Abstract
A systematic phylogenetic footprinting approach was performed to identify conserved transcription factor binding sites (TFBSs) in mammalian promoter regions using human, mouse and rat sequence alignments. We found that the score distributions of most binding site models did not follow the Gaussian distribution required by many statistical methods. Therefore, we performed an empirical test to establish the optimal threshold for each model. We gauged our computational predictions by comparing with previously known TFBSs in the PCK1 gene promoter of the cytosolic isoform of phosphoenolpyruvate carboxykinase, and achieved a sensitivity of 75% and a specificity of approximately 32%. Almost all known sites overlapped with predicted sites, and several new putative TFBSs were also identified. We validated a predicted SP1 binding site in the control of PCK1 transcription using gel shift and reporter assays. Finally, we applied our computational approach to the prediction of putative TFBSs within the promoter regions of all available RefSeq genes. Our full set of TFBS predictions is freely available at http://bfgl.anri.barc.usda.gov/tfbsConsSites.
Key words: phylogenetic footprinting, phosphoenolpyruvate carboxykinase, transcription factor binding sites, mammalian gene promoters
Introduction
A major challenge of the post-genome era is the characterization of functional elements in genomic sequences (1). Comparisons between the human and mouse genomes have indicated that ~5% of mammalian genomes is conserved due to evolutionary constraints (2). Aside from being protein-coding (~1.5%), these conserved regions are likely to act as cis-regulatory elements, non-coding RNA genes and structural elements controlling biological processes such as gene transcription, translation, and chromosomal replication and condensation. However, due to the high complexity of the mammalian genome and gene regulation in mammals, many of these conserved non-coding elements remain unidentified, including cis DNA elements acting as transcription factor binding sites (TFBSs). Therefore, comparative genomics has emerged as a popular method for the discovery of these putative regulatory elements.
The binding of transcription factors (TFs) is important in tissue- and temporal-specific control of gene transcription. Because TFBSs are short and degenerate, their systematic discovery is a difficult problem. Of the approximately 2,000 TFs predicted in the human and mouse genomes 2., 3., known TFBS binding specificity models are only available for about 500 of them 4., 5.. It is estimated that only ~5,000 genomic TFBSs are known for less than 3,000 genes in vertebrates (6).
The binding specificities of TFBSs are often represented by a position weight matrix (PWM), a model based on the biophysical considerations of protein-DNA interactions (7). JASPAR is the most complete open-access TFBS matrix database with a total of 308 matrices up to the year 2006 (5). In contrast to other TFBS collections such as the TRANSFAC matrix database (4), the original JASPAR CORE collection (MA, 123 matrices) is a non-redundant dataset of high quality matrices supported by experimental evidence (8). Version 2 of JASPAR (5) introduced two more distinct collections: FAM (MF, 11 matrices) and phyloFACTS (PF, 174 matrices). MF is a collection of TFBS family models with TFBSs in each class sharing a similar protein structure in their TF DNA binding domains (9). The PF collection is a set of conserved and overrepresented regulatory motifs computationally derived from aligned mammalian promoter regions using a statistical approach (10).
Many existing functional element discovery approaches are based on sequence conservation and/or motif overrepresentation. For example, phylogenetic footprinting is a method for the discovery of regulatory elements through the identification of conserved motifs in a set of homologous regions (11). Several comparative genomics methods have been proposed for the identification of conserved features among orthologous sequences and co-regulated genes 12., 13., 14., 15., 16., 17.. Available software for the prediction of conserved TFBSs includes TFBS (18), MatInspector (19), ConSite (20), rVISTA (21) and Mulan/multiTF (22). Statistical methods have also been developed to detect conserved and overrepresented motifs within promoter regions (23). However, aside from the most prominently conserved TFBSs, there is a general lack of benchmarking of in silico predictions with experimental results. Particularly, a detailed quality control of in silico prediction of weakly conserved functional elements is currently lacking.
Phosphoenolpyruvate carboxykinase (PEPCK-C, EC 4.1.1.32) is a key enzyme in both hepatic and renal gluconeogenesis as well as in glyceroneogenesis in many mammalian tissues. PCK1 (RefSeq accession: NM_002591, GeneID: 5105) is a gene for the cytosolic isoform of PEPCK-C. The factors that control the transcription of PCK1 have been extensively studied 24., 25., 26., 27.. Transcription of PCK1 is induced by hormones such as glucagon (acting via cAMP), glucocorticoids and thyroxine, and is inhibited by insulin. In addition, nutrients such as glucose and fatty acids also modulate transcription of PCK1 in both the liver and the adipose tissue. Transcription of hepatic PCK1 is initiated at birth in coordination with the onset of gluconeogenesis in newborns. Finally, alterations in acid-base balance control the rate of transcription of PCK1 in the kidney cortex. Transcription of PCK1 has medical and economical significance, as PEPCK-C is the key enzyme in the control of hepatic glucose output and is thus a potential target for the regulation of blood glucose in human health and animal production.
