Abstract
We have recently cloned a pathogen inducible blast resistance gene Pi-kh from the indica rice line Tetep using a positional cloning approach. In this study, we carried out structural organization analysis of the Pi-kh locus in both indica and japonica rice lines. A 100 kb region containing 50 kb upstream and 50 kb downstream sequences flanking to the Pi-kh locus was selected for the investigation. A total of 16 genes in indica and 15 genes in japonica were predicted and annotated in this region. The average GC content of indica and japonica genes in this region was 53.15% and 49.3%, respectively. Both indica and japonica sequences were polymorphic for simple sequence repeats having mono-, di-, tri-, tetra-, and pentanucleotides. Sequence analysis of the specific blast resistant Pi-kh allele of Tetep and the susceptible Pi-kh allele of the japonica rice line Nipponbare showed differences in the number and distribution of motifs involved in phosphorylation, resulting in the resistance phenotype in Tetep.
Key words: comparative genomics, blast resistance gene, genome analysis, microcolinearity
Introduction
The genetic make-up and genome organization of related species is often sufficiently conserved, allowing alignments of the genomes. Genome alignment enables research communities to predict the presence of genes, build physical maps, and conduct comparative genome analysis among and between species. The recent genome sequencing of various organisms has enhanced the rate of new gene identification, annotation, and functional validation. Genome information available in the public domain has been used extensively in comparative genome studies with the help of bioinformatics tools.
Rice is considered as a model crop for genetic and molecular biology studies largely because of its small genome size (389 Mb) in cereals (1). The rice genome has been sequenced from two subspecies, indica cultivar 93-11 2., 3. and japonica cultivar Nipponbare 1., 4.. These two rice subspecies are thought to have diverged more than one million years ago (5). Sequence availability for each of the two rice subspecies has made comparative genomics an easy task. Genome alignment helps in carrying out comparative genome analysis, leading to the study of similarity and variation between two genomes or gene sequences, which is useful for functional studies, genetics, and crop breeding.
Among various biotic stresses like bacterial leaf blight, sheath blight, and stem borer that limit rice productivity, the blast disease caused by Magnaporthe grisea (hebert) Barr is a serious constraint in rice production at the global level. In our previous study, we have tagged a durable blast resistance gene Pi-kh from the indica rice line Tetep by using cleaved amplified polymorphic sequence (CAPS) and sequence tagged microsatellite site (STMS) markers at 1.6 cM and 1.1 cM distance, respectively (6). This gene was further fine mapped by using simple sequence repeat (SSR) markers at 0.7 cM and 0.5 cM distance, respectively, and its physical location was determined on the long arm of rice chromosome 11 (7). SSR markers identified in Tetep were used to locate the homologous region in the genomic sequence of the japonica rice line Nipponbare. Bioinformatics tools were used to identify candidate blast resistance genes from a physical map consisting of two overlapping chromosomes, namely bacterial artificial chromosome and P1 artificial chromosome, spanning a region of 143,537 bp on the long arm of rice chromosome 11. Consequently, a homologous sequence of 1.5 kb was cloned from Tetep. The cloned Pi-kh gene has a single open reading frame (ORF) and belongs to the nucleotide binding site (NBS)–leucine-rich repeat (LRR) class of disease resistance genes (7). However, further molecular analysis of the Pi-kh gene and its functional complementation has yet to be deduced.
Keeping in view the socio-economic importance of the blast resistance gene Pi-kh in sustainable management of rice blast in the northwestern Himalayan region of India, we carried out the present investigation to analyze the structural organization of candidate blast resistance Pi-kh locus and to perform microsynteny analysis of indica-japonica sequences on this specific locus.
Results and Discussion
Gene prediction and annotation
Sequence analysis was carried out to find the number and order of genes in a 100 kb region containing 50 kb upstream and 50 kb downstream sequences flanking to the Pi-kh locus in both indica and japonica rice lines. A total of 16 genes in indica and 15 genes in japonica were predicted in this region (Table 1, Table 2). In the indica variety, 8 genes are present in the plus strand and another 8 are in the minus strand. The longest ORF is 4,722 bp in the minus strand, followed by ORFs of 2,820, 2,178, and 1,020 bp. The other 12 ORFs are less than 1,000 bp (Table 1). In the case of japonica, 6 genes are present in the plus strand and 9 are in the minus strand. Out of the 15 ORFs, the sizes of 4 ORFs are longer than 2,000 bp, in which the longest ORF is 2,808 bp in the minus strand; 3 ORFs are in the range of 1,000 to 2,000 bp; the remaining 8 ORFs are less than 1,000 bp (Table 2).
Table 1.
