It provides a better probabilistic model interpretation of the data, which better identifies the position where the data are to concentrate on n-dimensional space; (2) It uniquely identifies the mixing matrix A; (3) It finds a not-necessarily orthogonal basis that may reconstruct the data better than PCA do in the presence of noise; (4) It is sensitive to high-order statistics in the data, not just the covariance matrix (23).
However, when the source models are sub-Gaussian, the relationship between these techniques is less clear. Please see Lee et al. (24) for a discussion of ICA in the context of sub-Gaussian sources.
ICA models of gene expression data
Now let the n × p matrix X denote the gene expression data (generally n<<p), and xij is the expression level of the jth gene in the ith assay. ri (a p-dimensional vector), the ith row of X, denotes the snapshot of the ith assay. Alternatively, cj (an n-dimensional vector), the jth column of X, is the expression profile of the jth gene. We suppose that the data have already been preprocessed and normalized, that is, every gene expression profile has mean zero and standard deviation one.
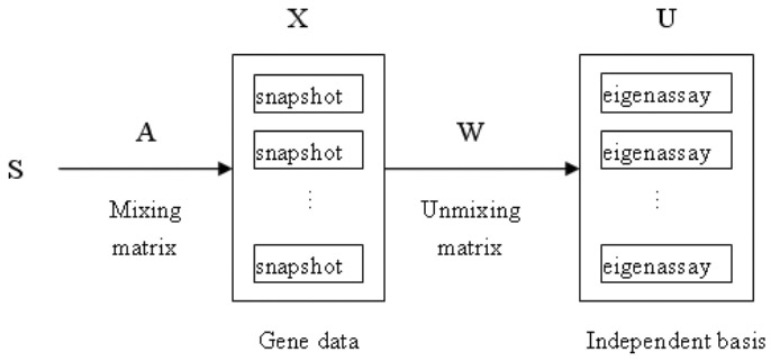
Regardless of which algorithm is used to compute ICA, we can apply ICA to model gene expression data as shown in Figure 1. In this model, the snapshots ri in X are considered to be a linear mixture of statistically independent basis snapshots (eigenassay) S combined by an unknown mixing matrix A. The ICA algorithm learns the weight matrix W, which is used to recover a set of independent eigenassays in the rows of U. In this architecture, the snapshots are variables and the gene expression profile values provide observations for the variables. Essentially, this method coincides with traditional ICA, such as the model of cocktail problem (16). Projecting the input snapshots onto the learned weight vectors produces the independent basis snapshots. As a result, the corresponding mixing and unmixing models can be represented as in (1), (2).
Fig. 1.
The gene expression data synthesis model. To find a set of independent basis snapshots (eigenassay), the snapshots in X are considered to be a linear combination of statistically independent basis snapshots (the rows in S), where W is the unmixing matrix and A is an unknown mixing matrix. The independent eigenassay is estimated as the output U of the learned ICA.
In this approach, ICA is used to find a matrix W such that the rows of U are as statistically independent as possible. The independent eigenassays estimated by the rows of U are then used to represent the snapshots. The representation of the snapshots consists of their corresponding coordinates with respect to the eigenassays defined by the rows of U, as shown below:
| (4) |
These coordinates are contained in the rows of the mixing matrix, A = W−1. Clearly, every coordinate aj (row of A) is an n-dimensional vector while the snapshot rj is a p-dimensional vector. In general, the number of genes in a single assay is in thousands while the number of assays is up to hundreds. So the above procedure can be used to compress the gene expression data. In this paper, we just use this idea to find a good set of basis snapshots (eigenassays) to represent gene expression data so that they can be reasonably classified.
