Abstract
To further study the characteristics of changes on the molecular level of rice mutants induced in space environment, we analyzed proteins in leaves and seeds of four rice mutants (two high-tillering and two low-tillering) in the 8th and 9th generations after a 15-day spaceflight, and compared with their ground controls by two-dimentional polyacrylamide gel electrophoresis and reverse phase liquid chromatography (RPLC). In addition, the albumin, globulin, prolamine, glutelin, and amylose of the mutant seeds were analyzed by RPLC and ultra-violet spectrometry. The results showed that the low-abundance proteins of leaves in the peak tillering stage are more likely to be induced compared with their corresponding controls. The albumin, globulin, and prolamine of the mutant seeds revealed changes when compared with their controls, and the characteristics of changes in different mutants were stably inherited in the 8th and 9th generations, suggesting that they can be used as biomarkers to identity the mutants induced by spaceflight. Moreover, two proteins (SSP9111 and SSP6302) were found to be expressed with high intensity (two-fold change) in different mutants, which were both correlated with photosystem according to mass spectrometry and database searching.
Key words: space environment, rice, mutant, proteomics
Introduction
Rice is one of the most important cereal crops in the world with a great need to be researched. As a model cereal plant for genetic and molecular studies, the rice genome has been sequenced from two subspecies (Oryza sativa L. ssp. indica and japonica) 1., 2., 3., 4.. Fifteen types of rice tissues and eleven types of rice organelles have been analyzed by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and the information is now available in the Rice Proteome Database (5). To date, rice proteomic studies have focused mainly on the alternation in genome expression induced by environmental factors such as stress conditions. Coldness, ozone, drought, salinity, and pathogen infection are common stress conditions that affect the growth and development of rice. The stress condition of interest in the present study is spaceflight.
In our previous studies, we have found that spaceflight can induce different rice mutants, and demonstrated that changes can be induced on the genome, proteome, and cellular levels 6., 7., 8., 9., 10., 11., 12., 13., 14., 15., 16., 17., 18.. The mutative rice resulted in a formidable challenge to identify the function and regulation of every encoded protein. Proteomics, owing to the latest developments in mass spectrometry (MS), is expected to fulfill this potential (19). However, when performing proteomic studies, we found that low-abundance proteins, including regulatory proteins and rare membrane proteins, were out of the scope of most proteomic techniques unless specific (20). In general, the methods involved for low-abundance proteins are protein profiling by 2D-PAGE and image comparison by specific software (21).
To further study the proteome characteristics and heredity of different mutants induced by spaceflight, four stably inherited rice mutants, including two high-tillering mutants 971-5 and 974-5, and two low-tillering mutants 971-4 and 974-1, were generated by continuous breeding and selection after a 15-day spaceflight on a recoverable satellite in 1996. To understand the proteome changes of stably inherited mutants, we scanned the proteome of leaves in the peak tillering stage of the 8th and 9th mutant generations using 2D-PAGE and reverse phase liquid chromatography (RPLC).
Proteins in seeds are divided into housekeeping proteins and storage proteins; the contents of housekeeping proteins are less in seeds, but they are necessary for seeds’ metabolization (22). Albumin and globulin are mostly active enzymes and are distributed in the aleurone layer of seeds. Prolamine and glutelin in endosperm (23) manage the metabolism, information transfer, and energy transformation of seeds, offering deoxidized nitrogen when seeds germinate (24). Amylose is the main substance in endosperm, reflecting the composition, shape, and quality inheritance of seeds (25). Therefore, in the present study, we analyzed heredity characteristics of albumin, globulin, prolamine, glutelin, and amylose on the molecular level of mutant seeds in the 8th and 9th generations.
So far, fingerprinting has gained more and more attentions and has been internationally accepted as a feasible means for quality control. Correlation coefficient between two fingerprints has been widely used to assess the similarity of fingerprints in chemical and medical studies (26). In this study, we also employed 2D-PAGE and RPLC fingerprinting to analyze the seed proteins and amylose.
Results
Leaf proteome study
Proteins obtained from individually selected leaves were compared with those from 30 randomly selected leaves in 2D-PAGE images. The result showed that there was no significant variation in protein expression between the randomly and individually selected leaves (significance level p > 0.05 based on t-test). Therefore, the following results were based on 30 randomly selected leaves from different mutants and their controls in the 8th and 9th generations.