Predicted genes and their annotations in the 100 kb region of the indica rice line
| Gene ID | Start (bp) | End (bp) | cDNA size (bp) | DNA strand | BLAST hit | Gene function |
|---|---|---|---|---|---|---|
| osi01 | 548 | 2,251 | 762 | + | ABA94924.1 | expressed protein |
| osi02 | 3,637 | 3,897 | 261 | + | ABA94925 | hypothetical protein LOC_Os11g41930 |
| osi03 | 6,691 | 7,218 | 528 | + | ABA94926.1 | hypothetical protein LOC_Os11g41940 |
| osi04 | 8,799 | 9,110 | 312 | + | OSJNBa0041A02.28 | hypothetical protein |
| osi05 | 10,079 | 10,666 | 588 | – | ABA94927.1 | hypothetical protein LOC_Os11g41950 |
| osi06 | 12,127 | 12,441 | 315 | + | ABA94928.1 | hypothetical protein LOC_Os11g41960 |
| osi07 | 13,653 | 15,123 | 525 | – | ABA94924.1 | hypothetical protein |
| osi08 | 15,991 | 30,983 | 4,722 | – | ABA94930.1 | hypothetical protein LOC_Os11g41980 |
| osi09 | 36,253 | 38,219 | 324 | + | ABA94971.1 | Phyb1, putative |
| osi10 | 39,515 | 40,644 | 1,020 | – | BAA75541.1 | L-zip+NBS+LRR |
| osi11 | 45,161 | 49,476 | 699 | + | ABA94971.1 | Phyb1, putative |
| osi12 | 50,252 | 51,649 | 630 | – | ABA94972.1 | L-zip+NBS+LRR, putative |
| osi13 | 54,698 | 55,033 | 336 | – | ABA94973.1 | hypothetical protein LOC_Os11g42020 |
| osi14 | 58,932 | 61,109 | 2,178 | + | ABA94974.1 | expressed protein |
| osi15 | 75,984 | 78,803 | 2,820 | – | ABA94975.1 | NB-ARC domain, putative |
| osi16 | 82,010 | 82,495 | 486 | – | NP_910833.1 | hypothetical protein |
Table 2.
Predicted genes and their annotations in the 100 kb region of the japonica rice line
| Gene ID | Start (bp) | End (bp) | cDNA size (bp) | DNA strand | BLAST hit | Gene function |
|---|---|---|---|---|---|---|
| osj01 | 33 | 3,693 | 1,257 | + | ABA94923.1 | GTP-PI-KH-binding protein, putative |
| osj02 | 5,596 | 9,614 | 1,623 | + | ABA94924.1 | expressed protein |
| osj03 | 12,488 | 14,583 | 588 | + | ABA94926.1 | hypothetical protein LOC_Os11g41940 |
| osj04 | 17,626 | 18,180 | 555 | – | ABA94927.1 | hypothetical protein LOC_Os11g41950 |
| osj05 | 19,549 | 19,986 | 438 | + | ABA94928.1 | hypothetical protein LOC_Os11g41960 |
| osj06 | 21,998 | 22,448 | 204 | – | ABA94929.1 | expressed protein |
| osj07 | 23,438 | 26,842 | 2,301 | – | ABA94930.1 | hypothetical protein LOC_Os11g41980 |
| osj08 | 31,077 | 39,956 | 2,415 | – | ABA94931.1 | hypothetical protein LOC_Os11g41990 |
| osj09 | 45,208 | 49,523 | 699 | + | ABA94971.1 | Phyb1, putative |
| osj10 | 50,513 | 51,533 | 810 | – | ABA94972.1 | L-zip+NBS+LRR, putative |
| osj11 | 54,624 | 54,959 | 336 | – | ABA94973.1 | hypothetical protein LOC_Os11g42020 |
| osj12 | 63,371 | 65,551 | 2,181 | + | ABA94974.1 | expressed protein |
| osj13 | 76,976 | 79,783 | 2,808 | – | ABA94975.1 | NB-ARC domain, putative |
| osj14 | 83,579 | 84,077 | 339 | – | ABA94976.1 | hypothetical protein LOC_Os11g42050 |
| osj15 | 103,360 | 105,306 | 1,947 | – | ABA94977.1 | Leucine-rich repeat, putative |
To know the functions of these predicted genes, BLASTX was performed with the NCBI nr protein database (Table 1, Table 2). Nine genes in indica and seven in japonica were predicted as hypothetical proteins, while two genes in indica and three in japonica were annotated as expressed proteins. Two genes (osi10 and osi12) in indica were annotated as encoding the putative L-zip+NBS+LRR domain, compared with one (osj10) in japonica. Similarly, two genes (osi09 and osi11) in indica were annotated as encoding the putative Phyb1 protein, compared with one (osj09) in japonica. The gene osi15 in indica was found similar to osj13 in japonica for encoding the putative NB-ARC domain [nucleotide-binding adaptor shared by APAF-1 (apoptotic peptidase activating factor), resistance (R) proteins, and CED-4]. The genes osj01 and osj15 in japonica were annotated as the putative GTP-binding protein and the putative LRR domain, respectively. However, their counterparts were not found in indica.
