Abstract
A concept of unique peptides (CUP) was proposed and implemented to identify whole-cell proteins from tandem mass spectrometry (MS/MS) ion spectra. A unique peptide is defined as a peptide, irrespective of its length, that exists only in one protein of a proteome of interest, despite the fact that this peptide may appear more than once in the same protein. Integrating CUP, a two-step whole-cell protein identification strategy was developed to further increase the confidence of identified proteins. A dataset containing 40,243 MS/MS ion spectra of Saccharomyces cerevisiae and protein identification tools including Mascot and SEQUEST were used to illustrate the proposed concept and strategy. Without implementing CUP, the proteins identified by SEQUEST are 2.26 fold of those identified by Mascot. When CUP was applied, the proteins bearing unique peptides identified by SEQUEST are 3.89 fold of those identified by Mascot. By cross-comparing two sets of identified proteins, only 89 common proteins derived from CUP were found. The key discrepancy between identified proteins was resulted from the filtering criteria employed by each protein identification tool. According to the origin of peptides classified by CUP and the commonality of proteins recognized by protein identification tools, all identified proteins were cross-compared, resulting in four groups of proteins possessing different levels of assigned confidence.
Key words: protein identification, unique peptide, tandem mass spectrometry
Introduction
Mass spectrometry (MS) based protein identification experiments have been the major resource for large-scale proteomic studies of a cell or an organism 1, 2, 3, 4, 5. Presently, there are numerous protein identification packages available such as MS-Tag (6), Mascot (7) and SEQUEST 8, 9. Reviews on these various protein identification tools were reported recently 10, 11.
The critical complexity in protein identification lies in the need to provide confidence levels for the results obtained using the above mentioned tools. A set of positive protein results can help derive accurate conclusions and develop an appropriate plan for further study. However, the practice of using a specific set of MS data to predict several peptides necessitates the separation of the “real” proteins by showing their high confidence. This protracted step is one of the most complicated in protein identification. The major difficulties in using these protein identification tools include multi-identification (i.e., a series of identified peptides may be used to identify two or more proteins), low-confidence identification (i.e., the Mowse score of each peptide is lower than the threshold Mowse score, though the total Mowse score may be greater than the threshold value), and pre-set threshold values used to determine the “true” peptide (e.g., Xcorr in SEQUEST).
An apparent downside in protein identification using SEQUEST is the determination of the Xcorr value. Under diverse Xcorr settings, the searched results, based on the same MS/MS data, may show great variation leading to ambiguity among biological researchers. For instance, from the MS/MS data of Saccharomyces cerevisiae (12), 1,227 proteins were recognized for Xcorr value set to 2.0 or greater while only 347 proteins were identified for Xcorr value greater than or equal to 2.5. These two sets of “identified” proteins were derivatives of the same MS/MS spectral dataset using the same protein identification tool. Consequently, these deviant protein results convey confounding messages to scientists when applied to interpreting phenotypic observations.
Comparisons among various protein identification tools were also reported 13, 14. For example, Chamrad et al. (13) applied different protein identification tools to the same set of MS and MS/MS spectral data and observed that only 30%-50% of the results were consistent. This underscores the fact that searched proteins from each protein identification tool generate different confidences, and only those proteins with high confidences can be recognized by these tools. Accordingly, a strategy to analyze the confidence of searched proteins is required.
Based on the concept of unique peptides (CUP) and the cross-comparison among identified proteins, a two-step strategy to study the confidence of whole-cell protein identification was developed in this study. The CUP filters first classify peptides into unique and non-unique clusters, and the step of cross-comparison adds the levels of assigned confidence to proteins identified by means of different protein identification tools. Depending on the accessibility of additional protein identification tools, the proposed dual step approach can be applied independently or in a combined mode. To demonstrate effect of the strategy, two extensively used protein identification packages, namely SEQUEST and Mascot, were employed to identify proteins from publicly available MS data, and the recognized proteins from these tools were investigated using the proposed two-step protein identification strategy.
Results
Concept of unique peptides
A unique peptide is defined as a peptide, irrespective of its length, that exists only in one protein of a proteome of interest, despite the fact that this peptide may appear more than once in the same protein. For example, for Proteins 1 and 2 digested by trypsin, the expected peptides with zero missed cleavage are illustrated in Figure 1.