From another viewpoint, the gene expression profiles (columns of X) can be regarded as points in a multidimensional space with dimensions corresponding to the number of samples. The linear ICA model X = AS represents the gene expression profiles (the columns of X) by a new set of basis vectors (the columns of A). This idea is based on two assumptions. First, the gene expression profiles are determined by a combination of hidden regulatory variables, which are called “expression modes”. Second, the genes’ responses to these variables can be approximated by linear functions 25., 26.. Expression mode k is characterized by its profile over the samples (kth column of A) and by its linear influences on the genes (kth row of S).
Interpretation of ICA model
The ICA model states that different modes can exert independent influences on the genes. To interpret this point in more details, the first step of the analysis is the study of the mixing matrix A. For a fixed eigenassay i, the coefficients aji represent the projection of snapshot j on source i, or the “importance” of eigenassay i in snapshot j. If one believes in the “linear mixture of independent eigenassay” model, and accepts identifying a source with a regulation pathway in first approximation, the coefficients aji would allow one to assert to which extent the eigenassay i is (positively or negatively) “active” in snapshot j.
In addition, the distribution of the column values of the mixing matrix A is often interesting and may reveal specific features of the dataset. Particularly interesting is the situation where the distribution of mixing coefficients for a given eigenassay exhibits a bimodal or multimodal behavior. This indicates that the source under consideration has a good discriminating power between two or more different classes of conditions. However, as Chiappetta et al. (27) have pointed out, even though bimodal distributions yield spectacular results, good discrimination may also be obtained without such a behavior.
A second step in the interpretation of the ICA results is to analyze carefully the behavior of specific genes in different eigenassays. It generally happens that a given independent eigenassay is characterized by a number of significantly overexpressed (or underexpressed) genes. Putting such genes into correspondence with snapshots, or clinical data, may happen to be extremely informative. Because the main aim of this paper is not to study biological interpretation of ICA results for microarray data, moreover, there have been many literatures involved in this issue, so we will not discuss it in details here. Readers who are interested in this issue can further see the literatures 25., 26., 27., 28. for the details.
Searching for the consensus eigenassays
Chiappetta et al. (27) have pointed out that unlike PCA, ICA requires searching for the maxima of a target function in a large-dimensional configuration space. Therefore, one often encounters difficulties with local maxima in which most algorithms may get stuck, and the result may be sensitive to initialization. We also found in experiment that compared with PCA, ICA is not always reproducible when used to analyze gene expression data. This problem had also been found by previous studies 26., 27.. In addition, the results obtained from an ICA algorithm are not “ordered”. Chiappetta et al. (27) concluded that, the reason of this phenomenon is that the ICA algorithm may converge to local optima. Moreover, they have proposed a “consensus source” (eigenassay) search algorithm, which yields extremely stable and robust estimates for the eigenassays as well as indications relative to their stability.
In this paper, we use the method advised by Chiappetta et al. (27) to overcome these difficulties with the following procedure. The independent source estimate is run for certain times (say, 100 times) with different random initializations, and “consensus sources” are recorded. In other words, the eigenassays obtained with a frequency larger than a certain threshold are conserved, and their frequencies of appearance are recorded and used as “credibility indices”. As a result, one is led to a (variable, data-driven) number of average consensus eigenassays .
Finally, the corresponding consensus mixing matrix A is computed as:
| (5) |
where V is the inverse of the n × n matrix C of the scalar product of the consensus eigenassays . More details can be found in the literature (27).
Feature selection
Generally, not all of the features are used for classification. To achieve good classification results, some features should be discarded. As a matter of fact, one goal of this study is the automatic selection of the best feature subset from a given ICA feature vector for classification. In contrast to the PCA method, where feature subset selection is based on an energy criterion, the selection of an ICA basis subset is not immediately obvious since the energies of the independent components cannot be determined in advance. Furthermore, it is conjectured that some feature selection schemes focusing on “recognition” rather than on “reconstruction” could augment the classification performance. With this goal in mind, we use the SFFS technique (18) to find the most discriminating ICA features.