Our analysis supports the evidence that there exists conservation of gene order across regions spanning many megabases (macrocolinearity) (8), but the colinearity of gene order and content at the level of local genome structure (microcolinearity) has also been observed (9). The LRR domain is implicated in interactions between proteins, ligands, and carbohydrates 10., 11.. Its role as a major determinant of recognition specificity is supported by studies on domain swaps among alleles of L and P genes in flax 12., 13.. In addition to recognition, LRR has the tendency to form horse-shoe-shaped molecules with β-sheet on the concave side. A central “xxLxLxx” motif, where “x” represents any amino acid, forms the β-sheet with leucine burried in the center of the protein and the adjacent residue, which is a hypervariable region exposed to solvents (10). Each R protein contains a conserved NBS that probably binds ATP or dATP (14). The NBS region, which is around 320 amino acids long, has several R proteins, including kinase 1a, 2, and 3a domains, as well as short motifs known as NB-ARC domains. The NB-ARC domain, which generally controls cell death, might be involved in ATP-dependent oligomerization or histidine aspartic acid phosphotransfer without nucleotide bindings.
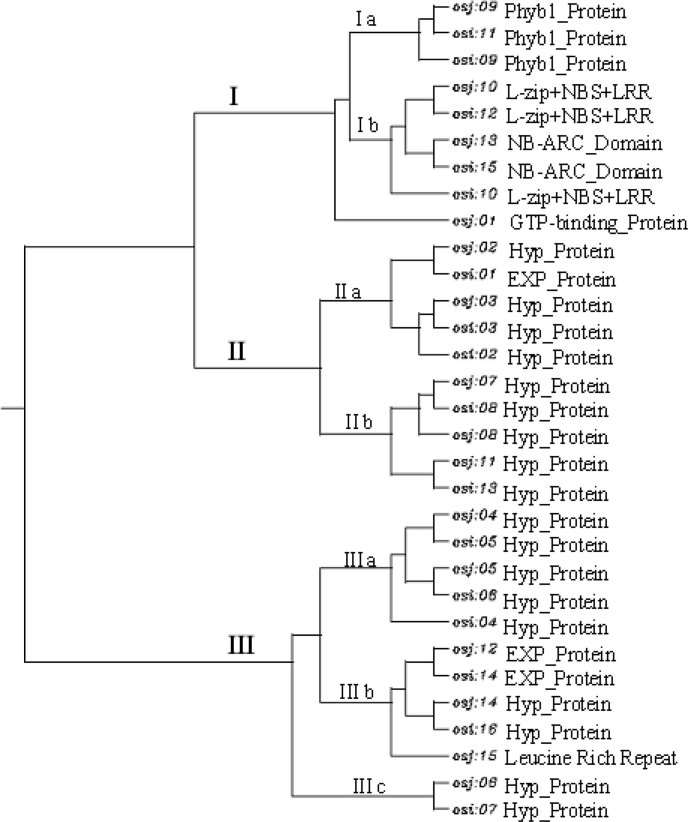
Analysis of the 100 kb target region revealed that the gene density in this region is one gene per 6.25 kb in indica and one gene per 6.67 kb in japonica, while the overall gene density of rice is one gene per 5.7 kb (15). The International Rice Genome Sequencing Project (IRGSP) detected a total of 37,544 non-transposable-element-related protein-coding sequences in rice, with a lower gene density of one gene per 9.9 kb (1). To further investigate the relationship among similar predicted gene functions of indica and japonica, multiple sequence alignments were performed using the ClustalW program (www.align.genome.jp) and a phylogenetic dendrogram was constructed. All the 16 predicted genes of indica and 15 genes of japonica were classified into three large groups (Figure 1). Groups I and II have two subgroups for each, while Group III has three subgroups, which is the largest group containing 51.6% of the predicted genes. The dendrogram shows that the predicted genes with similar functions between both cultivars are grouped together. This was expected since indica and japonica subspecies have a common ancestor and there is a high degree of gene conservation between both subspecies (16). In Group I, the genes encoding the putative Phyb1 protein, the L-zip+NBS+LRR domain, and the NB-ARC domain are clustered together, respectively. However, the gene osi10 in indica encoding for the L-zip+NBS+LRR domain did not cluster with the functionally similar predicted genes osi12 and osj10. This might be due to the long cDNA sequence (1,020 bp) of gene osi10 compared with osi12 (630 bp) and osj10 (816 bp). As a result, the alignment score between osi10 and osi12 as well as that between osi10 and osj10 was 8.64 and 11.11, respectively, whereas the alignment score between osi12 and osj10 was 32.85, indicating a better alignment. Groups II and III contain clusters of genes similar to the hypothetical proteins and expressed proteins from both indica and japonica rice lines.