Figure 1.
Illustration of the concept of unique peptides.
According to the definition, the peptide ANDR shown in Figure 1 is regarded as unique since it appears once in Protein 1 but not in Protein 2. The peptide NQEGHK is also considered unique based on the same standard. Neither MFPSTK nor WYVTR are unique peptides as they appear in both Proteins 1 and 2. Other unique peptides include MFPSR and CEGIK, found only in Protein 2. The definition of unique peptide is essential in protein identification. It is intuitive to identify Protein 1, if ANDR, NQEGHK, or both are identified. On the contrary, it becomes challenging to conclude whether Protein 1 or 2 exists if only MFPSTK is identified from the MS/MS data. Therefore, a unique peptide can act as a “protein tag” in protein identification.
Whole-cell protein identification
The general procedure implemented in a protein identification tool contains three steps: peptide ranking, peptide filtering and protein identification, which are equivalent to Steps 1, 3 and 4 shown in Figure 2 (the leftmost column). The MS and MS/MS ion spectra are combined to reconstruct the amino acid sequence of peptides. It is typical that not all experimentally obtained mass spectral data are used during protein identification. For example, Peng et al. (4) reported in their yeast proteome experiments that among 162,000 MS/MS ion spectra, only 26,815 peptides were reconstructed and considered confident, representing a 16.5% utilization of MS/MS information.
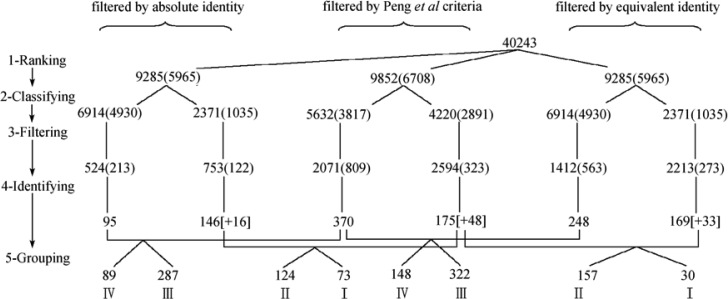
Figure 2.
Application of the concept of unique peptides and the effect of peptide filtering criteria on the whole-cell protein identification. The number shown in the parentheses represents non-redundant peptides, and the number surrounding by square brackets stands for proteins derived from the non-unique peptide database that were also found in the unique peptide database.
Steps 1, 3 and 4
Using MS/MS spectral datasets (12), 9,285 top-ranking peptides were derived from 40,243 MS/MS ion spectra by means of Mascot, and 9,852 top-ranking peptides were obtained by SEQUEST (Figure 2, Step 1). By applying the identity threshold score based on the absolute probability implemented in Mascot, 335 (213+122) peptides were satisfied; when SEQUEST-filtering criteria proposed by Peng et al. (4) were adopted, 1,132 (809+323) peptides were found (Figure 2, Step 3). As a result, 241 (95+146, using Mascot) and 545 (370+175, using SEQUEST) proteins were deemed identified (Figure 2, Step 4). It can be seen that the proteins identified by SEQUEST are 2.26 fold of those identified by Mascot. On average, each protein recognized by SEQUEST requires 2.08 peptides, whereas 1.39 peptides per protein are needed for Mascot. When an equivalent identity threshold (see Materials and Methods) was taken as the filtering criterion in Mascot, 836 (563+273) peptides satisfied the requirement, resulting in 417 (248+169) identified proteins (2.01 peptides per protein). Even though, the proteins identified by SEQUEST are still 1.31 fold of those identified by Mascot.
Step 2
Our proposed two-step strategy includes two additional yet critical steps: Steps 2 and 5 (Figure 2); that is, peptide classifying: classify top-ranking peptides into unique and non-unique peptide cluster, and protein regrouping: regroup identified proteins into four different levels of assigned confidence. In Step 2, a total of 6,708 top-ranking non-redundant peptides (from Step 1) reconstructed by SEQUEST were classified into 3,817 non-redundant unique and 2,891 non-redundant non-unique peptides, in which only 809 unique peptides satisfied filtering criteria (4), representing 21.19% of total unique peptides, from which 370 proteins were deduced. The ratio of unique peptide to protein was 2.19 (see supplementary file “sequest.xls” for relevant unique and non-unique peptides).