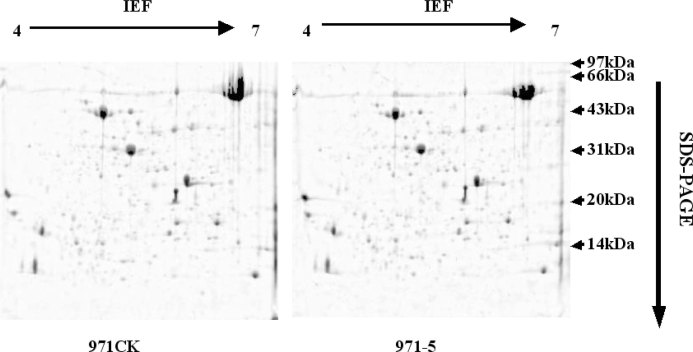
Isolated proteins were separated by 2D-PAGE with isoelectric focusing (IEF) of pH 4–7 and 3–10 (Figure 1). 2D-PAGE was performed three times for samples of each mutant and their ground controls, and the results were similar (p > 0.05). Approximately 300 protein spots were revealed from the gels of mutants and their corresponding controls using Bio-Rad PDQuest software. Expression levels of the same protein in mutants and their controls were compared to obtain a ratio. Only spots with a ratio less than 0.5 or more than 2 were considered as significantly differentially expressed proteins. Since 2D-PAGE is not sensitive enough to detect low-abundance proteins, RPLC was employed to further investigate the expression changes of low-abundance proteins (Figure 2).
Fig. 1.
2D-PAGE images of control 971CK and mutant 971-5.
Fig. 2.
RPLC fingerprints of control 971CK and mutant 971-5.
The matching ratio of proteins in Table 1 exhibits the differences of the leaf proteome between mutants and their controls. The number of significantly down-regulated proteins was more than that of significantly up-regulated proteins in all four different mutants detected by both 2D-PAGE and RPLC.
Table 1.
Comparison of protein expression between mutants and their ground controls
| Group | Match ratio of 2D-PAGE | Similitude degree of RPLC | Reduced over 2 fold |
Increased over 2 fold |
|||
|---|---|---|---|---|---|---|---|
| 2D-PAGE | RPLC | 2D-PAGE | RPLC | ||||
| 1 | 971CK | 84% | 56.1% | – | – | – | – |
| 971-5 | 79% | 17 | 27 | 3 | 15 | ||
| 2 | 971CK | 65% | 69.6% | – | – | – | – |
| 971-4 | 86% | 12 | 33 | 10 | 16 | ||
| 3 | 974CK | 71% | 69.7% | – | – | – | – |
| 974-5 | 87% | 29 | 22 | 15 | 11 | ||
| 4 | 974CK | 70% | 64.3% | – | – | – | – |
| 974-1 | 82% | 27 | 26 | 23 | 17 | ||
Statistical analysis of the protein expression variation between mutant 971-5 and its control 971CK is shown in Figure 3. Analysis in Figure 3A reveals that low-abundance proteins are more likely to be induced in mutants compared with high-abundance proteins. Proteins with altered expression levels in Figure 3A were also detected by RPLC (Figure 3B). Statistical analysis in Figure 3B also reveals that most of the proteins with altered expression levels are low-abundance proteins, which might be regarded as hydrophilic proteins, since they were eluted faster from the chromatogram column.
Fig. 3.
Statistical analysis of the protein expression variation between mutant 971-5 and its control 971CK. A. Statistical analysis on the proteins detected by 2D-PAGE. X-axis is the trend of protein expression, and Y-axis is the intensity of protein expression. The proteins between –0.5 to 0.5 in Y-axis are the matching proteins between control and mutant; the proteins between 0.5 to 1 in Y-axis have increased expression after spaceflight; the proteins between –1 to –0.5 in Y-axis have reduced expression after spaceflight; and the proteins close to 1 or –1 in Y-axis are extremely expressed or undetected in the mutant, respectively. B. Statistical analysis on the proteins detected by RPLC. X-axis is the relative retention time (RRT) (h), and Y-axis is the area percentage (A%). “▲”: Proteins close to 1 in Y-axis of Panel A; “□”: Proteins between 0.5 to 1 in Y-axis of Panel A; “■”: Proteins between −1 to −0.5 in Y-axis of Panel A; “Δ“: Proteins close to −1 in Y-axis of Panel A. Most of the changed proteins are lowly expressed, and the undetected proteins are dense between 0–0.4 h, outflowed fast from the chromatogram column, which might be hydrophilic proteins (in the red circle).
We also performed the same statistical analysis on the other three mutants and their controls, respectively (data not shown). Overall, the four mutants show the same pattern of protein expression variation when compared with their corresponding controls.