For this SFFS method, features are selected successively by adding the locally best feature points, which provides the highest incremental discriminatory information, to the exiting feature subset. In addition, the SFFS method goes through cleaning periods, in which features are removed systematically so long as the performance is improved after pruning. We use the leave-one-out cross-validation (LOOCV) in the training dataset to determine the number of components to be included in the model. In each LOOCV iteration (the number of iterations equals the sample size), one sample is left out of the data, a classification model is trained on the rest of the data, and this model is then evaluated on the left out data point. As an evaluation measure, the LOOCV performance is used. More details about SFFS can refer to the literature (29).
Classifier
After processing the gene expression data using ICA, the final step is to classify the dataset. There have been many methods for performing the classification tasks so far, such as radial basis function neural network (30), logistic discrimination, and quadratic discriminant analysis (11). Because the dimension of DNA microarray gene expression data is higher even after they are processed by ICA, and there are only few samples of the data achieved in general, we use SVM 31., 32., 33., which has been proved to be very useful and robust 34., 35., 36., to classify the gene expression data.
When it is used for classification, SVM can separate a given set of binary labeled training data with a hyper-plane that is maximally distant from them (the maximal margin hyper-plane). For the cases in which no linear separation is possible, they can work in combination with the technique of “kernels”, which automatically realizes a nonlinear mapping to a feature space. Generally, the hyper-plane founded by the SVM in a feature space corresponds to a nonlinear decision boundary in the original space.
Without loss of generality, let the ith input sample be the realization of the random vector β, and this input sample is labeled by the random variable γ ∈ {−1, +1}. Assume that ϕ : U ⇒ V (U ⊆ Rp, V ⊆ Rq) is a mapping from the input space U to a feature space V, and that we have a set of samples θ of m labeled data points: θ = {(β1, γ1), …, (βm, γm)}.The SVM learning algorithm is to find a hyper-plane (ω, b) such that the quantity
| (6) |
is maximized, where 〈〉 denotes an inner product, the vector ω has the same dimensionality as V, ‖ω‖2 is held as a constant, b is a real number, and χ is called the margin. The quantity corresponds to the distance between the point βi and the decision boundary. When multiplied by the label γi, it gives a positive value for all correct classifications and a negative value for all the incorrect ones. The minimum of this quantity over all the data is positive if the data is linearly separable, which is called “margin”. Given a new data sample β to be classified, a label is assigned according to its relationship to the decision boundary, and the corresponding decision function is:
| (7) |
Evaluation
To validate the efficiency of the proposed method, we applied it to classify three different DNA microarray datasets of colon cancer (3), acute leukemia (2), and high-grade glioma (20). All data samples have already been assigned to a training set or test set. An overview of the characteristics of all the datasets can be found in Table 1. The acute leukemia dataset (1) has already been used frequently in previous microarray data analysis studies. In our experiment, this dataset is preprocessed by setting threshold and log-transformation on the original data, similar to the way in the original publication. Threshold technique is generally achieved by restricting gene expression levels to be larger than 20. In other words, the expression levels that are smaller than 20 will be set to 20. Regarding the log-transformation, the natural logarithm of the expression levels is usually taken. In addition, no further preprocessing is applied to the rest of the datasets.
Table 1.
Overview of the three datasets for classification
| Dataset | No. of training set |
No. of test set |
No. of genes | ||
|---|---|---|---|---|---|
| Class 1 | Class 2 | Class 1 | Class 2 | ||
| Colon cancer | 14 | 26 | 8 | 14 | 2,000 |
| Acute leukemia | 11 | 27 | 14 | 20 | 7,129 |
| High-grade glioma | 21 | 14 | 14 | 15 | 12,625 |
Since all data samples in the three datasets have already been assigned to a training set or test set, we built the classification models using the training samples, and estimated the classification correct rates using the test set.