Fig. 1.
Phylogenetic analysis of the predicted genes of indica and japonica rice lines.
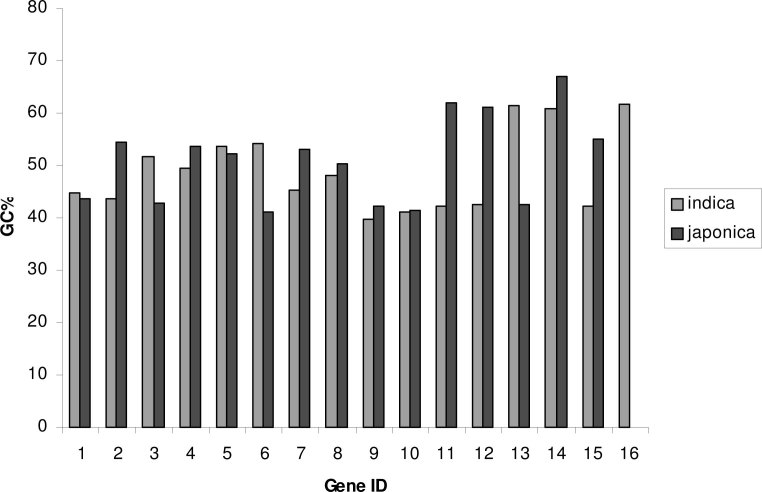
The GC content of the 16 indica genes varies from 41.1% to 61.7%, while it is 41.2% to 67.0% for the 15 japonica genes (Figure 2). When performing gene to gene comparison, 10 genes predicted in the japonica rice line (excluding osj01, osj03, osj05, osj06, and osj13) have slightly higher GC content than those of indica. The average GC content of indica and japonica genes in the 100 kb region is 53.15% and 49.3%, respectively. The average GC percentage is higher in indica due to the presence of one extra gene (osi16) as compared with the 15 genes predicted in this region of japonica. The average GC content of the indica genes excluding osi16 is 43.4%, compared with 49.3% of the japonica genes. The higher GC content in monocot genes than in eudicot genes has been reported before (17). In Gramineae genes, the gradients in GC content, codon usage, and amino acid usage have been reported along the direction of transcription (18). Our analysis showed that variations in GC content do exist between the genes of two subspecies of the genus Oryza.
Fig. 2.
Gene-wise distribution of GC content in indica and japonica rice lines.
Physical mapping of the predicted genes
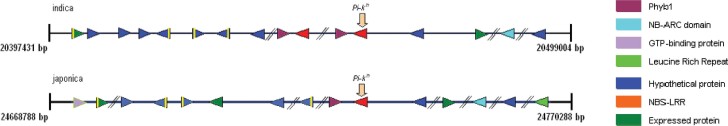
Microsynteny analysis was performed on the Pi-kh locus in both indica and japonica sequences (Table 3, Table 4). All the predicted genes from both subspecies showed 100% sequence similarity (except gene osj01 in japonica that showed 99% homology) with respective chromosome database. Based on the above information, all the genes were classified and plotted with respect to their exact physical positions and directions on chromosome 11 of both indica and japonica rice lines (Figure 3). In both rice lines, the predicted Pi-kh gene was flanked by the Phyb1 gene at the left side in reverse orientation. This analysis further showed macrocolinearity as well as rearrangement and duplication since there are two genes encoding the putative Phyb1 protein and two genes encoding the L-zip+NBS+LRR domain in indica. Therefore, on careful observation of the genome sequences, a narrower region of divergence can be found (19). This region relates to the area of divergence between two rice subspecies, and the alignment of the two rice subspecies may help in identifying regions of cereal genomes that are prone to rapid evolution. Similar results were obtained by Han and Xue (20), where they showed extensive conservation of microcolinearity in the gene order and gene content between indica and japonica, but they also discovered significant number of rearrangements and polymorphisms when comparing the two genomes. Whole genome analysis of indica and japonica rice revealed 18 distinct pairs of duplicated segments that cover 65.7% of the genomes (5). It was concluded that ongoing individual gene duplications provide a continuous source of new material for the genesis of genes in rice (5). Song et al. (19) identified orthologous regions from maize, sorghum, and the two rice subspecies. They found that gross macrocolinearity is maintained but microcolinearity is incomplete among these cereals. Deviation from gene colinearity is attributed to the changes such as gene insertion, deletion, duplication, or inversion.