Two special proteins in leaves
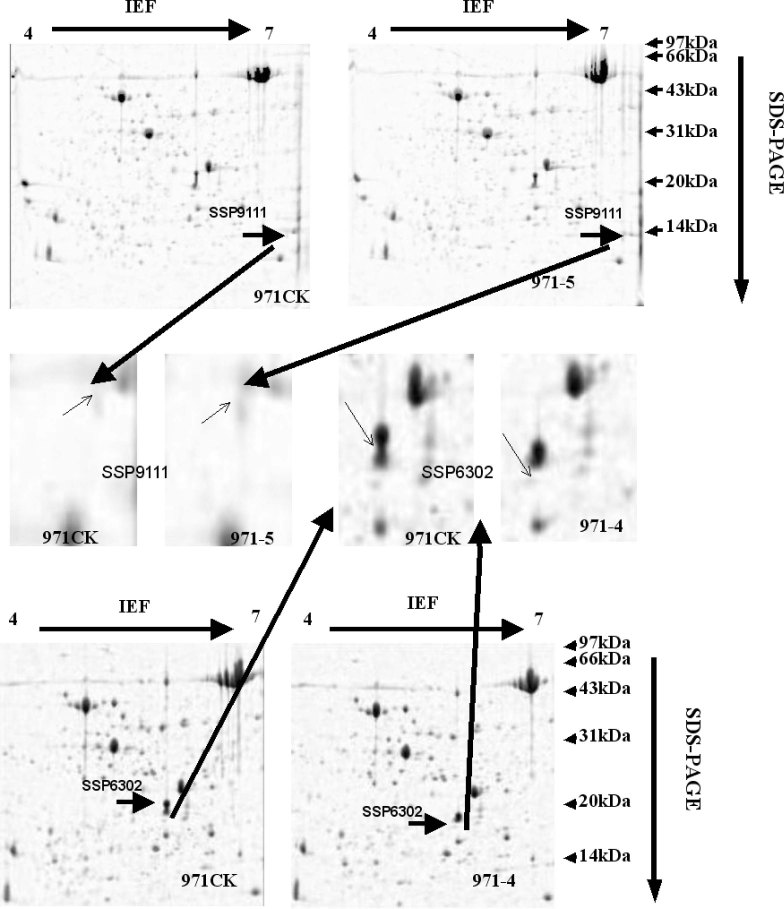
When comparing the 2D-PAGE images of 971CK/971-5 and 971CK/971-4, two proteins, named as SSP9111 and SSP6302, were found to be expressed with high intensity (>two-fold change) by PDQuest analysis software (Figure 4).
Fig. 4.
2D-PAGE expression of proteins SSP9111 and SSP6302.
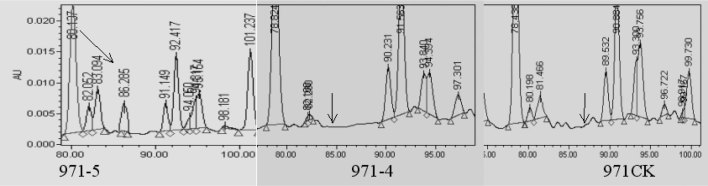
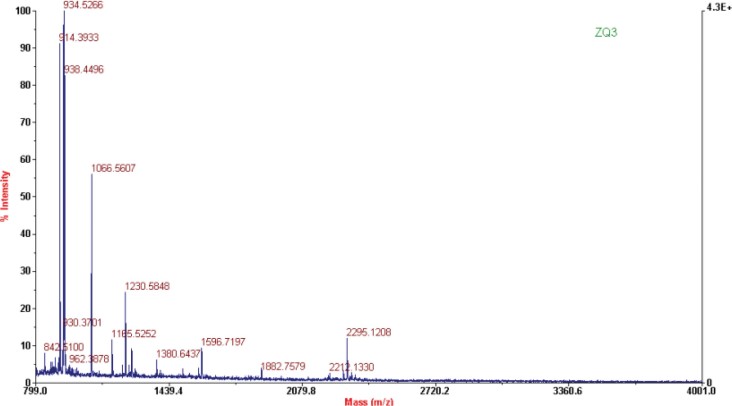
The protein SSP9111 was expressed with high intensity in 971-5 but was undetected in 971-4 and 971CK by 2D-PAGE. In RPLC, the retention time of SSP9111 appeared at about 82–86 min (Figure 5). The peak of SSP9111 was strong in 971-5, while the peak was weak but could be found in both 971CK and 971-4, suggesting that SSP9111 is not a new protein induced by space environment but just significantly increased.