Many of the regulatory elements have been identified in the rat PCK1 promoter 24., 26., 28.. The major TFBSs in the PCK1 promoter include a cAMP regulatory element (CRE) at −87 to −74 in the rat PCK1 promoter (critical for cAMP control of gene transcription, chr20: 55,569,486–55,569,499), an adjacent NF1 site at −123 to −87 (chr20: 55,569,449–55,569,486), an HNF-1 site at −200 to −164 (required for renal-specific gene transcription, chr20: 55,569,372–55,569,408), a C/EBPɑ binding site at −248 to −230 (required for liver-specific gene transcription and for full induction by cAMP, chr20: 55,569,326–55,569,344), and a glucocorticoid and insulin control region (GRU) at −456 to −400 (chr20: 55,569,124–55,569,192). There is also an important regulatory region at −1,000 in the rat PCK1 promoter. This region binds PPARγ2 and is involved in the tissue-specific expression of PCK1 in brown and white adipose tissue. TFs that bind to virtually all of these key sites in the PCK1 promoter have been identified. A recent review of our current understanding of the interactions of the various TFs and their potential control co-regulatory proteins (such as PGC-1ɑ and CBP) and co-repressors (histone deacetylases) can be found in the literature (28).
In this study, a systematic approach combining PWM from the JASPAR database and a phylogenetic footprinting algorithm TFLOC (Transcription Factor binding site LOCater) was optimized to detect weakly conserved TFBSs in mammalian gene promoters using an empirical matrix-specific threshold. The TFLOC program was originally developed for the UCSC Genome Browser (29) to identify conserved TFBSs within human-mouse-rat (HMR) alignments using the TRANSFAC matrix database. This approach originally used a Gaussian-based method to determine cutoffs to identify conserved binding sites. We further improved the predictive power of this approach by considering non-Gaussian distributions of matrices and by fine tuning the threshold of each PMW. The sensitivity and specificity of our in silico approach were assessed by comparing computational predictions with previously known binding sites in the PCK1 promoter. A newly discovered SP1 binding site was subjected to experimental verification via gel shift and reporter assays. Additionally, this study provides an easy access resource for researchers to develop new working hypotheses for transcriptional regulation studies. The full set of conserved TFBS predictions is freely available at http://bfgl.anri.barc.usda.gov/tfbsConsSites.
Results
Distribution of raw scores of JASPAR PWMs in mammalian promoter regions
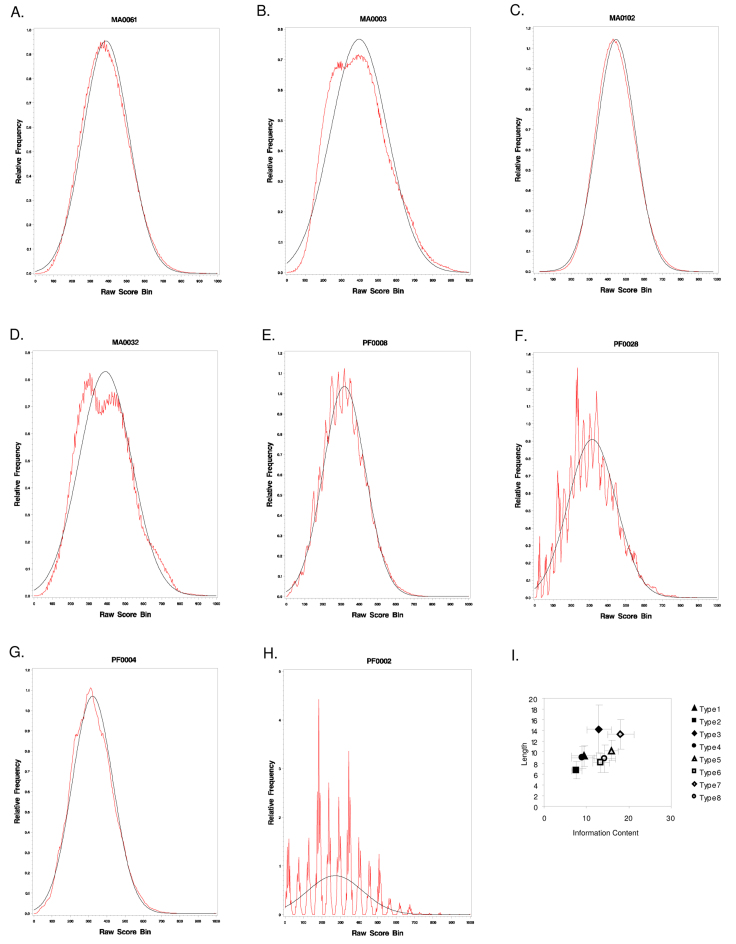
Many TFBS prediction programs depend on the assumption that matching scores follow a Gaussian distribution to determine their thresholds. Accordingly, we performed a standard normality test to determine whether the distribution of scores for each PWM follows a Gaussian distribution. We obtained raw scores for all JASPAR PWMs for every position in all available RefSeq promoter regions using TFLOC. TFLOC outputs a matrix similarity score that is scaled such that 1 represents a perfect match to the PWM and 0 represents the worst possible match. We chose the rat genome