To simplify the computation, we normalized the expression values for each of the genes such that each sample has zero mean and unit variance. We first performed ICA on Xtn to produce two matrixes Atn and U such that
| (8) |
| (9) |
Hence, the rows of Atn contain the coefficients (representations) of the linear combination of statistically independent sources (rows of S) that comprise Xtn. For the test set Xtt, we can achieve their representations by the following equation:
| (10) |
After the representations of the training and test data have been achieved, we then used SFFS and SVM to select independent features for experiment. The numbers of the selected features were determined by using LOOCV in the training dataset. What should be denoted is that the eigengenes (columns of A) and the eigenassays (rows of S) were not simply calculated by FastICA. In experiments, they were calculated by using ICA and consensus sources algorithm.
In this study, we used the SVM with RBF kernel as the classifier. Since building a prediction model requires good generalization towards making predictions for previously unseen test samples, tuning the parameters is an important issue, which requires optimization of the regularization parameter as well as the kernel parameter of SVM. This was done by searching a two-dimensional grid of different values for both parameters. Moreover, the small sample size characterizing microarray data restricts the choice of an estimator for the generalization performance. To solve these problems, the optimization criterion also used the LOOCV performance described above. The value of the regularization parameter corresponding to the largest LOOCV performance was then selected as the optimal value.
To obtain reliable experimental results showing comparability and repeatability for different numerical experiments, we not only used the original division of each dataset in training and test set, but also reshuffled all datasets randomly. In other words, all numerical experiments were performed with 20 random splits of the three original datasets. In addition, they are also stratified, which means that each randomized training and test set contains the same amount of samples of each class compared with the original training and test set.
We used our proposed method (ICA+SFFS+SVM) to analyze the three gene expression datasets. For comparison, we also used SVM, PCA+SVM, PCA+SFFS+SVM, and ICA+SVM methods respectively to do the same tumor classification experiment. The classification results for tumor and normal tissues using these methods are displayed in Table 2. For each classification problem, the experimental results gave the statistical means and standard deviations of accuracy on the original dataset and 20 randomizations as described above. Since the random splits for training and test set are disjoint, the results given in Table 2 are unbiased and can in general also be too optimistic.
Table 2.
Comparison of the classification performances of five methods on three datasets
| No. | Method | Colon cancer dataset |
||
| LOOCV performance (%) | Accuracy on training set (%) | Accuracy on test set (%) | ||
| 1 | SVM | 88.25±3.74 | 95.00±2.35 | 88.18±3.83 |
| 2 | PCA+SVM | 87.25±2.99 | 93.25±2.05 | 89.54±3.74 |
| 3 | PCA+SFFS+SVM | 89.25±3.13 | 91.00±4.28 | 89.54±3.74 |
| 4 | ICA+SVM | 83.50±4.44 | 96.00±3.37 | 89.09±4.39 |
| 5 | ICA+SFFS±SVM | 91.25±2.12 | 93.75±2.42 | 89.54±3.74 |
| No. | Method | Acute leukemia dataset |
||
| LOOCV performance (%) | Accuracy on training set (%) | Accuracy on test set (%) | ||
| 1 | SVM | 93.69±2.21 | 100±0.00 | 95.30±3.45 |
| 2 | PCA+SVM | 91.59±2.99 | 100±0.00 | 93.24±5.01 |
| 3 | PCA+SFFS+SVM | 96.58±2.78 | 97.90±2.78 | 93.53±4.55 |
| 4 | ICA+SVM | 90.82±3.79 | 100±0.00 | 95.30±3.45 |
| 5 | ICA+SFFS+SVM | 97.90±2.07 | 99.21±1.27 | 96.77±2.57 |
| No. | Method | High-grade glioma dataset |
||
| LOOCV performance (%) | Accuracy on training set (%) | Accuracy on test set (%) | ||
| 1 | SVM | 80.00±6.67 | 99.52±1.51 | 66.55±4.00 |
| 2 | PCA+SVM | 78.09±7.17 | 94.76±3.51 | 67.93±3.65 |
| 3 | PCA+SFFS+SVM | 88.10±5.61 | 97.62±3.37 | 67.24±6.35 |
| 4 | ICA+SVM | 77.64±6.51 | 99.52±1.51 | 71.38±4.31 |
| 5 | ICA+SFFS+SVM | 88.54±4.00 | 97.62±3.37 | 71.73±3.91 |
From Table 2, we can see that for the colon data, the LOOCV performance of every method is different, where the performance of our proposed method (Method 5) is the highest. Yet, for the accuracy on test set, the performances are very similar.