For Mascot, 4,930 out of 5,965 top-ranking peptides were unique according to the proposed CUP, and the remaining 1,035 peptides were non-redundant and non-unique. When the absolute identity threshold was applied, there were 213 unique peptides (corresponding to 4.32% of total unique peptides) that satisfied Mascot filtering criteria, from which 95 proteins were deemed identified, and the unique peptide-to-protein ratio was 2.24 (refer to supplementary file “mascotab.xls”). In parallel, 563 unique peptides satisfied the equivalent identity threshold criteria. This represents 11.42% of total unique peptides in this category, which translates to 248 proteins, and the ratio of unique peptide to protein was 2.27 (refer to supplementary file “mascot-eq.xls”).
Step 5
To further analyze the confidence of identified proteins obtained from different protein identification tools, these proteins were cross-compared (Figure 2, Step 5). Among 370 SEQUEST-recognized and 95 Mascot-recognized proteins using absolute identity threshold (both deduced according to CUP), there were 89 proteins found by both tools and were considered as a protein group with the highest confidence (Level IV). The remaining 287 proteins possessing unique peptides were assigned as Level III, a group of proteins with the second highest confidence. The above two levels of proteins represent 65.62% of total recognized proteins. Correspondingly, after cross-comparing 175 proteins from SEQUEST and 146 proteins from Mascot (both derived from non-unique peptide cluster), 124 common proteins and the remaining 73 non-common proteins were collected and assigned as Level II and Level I proteins, respectively, representing two low confident groups in this proposed strategy (see supplementary file “sequest-mascot-ab.xls” for a complete listing of re-grouped peptides).
When proteins were identified by means of equivalent identity threshold in Mascot and cross-compared to those identified by SEQUEST, four groups of proteins were collected. Each group contains 148, 322, 157, and 30 proteins ranging from Level IV to Level I, respectively. Both Level IV and Level III proteins occupy 71.54% of total identified proteins under this set of filtering criteria (see supplementary file “sequest-mascot-eq.xls” for a complete listing of re-grouped peptides).
The identified proteins with higher levels of confidence defined by our proposed strategy require no (Level IV proteins) or a lesser degree (Level III proteins) of human intervention, because these proteins were identified by means of CUP. In contrast, both Level II and Level I proteins must be carefully scrutinized when drawing conclusions.
Discussion
Characteristics of unique peptides and missed cleavage
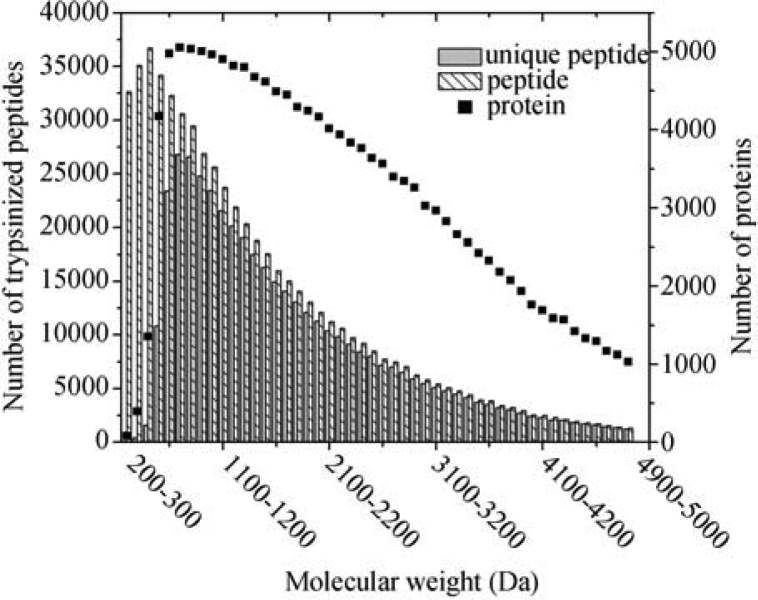
Figure 3 portrays distributions of the number of proteins, trypsinized peptides (including both unique and non-unique), and trypsinized unique peptides at different molecular weights. The number of matched proteins decreases monotonically as molecular weight increases, whereas a large portion of proteins are related to low-molecular-weight peptides. In the high-molecular-weight region, the numbers of peptides and unique peptides are nearly the same, illustrating that either peptide type could be used to identify proteins, and a highly accurate result could be obtained due to a smaller sample size (~1,000 proteins and ~10,000 peptides). Comparatively, the differences between the number of peptides and the number of unique peptides become noticeable in the low-molecular-weight region, indicating the difficulty of deducing true proteins from a pool of tens of thousands of peptides. When the characteristics of unique peptides are applied to protein identification, the uncertainty of deducing proteins from peptide fragments is minimized. The significance of CUP becomes obvious and effective, particularly for identifying proteins possessing short peptides. The CUP also simplifies the efforts exerted on the whole-cell protein identification, since no advanced and/or complicated mathematics or statistical reasoning is required. The drawback of CUP is that not all proteins possess unique peptides, meaning that different protein identification approaches are required to identify those proteins that do not possess unique peptides as defined by CUP.