Fig. 5.
Detection of SSP9111 in RPLC fingerprints of 971-5, 971-4, and 971CK.
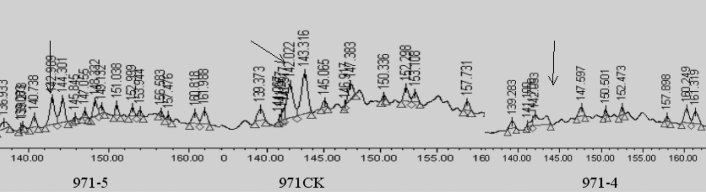
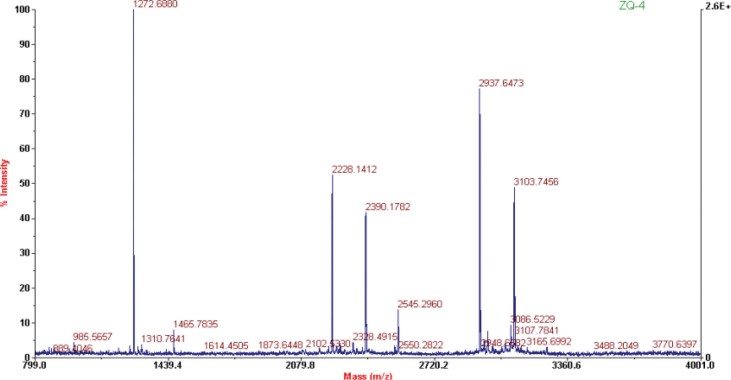
The protein SSP6302 was detected in 971CK and 971-5 but was undetected in 971-4 by 2D-PAGE. In RPLC, the retention time of SSP6302 appeared at about 142–143 min (Figure 6). Although the peak was very weak, it could be found in 971-4. In comparison, both proteins were undetected in 971-4 by 2D-PAGE, but their weak peaks could be found in RPLC, suggesting that RPLC could be an important complementary method to 2D-PAGE in analyzing low-abundance proteins.
Fig. 6.
Detection of SSP6302 in RPLC fingerprints of 971-5, 971-4, and 971CK.
Mass spectrometry of the two proteins
The two proteins SSP9111 and SSP6302 were further analyzed by MS (Fig. 7, Fig. 8). Searching results in the National Center for Biotechnology Information (NCBI) non-redundant (nr) protein database (Accession No. SSP9111: 56966765, 671740; SSP6302: 50936537) showed that both proteins have certain function in photosynthesis. SSP9111 is a Rubisco activase complexed with 2-carboxyarabinitol-1,5-bisphosphate or ribulose-bisphosphate carboxylase, which is the key enzyme for photosynthetic carbon metabolism efficiency. SSP6302 is the photosystem I (PSI) reaction center subunit IV, which is a thylakooid hypothallus surface protein participating in the electron transfer of PSI in photosynthesis. Identification of the two proteins suggests that the high-yield mutation might be correlated with the mutation of photosystem proteins.
Fig. 7.
MS of SSP9111.
Fig. 8.
MS of SSP6302.
Seed proteome and amylose study
Individually selected seeds of each variety (mutants and their ground controls) were analyzed by RPLC. The similitude degree of individually selected seeds was high (>90%) in each variety, but was lower (<70%) among different varieties (Table 1), and the mutants were different from their controls owing to the low similitude degree, suggesting that mutations occurred after spaceflight and each mutation is distinguishable from the other. Then randomly selected seeds were compared with individually selected seeds to test the similarity of two different selection modes. The high similarity (p > 0.05) suggests that the mode of selection had little effect on the analysis of different rice varieties. Therefore, the following results were based on the randomly selected seeds in the 8th and 9th generations.