Table 3.
Microsynteny analysis of the predicted genes in the 100 kb region of the indica rice line
| Gene ID | BLAST hit | Bit score | E-value | Homology | Start (bp) | End (bp) |
|---|---|---|---|---|---|---|
| osi01 | Chr11 2003-08-01 BGI | 808 | 0.0 | 420/420 (100%) | 20,399,132 | 20,399,551 |
| osi02 | Chr11 2003-08-01 BGI | 502 | E-142 | 261/261 (100%) | 20,400,937 | 20,401,197 |
| osi03 | Chr11 2003-08-01 BGI | 1,015 | 0.0 | 528/528 (100%) | 20,403,991 | 20,404,518 |
| osi04 | Chr11 2003-08-01 BGI | 600 | E-171 | 312/312 (100%) | 20,406,099 | 20,406,410 |
| osi05 | Chr11 2003-08-01 BGI | 1,131 | 0.0 | 588/588 (100%) | 20,407,966 | 20,407,379 |
| osi06 | Chr11 2003-08-01 BGI | 606 | E-173 | 315/315 (100%) | 20,409,427 | 20,409,741 |
| osi07 | Chr11 2003-08-01 BGI | 604 | E-172 | 314/314 (100%) | 20,412,423 | 20,412,110 |
| osi08 | Chr11 2003-08-01 BGI | 4,224 | 0.0 | 2,197/2,197 (100%) | 20,416,754 | 20,414,558 |
| osi09 | Chr11 2003-08-01 BGI | 202 | 7E-52 | 105/105 (100%) | 20,434,981 | 20,435,085 |
| osi10 | Chr11 2003-08-01 BGI | 1,017 | 0.0 | 529/529 (100%) | 20,437,343 | 20,436,815 |
| osi11 | Chr11 2003-08-01 BGI | 217 | 4E-56 | 113/113 (100%) | 20,446,229 | 20,446,341 |
| osi12 | Chr11 2003-08-01 BGI | 706 | 0.0 | 367/367 (100%) | 20,448,209 | 20,447,843 |
| osi13 | Chr11 2003-08-01 BGI | 646 | 0.0 | 336/336 (100%) | 20,452,333 | 20,451,998 |
| osi14 | Chr11 2003-08-01 BGI | 4,188 | 0.0 | 2,178/2,178 (100%) | 20,456,232 | 20,458,409 |
| osi15 | Chr11 2003-08-01 BGI | 5,422 | 0.0 | 2,820/2,820 (100%) | 20,476,103 | 20,473,284 |
| osi16 | Chr11 2003-08-01 BGI | 935 | 0.0 | 486/486 (100%) | 20,479,795 | 20,479,310 |
Table 4.
Microsynteny analysis of the predicted genes in the 100 kb region of the japonica rice line
| Gene ID | BLAST hit | Bit score | E-value | Homology | Start (bp) | End (bp) |
|---|---|---|---|---|---|---|
| osj01 | Chr11_pmol osa1 | 531 | E-150 | 278/279 (99%) | 24,671,510 | 24,671,788 |
| osj02 | Chr11_pmol osa1 | 1,269 | 0.0 | 660/660 (100%) | 24,674,714 | 24,675,373 |
| osj03 | Chr11_pmol osa1 | 608 | E-173 | 316/316 (100%) | 24,682,968 | 24,683,283 |
| osj04 | Chr11_pmol osa1 | 1,067 | 0.0 | 555/555 (100%) | 24,686,880 | 24,686,326 |
| osj05 | Chr11_pmol osa1 | 842 | 0.0 | 438/438 (100%) | 24,688,249 | 24,688,686 |
| osj06 | Chr11_pmol osa1 | 367 | E-102 | 191/191 (100%) | 24,691,148 | 24,690,958 |
| osj07 | Chr11_pmol osa1 | 4,109 | 0.0 | 2,137/2,137 (100%) | 24,695,542 | 24,693,406 |
| osj08 | Chr11_pmol osa1 | 1,396 | 0.0 | 726/726 (100%) | 24,705,726 | 24,705,001 |
| osj09 | Chr11_pmol osa1 | 217 | 5E-56 | 113/113 (100%) | 24,717,676 | 24,717,788 |
| osj10 | Chr11_pmol osa1 | 1,148 | 0.0 | 597/597 (100%) | 24,719,809 | 24,719,213 |
| osj11 | Chr11_pmol osa1 | 646 | 0.0 | 336/336 (100%) | 24,723,659 | 24,723,324 |
| osj12 | Chr11_pmol osa1 | 4,194 | 0.0 | 2,181/2,181 (100%) | 24,732,071 | 24,734,251 |
| osj13 | Chr11_pmol osa1 | 5,399 | 0.0 | 2,808/2,808 (100%) | 24,748,483 | 24,745,676 |
| osj14 | Chr11_pmol osa1 | 381 | E-105 | 198/198 (100%) | 24,752,777 | 24,752,580 |
| osj15 | Chr11_pmol osa1 | 3,744 | 0.0 | 1,947/1,947 (100%) | 24,774,006 | 24,772,060 |
Fig. 3.