as the reference sequence and obtained distributions based on the scores of all substrings in all upstream sequences. These distributions were plotted as histograms using a bin size of 0.001 (Figure 1A–H and Figure S1). Three parameters were chosen to measure the fit of a histogram to a Gaussian distribution: (1) the shift of the mean from the expected center (0.5); (2) the deviation from a Gaussian distribution using the Kolmogorov-Smirnov distance (KS distance); and (3) the asymmetry of the distribution, as measured by the skewness. To group similar score distributions, we chose three thresholds, one for each parameter, based on manual examination: (1) mean + standard deviation ≥ 0.5; (2) KS distance ≤ 0.1; and (3) skewness ≤ 0.2. Representative examples of eight histogram types are shown in Figure 1A–H. All 308 histograms of JASPAR TFBS matrices were assigned into one of these eight histogram types. We also plotted the distributions of the motif length and information content for each histogram type (Figure 1I). The distribution of these 308 histograms for each JASPAR collection is listed in Table 1. Properties of each JASPAR matrix can be found in Table S1.
Fig. 1.
Raw score distribution family types and their relationship with the length and information content of matrices. The occurrences of raw scores within each bin of size 0.001 were recorded and plotted (red). The corresponding normal distribution was fitted to the observed values and superimposed on the histogram plots (black). A representative histogram is presented for each family type. Histograms of Types 1–4 (A, B, C and D) have means centered close to 0.5. Type 1 histograms have symmetric Gaussian bell-shaped curves; Type 2 histograms have asymmetric Gaussian bell-shaped curves; Type 3 histograms are symmetric but do not follow a Gaussian bell curve; and Type 4 histograms are asymmetric and do not follow a Gaussian bell curve. Types 5–8 (E, F, G and H) correspond to Types 1–4, respectively, except that Types 5–8 histograms are shifted to the left. I. Distributions of the motif length and information content for each histogram type.
Table 1.
Types of JASPAR PWM raw score histograms
| Type | Mean+SD | KS distance | Skewness | MA | MF | PF |
|---|---|---|---|---|---|---|
| 1 | ≥0.5 | ≤0.1 | ≤0.2 | 14 | 6 | 0 |
| 2 | ≥0.5 | ≤0.1 |
>0.2 |
28 |
4 |
0 |
| 3 | ≥0.5 | >0.1 | ≤0.2 | 22 | 0 | 0 |
| 4 | ≥0.5 | >0.1 | >0.2 | 18 | 0 | 0 |
| 5 | <0.5 | ≤0.1 | ≤0.2 | 1 | 0 | 3 |
| 6 | <0.5 | ≤0.1 |
>0.2 |
11 |
1 |
92 |
| 7 | <0.5 | >0.1 | ≤0.2 | 4 | 0 | 0 |
| 8 | <0.5 | >0.1 | >0.2 | 25 | 0 | 79 |
The relative frequencies of each type within the three JASPAR collections are dramatically different (Table 1). In the MA and MF collections, the majority of PWMs do not shift to the left (MA: 82/123 and MF: 10/11 in Types 1–4 histograms). Conversely, all PF matrices shift to the left (Types 5–8, 174/174). Additionally, over 98% of PF histograms (171/174) are negatively skewed (Types 6 and 8). Such differences may be related to the fact that many PF matrices have highly degenerate consensus sequences, because their frequency matrices are made of a large number of motifs extracted from mammalian genome alignments 5., 10..