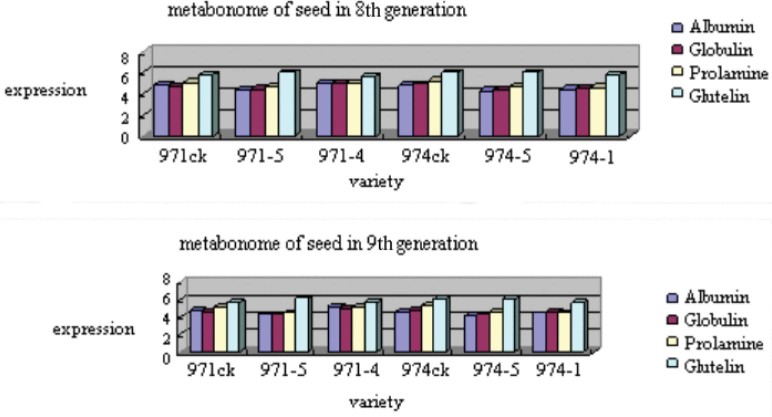
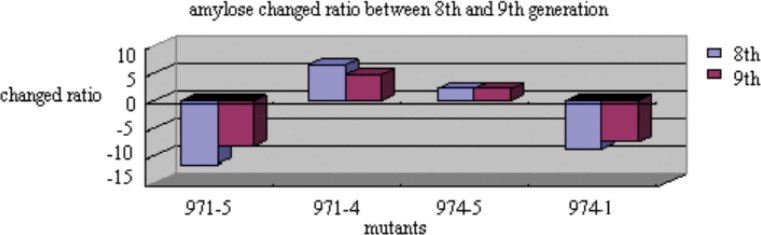
Seed proteome and amylose were investigated to study the stability of the mutants after spaceflight. Table 2 shows the protein contents altered in mutants compared with their controls detected by ultra-violet (UV). The protein contents in mutant seeds were altered, but the alternation was stable in the 8th and 9th generations (Figure 9). In addition, scanning the amylose in mutant seeds by RPLC and UV, we found that the change of amylose was also stable in the 8th and 9th generations (Figure 10).
Table 2.
Protein contents altered in mutant seeds compared with their controls
| Content | Changed ratio (%) |
|||
|---|---|---|---|---|
| 971-5 | 971-4 | 974-5 | 974-1 | |
| Albumin | −10.08 | +5.52 | −10.92 | −7.73 |
| Globulin | −6.03 | +5.42 | −11.83 | −6.54 |
| Prolamine | −9.86 | −1.03 | −12.49 | −13.74 |
| Glutelin | +5.95 | −2.08 | −0.66 | −5.52 |
Fig. 9.
The stable content of seed proteome in the 8th and 9th generations.
Fig. 10.
The changed ratio of amylose between the 8th and 9th generations.
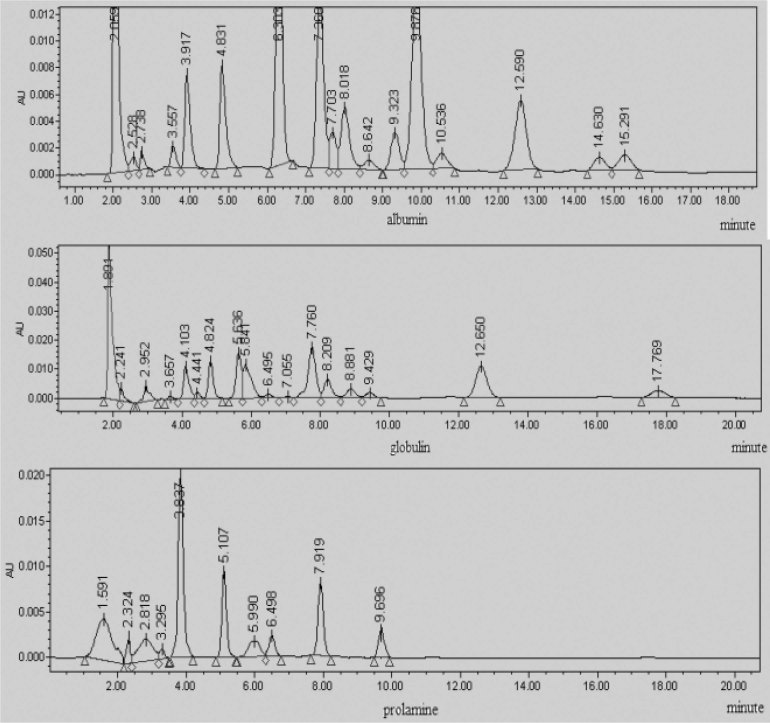
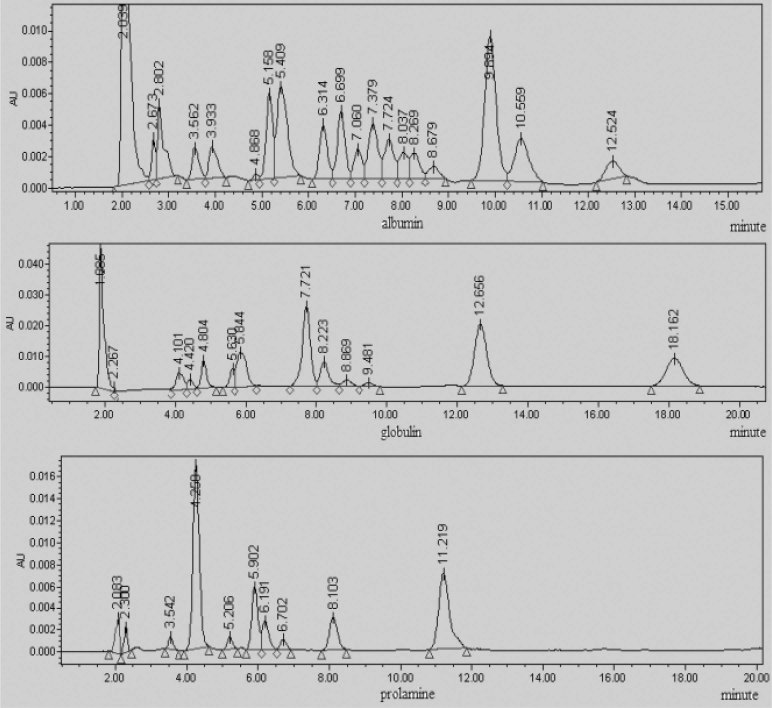
We then compared and analyzed the fingerprints of albumin, globulin, prolamine, and glutelin between mutants and their controls by RPLC. The fingerprints of albumin, globulin, and prolamine in 971CK and 971-5 are shown in Fig. 11, Fig. 12. The similitude degree of glutelin in all the mutants and their controls had little variation (p >0.05), suggesting that the space environment had little effect on glutelin. However, albumin, globulin, and prolamine showed variations between mutants and their controls (p <0.05). According to the results, we can draw the conclusion that some contents of proteins were changed in the seeds of the mutant varieties, and the mutations (the peaks) were stably inherited in the 8th and 9th generations.
Fig. 11.
RPLC fingerprints of albumin, globulin, and prolamine in 971CK.
Fig. 12.
RPLC fingerprints of albumin, globulin, and prolamine in 971-5.
Discussion
The present study has indicated that spaceflight induces mutants of the rice seeds, expressing different proteomic changes with different heritable phenotypes. More down-regulations of protein expression, which were low-abundance proteins, were induced in all the four mutants. As many vital processes are controlled not only by relative-abundance proteins but also by low-abundance proteins, it is important to comprehend complex biological functions of proteins after spaceflight.