Physical map of the genes predicted in the 100 kb region of chromosome 11 of both indica and japonica rice lines. The position of arrow head indicates the direction of the gene. Vertical yellow lines in the gene represent repeat elements present within the gene. The position of the Pi-kh gene is shown with the vertical arrow.
Identification of SSRs in the Pi-kh locus
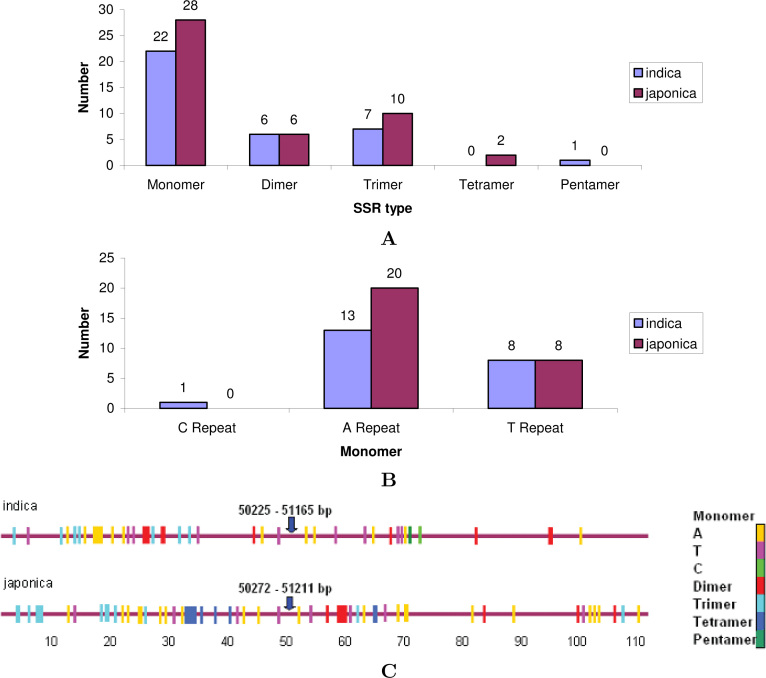
The frequency of SSRs in the 100 kb region was calculated on both japonica and indica rice lines (Figure 4). In this region, there are more monomers (A, C, and T repeats) in japonica than in indica, whereas dimers are equal in both sequences (Figure 4A). The number of trimers in japonica and indica sequences is 10 and 7, respectively. C repeat and pentamer are absent in japonica, whereas tetramer is absent in indica (Figure 4A and B). All together japonica and indica has 46 and 36 SSRs in this region, respectively. In the 100 kb region of both rice lines, the first 70 kb region is rich in SSRs compared with the rest of the region (Figure 4C). The Pi-kh locus is flanked by monomer T and A repeats in both rice lines at the left and the right side, respectively. We found that 76.0% and 91.0% of the repeats were present in the intergenic region of japonica and indica, respectively, whereas 23.9% and 8.6% repeats were also detected within the genes encoding for hypothetical and expressed proteins.
Fig. 4.
Distribution of SSRs in the 100 kb region of chromosome 11 of indica and japonica rice lines. A. The number of SSR types present in both sequences. B. The number of monomer repeats present in this region. C. Physical mapping of SSRs in the 100 kb region of indica and japonica rice lines. The position of the Pi-kh gene is indicated with the blue arrow. The types of repeats are shown in different colors.