For the leukemia data, Method 5 performs better than other four methods on the LOOCV performance and the accuracy on test set. The performances of Methods 1 and 4 for the test set are similar, while those of Methods 2 and 3 are relatively low. For the glioma data, Method 5 is better than other methods on the LOOCV performance and the accuracy on test set. The accuracy of Method 4 on test set is high, yet its LOOCV performance is the lowest one. From the analysis above, we can see that our method is indeed efficient and feasible. Of course, this only comes from the three datasets used in this paper. In fact, there is no method whose classification effect is always the best for all the datasets.
In addition, for all the three datasets, the standard deviations of accuracy on test set obtained by Method 5 are relatively small, which means that Method 5 is more stable than other methods. Another thing we can see from Table 2 is that, for a given dataset, when the LOOCV performance is high, the accuracy on test set is not definitely high. This is embodied especially in the colon dataset.
Conclusion
In this paper, we presented ICA methods for the classification of tumors based on microarray gene expression data. The methodologies involve dimension reduction of high-dimensional gene expression data using ICA, followed by feature selection using SFFS and classification applying SVM. We compared the experimental results of our method with those of other four methods on three datasets, and the results show that our method is effective and efficient in predicting normal and tumor samples from three human tissues. Furthermore, these results hold under rerandomization of the samples.
Since currently we have no suitable gene expression data of multiclass at hand, we only studied binary tumor classification problem in the experiments. In fact, our method can be extended to address the problems with multiclass by using other appropriate classifiers such as neural networks. In future works, we will continue to study the ICA model of gene expression data, try to apply this method to solving multiclass problems of tumor classification, and make full use of the information contained in the gene data to restrict ICA models so that more exact prediction of tumor class can be achieved. In particular, we will also study the application of other ICA models (such as nonlinear ICA models) in the tumor classification, and investigate how to use the method proposed in this paper on the application of other gene datasets.
Authors’ contributions
CHZ conducted data analyses and prepared the manuscript. DSH and XZK conceived the idea of using this approach and assisted with manuscript preparation. XMZ collected the datasets. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 30700161), the National High-Tech Research and Development Program (863 Program) of China (No. 2007AA01Z167 and 2006AA02Z309), China Postdoctoral Science Foundation (No. 20070410223), and Doctor Scientific Research Startup Foundation of Qufu Normal University (No. Bsqd2007036).