Figure 3.
Distributions of in silico trypsinized unique peptides, peptides (including both unique and non-unique), and proteins at different molecular weights. This figure is constructed by allowing one missed cleavage.
A protease, such as trypsin, is used to digest a whole-cell protein sample, resulting in a pool of peptides at various lengths. When a yeast proteome is in silico digested by trypsin, 334,520 peptides are resulted from 5,863 proteins if a perfect trypsinzation is assumed (i.e., no missed cleavage is allowed). The peptide-to-protein ratio is 57. When only unique peptides are taken into consideration, the same ratio reduces to 31. If one missed cleavage on the amino acid sequence of a protein is allowed, the peptide-to-protein ratio becomes 113, and the ratio reduces to 78 when only considering unique peptides. This ratio depicts the average number of peptides required in order to match a protein out of a yeast proteome. A smaller ratio at each respective missed cleavage indicates a significant reduction of false positive protein identification from a whole-cell protein sample, and a high confidence of identified results would be expected because of the characteristics of unique peptides implemented in the proposed protein identification strategy.
Unique peptides and molecular weight
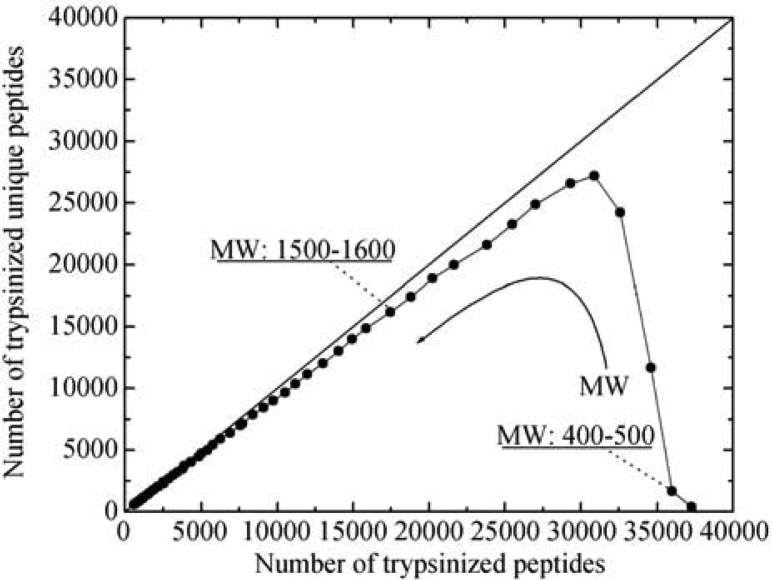
Figure 4 depicts that at low-molecular-weight region, there are more trypsinizied peptides than unique ones; for example, at molecular weights between 400 and 500 Da, there are 35,982 peptides compared to 1,672 unique peptides. The ratio of these two peptide groups is 21.52. As the molecular weight of a peptide increases, the peptide ratio approaches 1; for example, at molecular weight between 1,500 and 1,600 Da, the ratio is 1.08 (17,446 vs. 16,198). This observation clearly points out the difficulty of identifying proteins from a low-molecular-weight peptide pool. Without the implementation of CUP to whole-cell protein identification, a high false positive rate of protein identification would be expected, leading to erroneous conclusions.