Through the reports about Rubisco activase 27., 28., we know that SSP9111 is a coding protein of nuclear gene and is modified in chloroplast. The activity of Rubisco activase is increased with photosynthetic photon flux density when saturated in light intensity of 300 µmol/m.s. That maybe the first step of Rubisco’s active increase. SSP6302 (PSI reaction center subunit IV) is a peripheral protein concerned with Fe–S protein anchorage and electron transfer in PSI (29). SSP9111 increased in the high-tillering and high-yield mutant 971-5, while SSP6302 decreased markedly in the low-tillering and low-yield mutant 971-4, so it is very interesting to study the two proteins involved with tillering phenotype changes.
In the analysis of proteins and amylose of rice seeds, the changes of albumin, globulin, and prolamine varied among different mutants, indicating that the changes of mutants were from different aspects. These changes were stable in the 8th and 9th generations, so they can be used as biomarkers to identity the mutants induced by spaceflight.
Materials and Methods
Sample collection
Dry rice seeds were carried by a recoverable satellite JB-1 for a 15-day spaceflight in 1996. Before spaceflight, rice had been planted steadily in the Space Environment Biological Effect Base of Harbin Institute of Technology (Wuchang, China) for many years. After spaceflight, the seeds were returned and bred in the above base to obtain stable mutants with new traits. The recovered seeds were germinated and grown on the ground with their parallel controls. After breeding and selection, the following phenotypic mutants were identified: mutants 971-5 and 974-5 acquired new traits after spaceflight such as increased yield and high tillering compared with their controls 971CK and 974CK, respectively. In contrast, mutants 971-4 and 974-1 exhibited reduced yield and low tillering compared with their controls 971CK and 974CK, respectively. The rice leaves used in this study were from stable mutants in the 8th generation (materials picked on July 7, 2004) and the 9th generation (materials picked on July 10, 2005). In the peak tillering stage, leaves from the above mutants were collected and quickly frozen in liquid nitrogen. Rice seeds were also harvested and selected. All samples were stored at −80°C for further study.