Strikingly, about 48% (148/308) of the histograms are not bell-shaped (Types 3, 4, 6, 8; Figure 1). For example, 72 histograms (3 from MA collection and 69 from PF collection) had multiple frequent peaks (Figure 1H and Figure S1). This may be related to the conversion step of our raw score calculation.
In summary, only 20 out of 308 JASPAR matrices (6.5%) display a Gaussian distribution centered around 0.5 in upstream mammalian promoter regions. Other matrices have distributions that shift dramatically to the left, and/or do not follow a Gaussian distribution and/or are not symmetric. Therefore, these results demonstrate that the Gaussian distribution assumption required by many statistical tests is invalid for most of JASPAR PWMs.
Determination of PWM thresholds
Because the score distributions of most PWMs were non-Gaussian, an empirical test was performed to establish an optimal threshold for the prediction of conserved TFBSs in HMR alignments. Since many functional elements are evolutionarily constrained, it is expected that they would be enriched in the higher range of raw scores for conserved sites. For each species, a series of cutoffs (from the top 0.01% to 10% of all predictions) were set using all PWM raw scores across all available RefSeq genes. Because TFLOC only reports a conserved binding site meeting the threshold score in all three species, the lowest raw score among the three species was chosen for the final threshold. These individualized thresholds for each JASPAR matrix are given in Table S2 for each cutoff.
We assessed our predictions at each threshold by calculating their sensitivity and specificity with respect to 16 known binding sites in the PCK1 promoter (Figure 2, Table 2 and Table S3). We measured sensitivity as the percentage of known binding sites that overlapped our predictions by at least 50%. As the threshold increased from 0.01% to 0.03%, the sensitivity increased from 50.0% and eventually saturated at 75.0%. Most of the previously characterized binding sites were identified by our method at the correct position and orientation, including CREB1 (cAMP regulatory element binding protein, chr20: 55,569,486–55,569,499), which is important in the mediation of cAMP induction of PCK1 transcription, and C/EBP (CCATT/enhancer binding protein, chr20: 55,569,326–55,569,344), which is crucial for liver-specific gene expression, for the full induction of PCK1 by cAMP, and for the rapid increase in hepatic transcription of PCK1 at birth. Of the four previously characterized binding sites not identified by our method, CRE-2 (chr20: 55,569,417–55,569,437) is rodent-specific, and is thus expected to be discarded by our method; the other three missed sites include binding sites for GRE (chr20: 55,569,197–55,569,227), SREBP (chr20: 55,569,253–55,569,260) and NF1/CTF1 (chr20: 55,569,449–55,569,486) (Table S4). Although their individual overlaps were below 50%, NF1/CTF1 had two overlaps with MF and seven overlaps with PF predictions. A full coverage (100%) of the NF1/CTF1 binding site was achieved by merging these overlapping predictions. The GRE binding site had two overlapping predictions in the middle of core sequences covering 11 out of 47 bases. The SREBP binding site had the lowest coverage, but overlapped with known sites such as the RARE2 and ERRa regulatory elements, both of which were correctly predicted. Therefore, these three special cases were also identified as conserved regulatory elements by our approach.