Figure 4.
Correlation of trypsinized unique peptides and trypsinized peptides (including both unique and non-unique) at different molecular weights.
Figure 4 also illustrates that as the molecular weight of a peptide increases, the differentiation between trypsinized peptides and trypsinizied unique peptides becomes unremarkable (approaching to the 45° line). In other words, peptides having larger molecular weight possess characteristics as defined by CUP. The correlation of the average length of a unique peptide (Y) to its average molecular weight (X) can be expressed as Y = 0.0089X, which means that the longer a trypsinizied peptide, the higher the possibility of it being regarded as a unique peptide. As a result, a much higher confidence of the identified proteins could be drawn.
Filtering criteria
Different protein identification tools implement different peptide ranking and filtering criteria. SEQUEST generates in silico mass spectra, compares them to the experimentally obtained ones, and ranks the matches; whereas Mascot pre-processes intensities of mass signals in order to increase the signal-to-noise ratio, and uses a probability-based approach to rank the matches (7). The absolute probability in conjunction with a user-specified false positive rate (based on type II error) was adopted by Mascot and used to calculate an identity threshold to filter ranked peptides. Due to different degrees of stringency applied to removing low confident peptides, there are over 50% differences in identified proteins derived from the same MS/MS ion spectra (refer to Steps 1, 3 and 4 in Results section). To validate the hypothesis that the differentiation in the number of matched proteins results from different filtering criteria, an equivalent identity threshold using data reported in Peng et al. (4) was estimated and applied to the same spectra. The result clearly indicates that the peptide filtering criteria (absolute identity vs. equivalent identity; or, 241 vs. 417 proteins) hold enormous impact on the protein identification; the looser the filtering criteria, the larger number of matched proteins. By further examining proteins identified by Mascot and SEQUEST, a noticeable disagreement in matched proteins was observed, causing one to wonder which are true positive proteins.
It is meaningless to determine which peptide-filtering criteria implemented in each respective protein identification tool is superior to others, since each set of criteria has its own strength and weakness. To utilize the strength of those criteria built in different protein identification tools, one could cross-compare matched proteins deduced by each protein identification tool; as such, a high confident set of results could be obtained, and different levels of confidence of recognized proteins could be assigned.
Origin, commonality and confidence of identified proteins
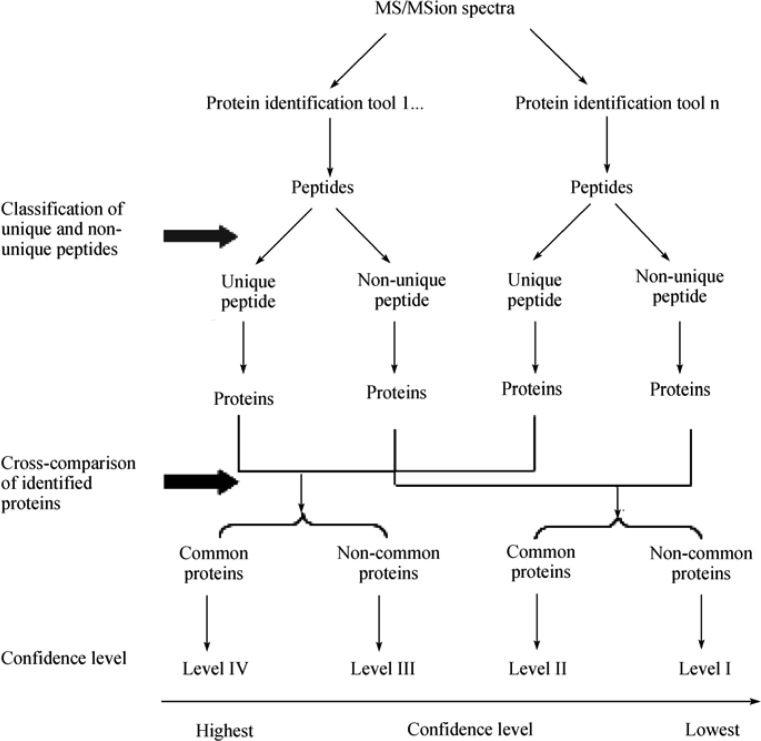
To minimize the inconsistency of identified proteins derived from different protein identification tools, and thus to increase the confidence of identified proteins, a cross-comparison step among matched proteins was implemented into the proposed protein identification strategy. Four levels of confidence were assigned according to the origin of a peptide (unique vs. non-unique) and the commonality of a matched protein (presence vs. absence) in all protein identification tools. As illustrated in Figure 5, a Level IV protein is a protein possessing unique peptides and found in all protein identification tools, whereas a protein comprised of unique peptides but not found in all protein identification tools is assigned as a Level III protein. Those proteins consisting of non-unique peptides and either found or not found in all protein identification tools are called Level II and Level I proteins, respectively. A higher level of confidence of matched proteins (such as Level IV and Level III proteins) implies that a true positive identification is concluded, and requires no or a lesser degree of manual validation. On the other hand, a false positive identification might result from those Level II and Level I proteins; therefore, careful scrutinization becomes unavoidable.