Protein preparation
After grinding, the powder of leaves was precipitated in acetone by adding ice-cold trichloroacetic acid, followed by incubation at –20°C for at least 1 h. After centrifuging, the acetone pellets were rotated and incubated. Fresh cold acetone was added into the pellets every 1 h until the supernatant was clear. After additional centrifuging, the pellets were dried and then stored at –20°C for further studies. Rice seeds were also made into powder and mixed with water followed by centrifugation at 4°C for 15 min. A solution with 10% NaCl, 50% glycerin, and 0.1 mol/L NaOH was added consequently to obtain albumin, globulin, prolamine, and glutelin.
2D-PAGE and RPLC conditions
In 2D-PAGE, approximately 500 µg/350 μL rehydration buffer of extracted proteins was loaded onto BioRad (Herculas, USA) IPG strips of pH 3–10 and 4–7 in length of 17 cm. The first-dimensional IEF was conducted at 20°C using a Bio-Rad Protean IEF Cell system. After the first-dimensional IEF, strips were equilibrated and transferred onto 13% polyacrylamide gels of 18 cm×20 cm. 2D-PAGE was performed in a Bio-Rad Protean II xi Multi-Cells system (50 mA per gel), and the gels were stained with Coomassie brilliant blue (CBB). The obtained images were analyzed by PDQuest software (Bio-Rad).
RPLC was employed to analyze the proteins as a complementary method. The proteins were separated by C18 column (symmetry 3.9 mm×150 mm) and C4 column (symmetry 3.9 mm×150 mm, 300 A), equilibrated with acidified water containing 0.12% trifluoroacetic acid (TFA) for 15 min. The sample was eluted in a linear gradient of 0–60% v/v acetonitrile in 0.1% TFA water over 60 min at a flow rate of 1,000 μL/min (40°C, 280 nm wavelength). The reagent for RPLC was from Sigma Co. (St. Louis, USA).
2D-PAGE and RPLC were employed to investigate the whole proteome and proteins of interest, focusing on the alternation of protein expression after spaceflight. The controls 971CK were prepared as samples in the precision test, repeatability test, and stability test of RPLC.
RPLC and UV were employed to investigate the mutant stability between the 8th and 9th generations focused on albumin, globulin, prolamine, glutelin, and amylose.
2D-PAGE and RPLC fingerprinting were employed to analyze the leaf proteins, seed proteins, and amylose. The ground controls were regarded as standard fingerprints. Fingerprint similitude analysis software (developed by Zhejiang University and Dalian Institute of Chemical Physics, CAS) was used to estimate the sampling distribution of the correlation coefficient between the control’s fingerprint and the mutant’s fingerprint.
Mass spectrometry
CBB-stained proteins were excised from gels, washed with 25% methanol, 7% acetic acid at room temperature for 12 h, and destained with 50 mM NH4HCO3 in 50% methanol at 40°C for 1 h. After drying under vacuum, gel spots were incubated in 50 μL reduction solution (10 mM EDTA, 10 mM DTT, and 100 mM NH4HCO3) at 60°C for 1 h. Then the gel spots were dried under vacuum and incubated in 50 μL alkylation solution (10 mM EDTA, 10 mM iodoacetamide, and 100 mM NH4HCO3) at room temperature for 30 min in the dark. After washing with water, the gel spots were minced, dried under vacuum, and digested in 50 μL of 10 mM Tris-HCl (pH 8.0) containing 1 pM trypsin at 37°C for 10 h. Acetonitrile (100 μL) containing 0.1% TFA was added to each gel piece and sonicated. Purification of the generated peptides was achieved by adding directly 10 mg/mL α-cyano-4-hydroxycinnamic acid, 0.3% TFA, 50% acetonitrile matrix, and air-dried onto a plate for analysis using MALDI-TOF MS. MS was performed by Genecore Co. Ltd. (Shanghai, China) under ABI’s Voyager-DE Pro Biospectrometry Workstation (Foster City, USA). The results were searched using MS-Fit Protein Prospector (http://prospector.ucsf.edu/) of University of California, San Francisco, USA.
Authors’ contributions
WHL collected the datasets, conducted data analyses, and prepared the manuscript. XZW conducted data analyses and co-wrote the manuscript. QZ collected the datasets. SHG and PX cultured and offered the materials. YQS conceived the idea of using this approach, supervised the project, and assisted with manuscript preparation. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank collaborators of the Space Environment Biological Effect Base of Harbin Institute of Technology for important suggestions. This work was funded by the Commission of Science Technology and Industry for National Defense (Grant No. 2001-895) and the National Hi-Tech Research and Development Program (863 Program) of China (Grant No. 20060802).