Repeat elements play a major role in gene duplication and amplification for generating new alleles in the population. IRGSP has identified and annotated a total of 18,828 Class I di-, tri-, and tetranucleotide SSRs, representing 47 distinctive motif families (1). They reported an average of 51 hypervariable SSRs per Mb, with the highest density occurring on chromosome 3 (55.8 SSR/Mb) and the lowest occurring on chromosome 4 (41.0 SSR/Mb). These repeat elements also act as SSR markers for specific regions of the genome. Thousands of such SSRs have already been shown to amplify well and are polymorphic in a panel of diverse cultivars, and thus are of immediate use for genetic analysis (1). Both of the sequences from japonica and indica are polymorphic for SSRs. The results on the SSR distribution in the 100 kb region showed that these SSRs are mono-, di-, tri-, tetra-, and pentanucleotides. Similar results were obtained on the SSR distribution in rice and Arabidopsis genomes, which also reported that the majority of the SSRs were mono-, di-, tri-, tetra-, and pentanucleotides, accounting for up to approximately 80% of all the SSRs found in various regions of the genomes (21). As described above, there are more SSRs in intergenic regions than in intragenic regions. This might be the reason that the sequences of rice chromosomes 11 and 12 are rich in disease resistance genes and recent gene duplications (22). Therefore, the resistance and defense response genes, enriched on these chromosomes relative to the whole genome, have evolved due to duplication, amplification, and reduplication. SSRs play a major role in this process of evolution. Within the gene, only trimer repeats were found to be present in both japonica and indica sequences. This is consistent with the study of Zhang et al. (23) in which a more comprehensive survey of SSRs was performed in Arabidopsis and showed that SSRs in general are more favored in upstream regions of the genes and trinucleotide repeats are the most common repeats found in the coding regions.
Sequence analysis of Pi-kh alleles isolated from Tetep and Nipponbare
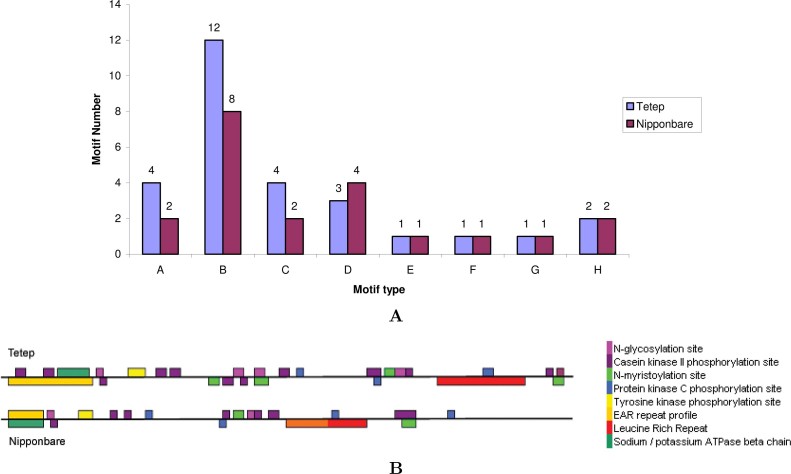
For the analysis and characterization of Pi-kh alleles in both indica (Tetep) and japonica (Nipponbare) rice lines, motif identification was performed using the motif search tool in EXPASY software (www.expasy.org). Eight types of common motifs were found in the Pi-kh locus of both rice lines with different number and spatial distribution (Figure 5). Four types of motifs, including tyrosine kinase phosphprylation site, EAR repeat profile, Na/K-ATPase β-chain, and LRR, are in the same number for each in Tetep and Nipponbare alleles, while others are variable in number (Figure 5A). There are 4 N-glucosylation sites and 4 N-myristorylation sites in Tetep allele compared with only 2 for each in Nipponbare allele. Similarly, Tetep allele has 12 casein kinase II phosphorylation sites while Nipponbare allele has only 8. The position of each motif is also different in both alleles as shown in Figure 5B.
Fig. 5.
Analysis of motifs and their physical positions in the Pi-kh gene isolated from indica (Tetep) and japonica (Nipponbare) sequences. A. The number of motifs predicted in Pi-kh alleles of indica and japonica. A: N-glucosylation site; B: casein kinase II phosphorylation site; C: N-myristorylation site; D: protein kinase C phosphorylation site; E:tyrosine kinase phosphorylation site; F: EAR repeat profile; G: Na/K-ATPase β-chain; H: leuecine-rich repeat. B. Physical positions of motifs distributed in Pi-kh alleles of indica and japonica sequences. The types of motifs are shown in different colors.
Protein kinases and phosphatases are crucial for the activation of early defense responses in plants. As reported by de Vries et al. (24), the tomato Pto gene encodes a functional N-myristoylation motif that is required for signal transduction in Nicotiana benthamiana. Similarly, the Pto, Xa21, and Rpg1 R genes and several R-mediated signalling components encode kinases, suggesting a major role for phosphorylation in R-specified signalling (25). Phosphorylation-related events and protein kinases participate in the R-gene-mediated pathogen recognition and downstream signalling as established for Arabidopsis PBS1 and RIN4 proteins 26., 27.. Thus, the difference in blast resistance and susceptibility of the two rice subspecies may be attributed to the different number of motifs and their spatial distribution.