Figure 5.
Illustration of the two-step strategy for whole-cell protein identification. The initial step (Step 2 in Figure 2) involves the classification of top-ranking peptides from each protein identification tool into unique and non-unique peptide pools, which are subsequently used for protein identification. The last step (Step 5 in Figure 2) cross-compares the identified proteins from previous step: common proteins identified in the cross-comparison step are more confident than the non-common ones.
Conclusion
Generally, a protein identification tool takes MS and/or MS/MS ion spectra to rank and deduce the most probable peptides, and the resulting proteins are regarded as identified. These proteins are then examined and validated by experienced MS staff. Since manual curation is involved, the efficiency of high-throughput technologies such as shotgun proteomic experiments is discounted. Frequently, different peptides are identified even when the same set of MS and MS/MS spectra is interpreted by a single software tool (12). Different proteins might be reported by various identification tools, even though the concept of unique peptides is incorporated to improve the confidence of identified proteins. Cross-comparison can, therefore, further enhance the level of confidence of matched proteins by finding common proteins in different protein identification tools from the same dataset of MS and MS/MS ion spectra. The proposed concept of unique peptides can be seamlessly integrated into the existing protein identification tools. The developers of these tools need only to incorporate the pre-built unique peptide database in Step 2 of the described two-step strategy. Although the peptide database is in silico constructed for trypsin, the proposed approach can be readily implemented and extended to other proteinases to cleave proteins if the cleavage sites were known.
Materials and Methods
The proposed two-step protein identification strategy was examined using MS/MS spectral dataset retrieved from http://bioinformatics.icmb.utexas.edu/OPD/ (12). The accession number of the dataset is “opd00034_YEAST”, which was compressed and named as “6-04-03-YPD_test.sequest.zip”. The dataset contains 11 fractions of whole-cell eluents of S. cerevisiae, including 40,243 MS/MS spectra (in “.dta” format) and the corresponding SEQUEST-processed results (in “.out” format). The proposed strategy is illustrated in Figure 5 and described below.
Construction of trypsinized unique/non-unique peptide database
Yeast protein sequences (“s.cerevisiae.pep”) were retrieved from Kyoto Encyclopedia of Genes and Genomes (ftp://ftp.genome.jp/pub/kegg/genes/organisms/sce/; April 2008), which contain 5,863 proteins. The enzymatic cleavage sites of trypsin tabularized by Snyder (15) were implemented to in silico construct a tryptic yeast peptide pool. Allowing one missed cleavage, 663,177 peptides were obtained. Based on the concept of unique peptides, 445,227 peptides were resulted.
Overall average identity threshold and homology threshold (Mab and Meq)
The overall average identity threshold (Mab) and homology threshold (Meq) were calculated by summing all respective identity and homology thresholds found in all 11 Mascot summary reports and divided by its respective total number of appearances. As a result, Mab = 27.665 and Meq = 15.962.