References
- 1.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 2.Goff S.A. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 3.Yu J. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu S. Rice Proteome Database based on two-dimensional polyacrylamide gel electrophoresis: its status in 2003. Nucleic Acids Res. 2004;32:D388–D392. doi: 10.1093/nar/gkh020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun, Y. 2001. Mutation effect of space environments on rice seeds. In Proceedings of the 52nd International Astronautical Congress. Toulouse, France.
- 7.Sun Y. Rice Mutation after Flown in Space. J. Harbin Inst. Technol. (New Series) 2002;9:7–11. [Google Scholar]
- 8.Xu, J., et al. 2004. Inheritance of induction radiation sensitivity of space flight environments and γ-radiation on rice. In Proceedings of the 35th COSPAR Conference, F4.4-0008-04. Paris, France.
- 9.Cheng, Z., et al. 2004. Molecular analysis of rice plant mutated after space flight. In Proceedings of the 35th COSPAR Conference, F4.4-0011-04. Paris, France.
- 10.Wang J.M. Study on application of aerospace techniques in rice mutation breeding. Acta Agriculturae Nucleatae Sinica. 2004;18:252–256. [Google Scholar]
- 11.Wang J.M. Mutagenic differences of space environment and ground γ-irradiation in rice. Acta Agronomica Sinica. 2006;32:1006–1010. [Google Scholar]
- 12.Wei L.J. A comparative study on mutagenic effects of space flight and γ-rays irradiation in rice. Scientia Agricultura Sinica. 2006;39:1306–1312. [Google Scholar]
- 13.Lu, W., et al. 2006. Protein fingerprint diversification of rice seeds after space environment. In Proceedings of the 36th COSPAR Conference, F4.6-0001-06. Beijing, China.
- 14.Wang, W., et al. 2006. Differential analysis in proteome of space induced rice and soybean varieties. In Proceedings of the 36th COSPAR Conference, F4.6-0007-06. Beijing, China.
- 15.Li Y. Space environment induced mutations prefer to occur at polymorphic sites of rice genomes. Adv. Space Res. 2007;40:523–527. [Google Scholar]
- 16.Yu X. Characteristics of phenotype and genetic mutations in rice after spaceflight. Adv. Space Res. 2007;40:528–534. [Google Scholar]
- 17.Ma Y. Proteomic analysis of high yield rice variety mutated from space flight. Adv. Space Res. 2007;40:535–539. [Google Scholar]
- 18.Cheng Z. Transcriptomic analyses of space-induced rice mutants with enhanced susceptibility to rice blast. Adv. Space Res. 2007;40:540–549. [Google Scholar]
- 19.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 20.Barbier-Brygoo H., Joyard J. Focus on plant proteomics. Plant Physiol. Biochem. 2004;42:913–917. doi: 10.1016/j.plaphy.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed N., Rice G.E. Strategies for revealing lower abundance proteins in two-dimensional protein maps. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;815:39–50. doi: 10.1016/j.jchromb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 22.Bewley J.D., Black M. second edition. Plenum Press; New York, USA: 1994. Seeds: Physiology of Development and Germination. [Google Scholar]
- 23.Tanaka T. The occurrence of phosphatidyl choline exchange protein in leaves. Biochem. Biophys. Res. Commun. 1980;96:394–399. doi: 10.1016/0006-291x(80)91228-0. [DOI] [PubMed] [Google Scholar]
- 24.Kishimoto T. Glutelin basic subunits have a mammalian mucin-type O-linked disaccharide side chain. Arch. Biochem. Biophys. 1999;370:271–277. doi: 10.1006/abbi.1999.1406. [DOI] [PubMed] [Google Scholar]
- 25.Shu Q. Correlation between mass fraction of apparent amylose and (CT)n microsatellite polymorphisms of waxy gene in rice progenies. Chin. J. Appl. Environ. Biol. 1995;5:464–467. [Google Scholar]
- 26.Fang K.T. Critical value determination on similarity of fingerprints. Chemometrics Intell. Lab. Syst. 2006;82:236–240. [Google Scholar]
- 27.Zhang G. Molecular biology of Rubisco activase. Plant Physiol. Commun. 2004;4:633–637. [Google Scholar]
- 28.Han Y. The progresses of studies on Rubisco activase. Chin. Bull. Bot. 2000;17:306–311. [Google Scholar]
- 29.Zhang H.B., Xu D.Q. Photosystem II protein phosphorylation and its physiological significance. J. Plant Physiol. Mol. Biol. 2003;29:487–493. [Google Scholar]