From the present investigation, it can be concluded that in the comparison of structural organization of the Pi-kh locus in both indica and japonica sequences, macrocolinearity is maintained but microcolinearity is incomplete. Both sequences from indica and japonica are polymorphic for SSRs. Sequence analysis of the specific blast resistant Pi-kh allele of Tetep and the susceptible Pi-kh allele of Nipponbare revealed the differences in the number and distribution of phosphorylation motifs that participate in the R-gene-mediated pathogen recognition and downstream signalling, thus causing the difference in blast resistance and susceptibility of the two subspecies.
Materials and Methods
The rice genome sequence database (www.ncbi.nlm.nih.gov) served as a basic resource during the present investigation. The 1.5 kb fragments of the Pi-kh gene from Tetep and Nipponbare varieties were aligned using the local BLAST tool with indica and japonica sequences of chromosome 11 on local database (www.nrcpb.org), respectively. The target Pi-kh locus was identified in both cultivars, and 50 kb upstream and 50 kb downstream sequences were extracted along with the desired locus from the sequences of both cultivars. Gene prediction in the 100 kb region of both cultivars was performed using the FGENESH tool (www.softbery.com) trained for monocot species. Then BLASTX with the NCBI nr protein database was performed to know the functions of these predicted genes. Multiple alignments of predicted genes were carried out using the ClustalW program (www.align.genome.jp). Gene-wise GC content was determined using the Accelrys gene software (Accelrys Software Inc., San Diego, USA). For the analysis of small variations at local genome level, we used the MISA tool (http://www.ipk-gatersleben.de/en/) to identify and recognize the distribution pattern of repeat elements in the 100 kb region of both indica and japonica subspecies.
The cDNA sequences were compared with the rice pseudomolecule chromosome 11 database (build 3) and the indica chromosome 11 database using the local mega BLASTN tool in order to know the physical position of predicted genes. Based on the above information, the number of genes was classified and plotted along a line with respect to their physical positions and directions on chromosome 11 of indica and japonica type, respectively.
Authors’ contributions
SPK carried out the sequence analysis, BLAST search, and drafted the manuscript. VD participated in the analysis and figure drawing. NKS participated in the design of the study. TRS conceived the study, participated in its design and coordination, and wrote the final manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank IRGSP and Beijing Institute of Genomics (formerly Beijing Genomics Institute) for making the rice genome sequence data available in the public domain. This work was supported by the Department of Biotechnology and Indian Council of Agricultural Research, Government of India (grants to TRS), as well as the Council of Scientific and Industrial Research, Government of India (Senior Research Fellowship to SPK).
References
- 1.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 2.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 3.Yu J. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goff S.A. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 5.Bennetzen J.L. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma T.R. Molecular mapping of rice blast resistance gene Pi-kh in the rice variety Tetep. J. Plant Biochem. Biotech. 2005;14:127–133. [Google Scholar]
- 7.Sharma T.R. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genomics. 2005;274:569–578. doi: 10.1007/s00438-005-0035-2. [DOI] [PubMed] [Google Scholar]
- 8.Gale M.D., Devos K.M. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennetzen J.L., Ramakrishna W. Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 2002;48:821–827. doi: 10.1023/a:1014841515249. [DOI] [PubMed] [Google Scholar]
- 10.Jones D.A., Jones J.D.G. The role of leucine-rich repeat proteins in plant defences. Adv. Bot. Res. 1997;24:89–167. [Google Scholar]
- 11.Kobe B., Kajava A. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 12.Dodds P.N. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis J.G. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tameling W.I. The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Q. Rice bioinformatics. Analysis of rice sequence data and leveraging the data to other plant species. Plant Physiol. 2001;125:1166–1174. doi: 10.1104/pp.125.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y., Zhang Q. New Directions for A Diverse Planet: Proceedings of the Fourth International Crop Science Congress. 2004. The rice genome: implications for breeding rice and other cereals. Brisbane, Australia. [Google Scholar]
- 17.Carels N., Bernardi G. Two classes of genes in plants. Genetics. 2000;154:1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong G.K. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song R. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 2002;12:1549–1555. doi: 10.1101/gr.268302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B., Xue Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 2003;6:134–138. doi: 10.1016/s1369-5266(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 21.Lawson M.J., Zhang L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol. 2006;7:R14. doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Rice Chromosomes 11 and 12 Sequencing Consortia The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3:20. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L. Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics. 2004;20:1081–1086. doi: 10.1093/bioinformatics/bth043. [DOI] [PubMed] [Google Scholar]
- 24.de Vries J.S. Tomato Pto encodes a functional N-myristoylation motif that is required for signal transduction in Nicotiana benthamiana. Plant J. 2006;45:31–45. doi: 10.1111/j.1365-313X.2005.02590.x. [DOI] [PubMed] [Google Scholar]
- 25.Martin G.B. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 26.Swiderski M.R., Innes R.W. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001;26:101–112. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- 27.Mackey D. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]