References
- 1.Alizadeh A.A. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Golub T.R. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 3.Alon U. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bittner M. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 5.Furey T.S. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee A. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selaru F.M. Artificial neural networks distinguish among subtypes of neoplastic colorectal lesions. Gastroenterology. 2002;122:606–613. doi: 10.1053/gast.2002.31904. [DOI] [PubMed] [Google Scholar]
- 8.van’t Veer L.J. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.West M. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shipp M.A. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2000;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen D.V., Rocke D.M. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002;18:39–50. doi: 10.1093/bioinformatics/18.1.39. [DOI] [PubMed] [Google Scholar]
- 12.Pochet N. Systematic benchmarking of microarray data classification: assessing the role of non-linearity and dimensionality reduction. Bioinformatics. 2004;20:3185–3195. doi: 10.1093/bioinformatics/bth383. [DOI] [PubMed] [Google Scholar]
- 13.Dudoit S. Comparison of discrimination methods for the classification of tumor using gene expression data. J. Am. Stat. Assoc. 2002;97:77–87. [Google Scholar]
- 14.Alter O. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comon P. Independent component analysis—a new concept? Signal Process. 1994;36:287–314. [Google Scholar]
- 16.Hyvärinen A. Wiley; New York, USA: 2001. Independent Component Analysis. [Google Scholar]
- 17.Hyväarinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 18.Frank I.E., Friedman J.H. A statistical view of some chemometric regression tools. Technometrics. 1993;35:109–148. [Google Scholar]
- 19.Ekenel H.K., Sankur B. Feature selection in the independent component subspace for face recognition. Pattern Recognit. Lett. 2004;25:1377–1388. [Google Scholar]
- 20.Nutt C.L. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 21.Zheng C.H. MISEP method for postnonlinear blind source separation. Neural Comput. 2007;19:2557–2578. doi: 10.1162/neco.2007.19.9.2557. [DOI] [PubMed] [Google Scholar]
- 22.Zheng C.H. Nonnegative independent component analysis based on minimizing mutual information technique. Neurocomputing. 2006;69:878–883. [Google Scholar]
- 23.Bartlett M.S. Face recognition by independent component analysis. IEEE Trans. Neural Netw. 2002;13:1450–1464. doi: 10.1109/TNN.2002.804287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T.W. Independent component analysis using an extended infomax algorithm for mixed sub-Gaussian and super-Gaussian sources. Neural Comput. 1999;11:417–441. doi: 10.1162/089976699300016719. [DOI] [PubMed] [Google Scholar]
- 25.Hori G. Proceedings of the Third International Conference on Independent Component Analysis and Blind Signal Separation. San Diego; USA: 2001. Blind gene classification based on ICA of microarray data; pp. 332–336. [Google Scholar]
- 26.Liebermeister W. Linear modes of gene expression determined by independent component analysis. Bioinformatics. 2002;18:51–60. doi: 10.1093/bioinformatics/18.1.51. [DOI] [PubMed] [Google Scholar]
- 27.Chiappetta P. Blind source separation and the analysis of microarray data. J. Comput. Biol. 2004;11:1090–1109. doi: 10.1089/cmb.2004.11.1090. [DOI] [PubMed] [Google Scholar]
- 28.Martoglio A.M. A decomposition model to track gene expression signatures: preview on observer-independent classification of ovarian cancer. Bioinformatics. 2002;18:1617–1624. doi: 10.1093/bioinformatics/18.12.1617. [DOI] [PubMed] [Google Scholar]
- 29.Ferri F.J. Pattern Recognition in Practice IV. Elsevier Science Inc.; New York, USA: 1994. Comparative study of techniques for large-scale feature selection; pp. 403–413. [Google Scholar]
- 30.Haykin S. second edition. Prentice Hall; Englewood Cliffs, USA: 1998. Neural Networks: A Comprehensive Foundation. [Google Scholar]
- 31.Cristianini N., Shawe-Taylor J. Cambridge University Press; Cambridge, UK: 2000. An Introduction to Support Vector Machines and Other Kernelbased Learning Methods. [Google Scholar]
- 32.Boser B.E. Proceedings of the Fifth Annual Workshop on Computational Learning Theory. ACM Press; Pittsburgh, USA: 1992. A training algorithm for optimal margin classifiers; pp. 144–152. [Google Scholar]
- 33.Vapnik V. Wiley; New York, USA: 1998. Statistics Learning Theory. [Google Scholar]
- 34.Brown M. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanevski M. Advanced spatial data analysis and modelling with support vector machines. Int. J. Fuzzy Syst. 2002;4:606–615. [Google Scholar]
- 36.Drucker H. Support vector machines for spam categorization. IEEE Trans. Neural Netw. 1999;10:1048–1054. doi: 10.1109/72.788645. [DOI] [PubMed] [Google Scholar]