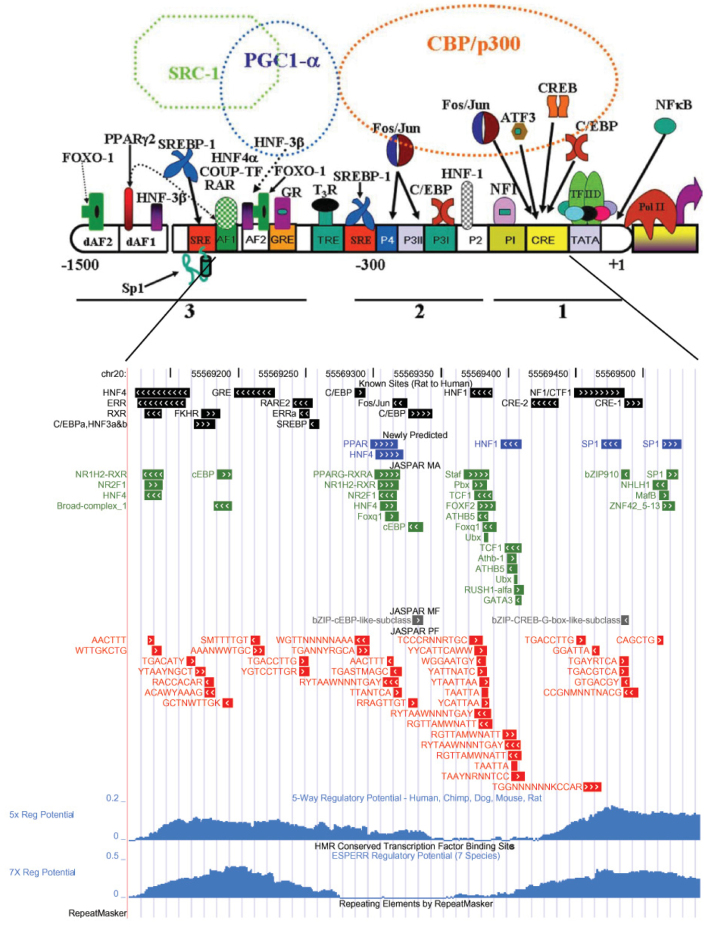
Fig. 2.
Known and predicted TFBSs as custom tracks on hg18 in the UCSC Genome Browser. Known and predicted TFBSs are represented as separate tracks in human genome assembly chr20: 55,569,120-55,569,542. Known TFBSs are in black and newly identified TFBSs are in blue. Predictions are organized into three tracks according to the JASPAR collection: MA (green), MF (gray), and PF (red). Displayed UCSC browser tracks include 5-way (5X) and 7-way (7X) regulatory potential, repeating elements by RepeatMasker, and Human/Mouse/Rat conserved TFBSs.
Table 2.
Prediction of TFBSs by TFLOC at various thresholds
| TFLOC | Threshold |
|||||
|---|---|---|---|---|---|---|
| 0.01% | 0.02% | 0.03% | 0.04% | 0.05% | 0.06% | |
| Known sites | 16 | 16 | 16 | 16 | 16 | 16 |
| Predicted known sites | 8 | 11 | 12 | 12 | 12 | 12 |
| Missed known sites | 8 | 5 | 4 | 4 | 4 | 4 |
| Total predicted sites | 27 | 52 | 78 | 98 | 111 | 128 |
| Overlapped predicted sites | 8 | 17 | 25 | 29 | 32 | 34 |
| Sensitivity | 50.0% | 68.8% | 75.0% | 75.0% | 75.0% | 75.0% |
| Specificity | 29.6% | 32.7% | 32.1% | 29.6% | 28.8% | 26.6% |
To estimate the false positive rate, specificity was defined as the percentage of predicted sites overlapping known TFBSs by at least 50%. As we increased the threshold, the specificity initially increased from 29.6% to 32.7% and then dropped steadily. At a threshold of 0.03%, our approach produced the best performance, with a sensitivity of 75% and a specificity of 32.1%. However, if the four special cases mentioned previously are considered, our sensitivity is 100%, with the rodent-specific site correctly excluded. Since a newly identified site was functionally verified in subsequent wet lab experiments (see functional verification section), these specificity and false positive rates might represent conservative estimates.
Using our optimal cutoff of 0.03%, we identified conserved TFBSs upstream of all available RefSeq genes (20,369 HMR alignments). The total number of putative TFBSs identified was consistent regardless of the reference species. Trivial differences were due to approximations made during the calculation of raw scores (see Methods). The rat genome was chosen as the reference sequence because: (1) the predictions were essentially identical among the three species; (2) rat coordinates made direct comparison with known sites straightforward; and (3) the PCK1 promoter is better characterized in rat than in other species. Our TFBS predictions for the PCK1 promoter are displayed in Figure 2 as tracks in the UCSC Genome Browser. In this figure, the top track depicts known rat TFBSs (black) mapped onto the human genome assembly using multiz alignments. The blue track represents newly discovered sites in this study. Our predictions are organized according to the JASPAR collections; that is, a separate track was created for each JASPAR collection (MA, green; MF, gray; PF, red). Seventy-eight matches were detected by our method in this region (MA 30, MF 2 and PF 46). In contrast, the default Human/Mouse/Rat conserved TFBS track in the Genome Browser has no matrix hits in this region at the default threshold, which is based on the Gaussian distribution assumption. As expected, our predictions also demonstrated high concordance with other related browser tracks, such as 5-way (5X) and 7-way (7X) regulatory potential (RP) 30., 31..