Estimation of equivalent identity threshold (λeq)
The equivalent identity threshod (λeq) was estimated from Mab. From Mascot’s Help – Results Interpretation, Mab = –10 log (fpr/x), in which fpr stands for false positive rate and x is the number of peptides falling within the mass tolerance window about the precursor mass. Given Mab = 27.665 and fpr = 0.05, x was then calculated to be 29.206. Incorporating x and fpr found in Figure 5A of Peng et al. (4), we have λeq = 15.471, which is close to Meq. Hence, when filtering peptides, λeq > 15 was implemented.
Whole-cell protein identification
Step 1: peptide ranking
A PERL script was developed to extract top-ranking peptides from all “.out” files. All “.dta” files were concatenated into 11 portions based on the eluent fractions, and each portion of spectral dataset was imported into Mascot to carry out MS/MS ion search. The search parameters used were: Type of search: MS/MS Ion Search; Database: NCBInr; Taxonomy: S. cerevisiae; Enzyme: Trypsin; Fixed modifications: Carbamidomethyl (C); Mass values: Average; Protein Mass: Unrestricted; Peptide Mass Tolerance: ±1 Da; Fragment Mass Tolerance: ±0.4 Da; Max Missed Cleavages: 1; Instrument type: ESI-TRAP. The top-ranking peptides from all 11 Mascot peptide summary reports were collected.
Step 2: peptide classification
According to the concept of unique peptides defined in the Results section, all top-ranking peptides from Step 1 were classified into two clusters: unique and non-unique peptides.
Steps 3 and 4: peptide filtering and protein identification
For SEQUEST, filtering criteria reported by Peng et al. (4) were used; that is, for singly charged ions (+1), the Xcorr value must be greater than 1.5; and for +2 and +3 ions, the Xcorr value must be greater than 2.0 and 3.3, respectively. In Mascot, each top-ranking peptide has an associated Mowse score and is accompanied by an (absolute) identity threshold (λab), which was calculated using absolute probability along with a pre-specified false positive rate. When a peptide had a Mowse score greater than its corresponding absolute identity, the protein harboring this peptide was then considered as “identified”.
Step 5: protein regrouping
Proteins identified in Step 4 from SEQUEST and Mascot were further cross-compared, and pooled into four groups according to their origins from unique and non-unique peptides. Refer to Figure 5 for the definition and the assigned level of confidence of these proteins.
Authors’ contributions
YZ and YHL contributed equally, including concept and strategy development, data analysis and interpretation. YHL supervised the work and co-wrote the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Supplementary Material
References
- 1.Link A.J. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 2.Mawuenyega K.G. Large-scale identification of Caenorhabditis elegans proteins by multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2003;2:23–35. doi: 10.1021/pr025551y. [DOI] [PubMed] [Google Scholar]
- 3.McCormack A.L. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 4.Peng J. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 5.Pflieger D. Systematic identification of mitochondrial proteins by LC-MS/MS. Anal. Chem. 2002;74:2400–2406. doi: 10.1021/ac011295h. [DOI] [PubMed] [Google Scholar]
- 6.Clauser K.R. Role of accurate mass measurement (+/−10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 7.Perkins D.N. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Eng J.K. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 9.Yates J.R., 3rd Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 10.Eng J.K. Computational tools for tandem mass spectrometry-based high-throughput quantitative proteomics. In: Srivastava S., editor. Informatics in Proteomics. CRC Press; Boca Raton, USA: 2005. pp. 335–351. [Google Scholar]
- 11.Nesvizhskii A.I. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 12.Prince J.T. The need for a public proteomics repository. Nat. Biotechnol. 2004;22:471–472. doi: 10.1038/nbt0404-471. [DOI] [PubMed] [Google Scholar]
- 13.Chamrad D.C. Evaluation of algorithms for protein identification from sequence databases using mass spectrometry data. Proteomics. 2004;4:619–628. doi: 10.1002/pmic.200300612. [DOI] [PubMed] [Google Scholar]
- 14.Elias J.E. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 15.Snyder A.P. Oxford University Press; New York, USA: 2000. Interpreting Protein Mass Spectra: A Comprehensive Resource; p. 166. Table 9.4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.