Comparison between MA and PF predictions
Several MA matrices have a similar corresponding matrix in the PF database. To determine the overall degree of similarity, an earlier study systematically compared all 174 PF matrices with all 123 MA matrices using the Pearson correlation coefficient (PCC), showing that 27% of MA matrices displayed strong correlation (PCC > 0.8) with a PF matrix (5). We examined our results to see if they display a similar overlap. In the PCK1 promoter, all MA predictions were covered by PF predictions with the exception of one MA prediction (SP1 at chr20: 55,569,517–55,569,526; Figure 2). Conversely, there were six regions covered by only PF predictions (Table 3), five of which (Regions 2–6) correspond to a known binding site. The distal Region 1 (chr20: 55,568,588–55,568,597) was the only region that did not overlap with any known site. Taken together, our results support the view that the PF collection serves as an extension to the MA collection, enhancing the coverage of the JASPAR database.
Table 3.
The six regions in the PCK1 promoter covered by only PF predictions
| Chr | Begin | End | Sequence | Strand | *1 | *2 | *3 | *4 | Score | JASPAR ID |
|---|---|---|---|---|---|---|---|---|---|---|
| Region 1 | ||||||||||
| chr20 | 55568588 | 55568594 | TGCCAAR | + | 43 | 49 | −905 | −899 | 955 | PF0047 |
| chr20 | 55568589 | 55568595 | GATTGGY | − | 44 | 52 | −904 | −896 | 876 | PF0031 |
| chr20 | 55568589 | 55568597 | GGGYGTGNY | − | 44 | 50 | −904 | −898 | 865 | PF0005 |
| Region 2 | ||||||||||
| chr20 | 55569162 | 55569168 | TGACATY | + | 545 | 551 | −403 | −397 | 909 | PF0042 |
| chr20 | 55569168 | 55569176 | YTAAYNGCT | + | 551 | 559 | −397 | −389 | 876 | PF0168 |
| Region 3 | ||||||||||
| chr20 | 55569209 | 55569216 | SMTTTTGT | − | 592 | 599 | −356 | −349 | 985 | PF0062 |
| chr20 | 55569211 | 55569219 | AAANWWTGC | + | 594 | 602 | −354 | −346 | 865 | PF0144 |
| Region 4 | ||||||||||
| chr20 | 55569245 | 55569252 | TGACCTTG | + | 645 | 652 | −303 | −296 | 880 | PF0038 |
| chr20 | 55569245 | 55569253 | YGTCCTTGR | + | 645 | 653 | −303 | −295 | 952 | PF0109 |
| Region 5 | ||||||||||
| chr20 | 55569286 | 55569297 | WGTTNNNNNAAA | − | 687 | 698 | −261 | −250 | 854 | PF0155 |
| chr20 | 55569288 | 55569297 | TGANNYRGCA | + | 689 | 698 | −259 | −250 | 854 | PF0067 |
| Region 6 | ||||||||||
| chr20 | 55569450 | 55569457 | TGACCTTG | + | 853 | 860 | −95 | −88 | 880 | PF0038 |
| chr20 | 55569456 | 55569469 | TGGNNNNNNKCCAR | + | 859 | 872 | −89 | −76 | 943 | PF0027 |
| chr20 | 55569462 | 55569467 | GGATTA | − | 865 | 870 | −83 | −78 | 843 | PF0093 |
Begin position in 1 kb rat promoter
End position in 1 kb rat promoter
Begin position relative to rat TSS
End position relative to rat TSS.
Newly discovered TFBSs
In addition to the concordance between known and predicted sites, seven novel sites were identified in the PCK1 promoter by our method (Figure 2). A promixal “SP1” site (chr20: 55,569,514–55,569,528) was supported by four predictions (Table S5), one of which was a properly oriented SP1 matrix. A second putative SP1 binding site (chr20: 55,569,469–55,569,484) has six overlapping predictions spanning both boundaries. A predicted HNF1 binding site (chr20: 55,569,394–55,569,410) is located adjacent to a well-characterized, upstream HNF1 site (chr20: 55,569,372–55,569,388), with an identical orientation. HNF1 is required for the renal-specific transcription of the PCK1 and is involved in the response of the gene promoter to changes in acid-base. This new HNF1 site is supported by eleven predictions. A putative HNF4 binding site (chr20: 55,569,302–55,569,322) and a PPAR binding site (chr20: 55,569,298–55,569,318) overlap with each other. Both are supported by five MA and four PF predictions. Other predictions include the distal PF-specific Region 1 (chr20: 55,568,588–55,568,597) and its upstream neighbor (chr20: 55,568,550–55,568,558), overlapping MYB.ph3 (Table S3).
Functional verification of a novel SP1 binding site
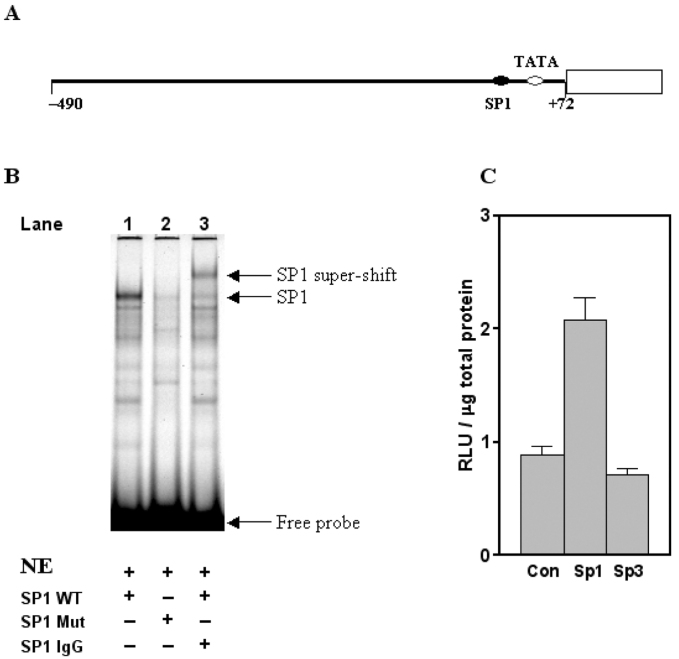
Since SP1 is ubiquitously expressed in many types of cells, including hepatocytes, we decided to test the proximal SP1 binding site (chr20: 55,569,514–55,569,528) to determine whether the predicted TFBS is functionally active (Figure 3A). Electrophoretic mobility shift assay (EMSA; that is, gel shift assay) showed that the endogenous SP1 interacted with the wild-type DNA fragment, which contains the putative SP1 binding site in the rat PCK1 promoter (Figure 3B, Lane 1). The binding of SP1 was abolished when a mutated DNA sequence was used in the assay (Figure 3B, Lane 2). The specificity of binding of SP1 to the wild-type DNA fragment was confirmed by an SP1-specific antibody in a super-shift assay (Figure 3B, Lane 3). To examine whether SP1 regulates the PCK1 promoter activity in vivo, a luciferase reporter p490-Luc (Figure 3A) was transiently transfected in HepG2 cells. When a plasmid over-expressing SP1 was co-transfected with p490-Luc, an increase in luciferase activity was detected (Figure 3C). As a control, co-transfection of SP3 with p490-Luc produced a marginal response. Based upon these results, we conclude that the predicted proximal SP1 binding site in the PCK1 promoter is functionally active under the tested conditions. However, more studies will be needed to fully decipher its biological role(s) in the regulation of the transcription of PCK1.
Fig. 3.
Experimental verification of newly identified proximal SP1 binding site in the PCK1 promoter. A. Schematic illustration of the location of the proximal SP1 binding site (chr20: 55,569,514–55,569,528) in the rat PCK1 promoter. The rat PCK1 promoter sequence (−490/+72) was linked to a luciferase gene to create a reporter construct, p490-Luc. B. Endogenous SP1 interacts with the proximal SP1 binding site. EMSA was performed using DNA fragments containing wild-type (SP1 WT) (Lane 1) or mutated (SP1 Mut) (Lane 2) binding site. The binding of SP1 to DNA fragment was confirmed via a super-shift with antibody against SP1 (SP1 IgG) (Lane 3). NE, nuclear extracts. C. Overexpression of SP proteins alters the PCK1 promoter activity. Control plasmid (Con) and plasmids over-expressing SP1 or SP3 were co-transfected with a luciferase reporter plasmid p409-Luc into HepG2 cells. The results are expressed as the means of relative luciferase activity ± S.E.M. for three experiments.
Discussion
The identification of cis-regulatory elements and the characterization of their interactions with respective TFs provide insight into tissue- and temporal-specific transcription of genes. The goal of this study is to provide a useful resource for less conserved but potentially functional TFBSs. We implemented a systematic approach to identify potential TFBSs genome-wide by searching for PWMs using a phylogenetic footprinting method (TFLOC). Our approach to the functional analysis of gene promoters features adjustable thresholds, expandable user-defined TFBS matrices, and the capability of whole genome analysis with minor changes. Computational predictions were evaluated against known TFBSs of the PCK1 promoter, and a newly identified SP1 site was verified for TF binding and transcriptional regulation activity. TFBSs in the rat PCK1 promoter were identified with acceptable sensitivity (75%) and specificity (about 32%) using a rigorous criterion.
The data we obtained were derived computationally, but were based on reasonable justifications and some experimental verification, therefore it is conceivable that a significant portion of our predictions will be biologically functional. Our approach thus provides an accessible resource for the comparative analysis of mammalian transcriptional regulation. Using the results of our genomic analysis, investigators who are interested in the regulation of a particular gene can prioritize their working hypotheses in subsequent experiments and discover new regulatory interactions. Additionally, researchers interested in identifying the targets of specific TFs or TF families can access a computational catalog of putative targets. Our predictions also provide the capability to probe for interactions between neighboring TFs. All of these features should be a valuable aid in the understanding of regulatory elements that are functionally conserved among mammalian species.
For this study, we restricted our analysis to the 1,000-base upstream regions of RefSeq genes, resulting in roughly 20.4 Mb of sequences. We chose to only search in these regions because 1,000-base upstream gene promoter regions are known to be strongly enriched for TFBSs. However, mammalian transcription start sites (TSSs) and surrounding regulatory elements are often poorly defined. We noted several TSS discrepancies between species. For example, many characterized rat TFBSs 32., 33. are located between the upstream mapped human TSS and the downstream rat endogenous TSS in the promoter region of the prolactin gene (NM_000948). Because multiz alignments use human sequence as a reference and stop at the human TSS, it was impossible to recover such sites in our final dataset. It is not known whether this discrepancy in TSS is due to the methods used to generate the alignments or a true difference between the species. Recent progress in the determination of mammalian TSS 34., 35. will provide a platform to further clarify such discrepancies in the future.
The JASPAR PF collection with lower information content and shorter length was found to be associated with histograms of Types 6 and 8. Due to the degenerate bases and short lengths of such PWMs, there is an increased likelihood of conserved matches to such PWMs to occur by chance. This highlights the importance of establishing proper thresholds for each matrix to filter such false positives. Compared with our results, the Human/Mouse/Rat conserved binding site track on the UCSC Genome Browser predicts far fewer sites. We believe that this might be attributed to the facts that (1) it assumes a Gaussian distribution, which is not true for most JASPAR matrices, and (2) it uses the best score obtained upstream of each RefSeq gene to compute its statistics (instead of all scores), which may lead to overstringent thresholds.
Many public websites help researchers to define and extract conserved sequences from multi-species alignments, such as Galaxy (36) and MCS Browser (37). In addition to providing alignments, the UCSC Genome Browser also provides predictive measures of regulatory regions such as 5X and 7X RP 30., 31. and PhastCons scores (16). We detected a high correlation of our predictions with 5X and 7X RP. These results were not surprising given that our HMR alignments were derived from alignments similar to those used by these tracks. However, by incorporating TFBS PWMs, our dataset offers an additional layer of information about trans-acting TFs.
Programs such as TFBS (18), MatInspector (19) and MATCH (38) search for motif patterns using PWMs within a single sequence. Conserved TFBS prediction programs such as ConSite (20) and rVISTA (21) can only search for conserved TFBSs within pairwise alignments. Mulan/multiTF (22) provides the most similar function to TFLOC for the detection of conserved TFBSs in multiple alignments. The “optimized for function” search option of multiTF is similar to our threshold strategies for limiting the density of TFBS matches in multiple alignments. Using the default settings (http://multitf.dcode.org/), Mulan/multiTF prediction was performed in the PCK1 promoter. While their specificities (ranging from 13.0% to 41.4%) were comparable to our approach, their sensitivities (ranging from 25.0% to 37.5%) were significantly lower (Table 4). Differences between the two approaches include: (1) different PWM databases (TRANSFAC vs. JASPAR); (2) the use of 1,000-base upstream promoter regions in this study; (3) the focus of this study on less conserved TFBSs; and (4) the use of independently adjusted thresholds in this study. The expected density of multiTF predictions in a random sequence is 3 or fewer sites per 10 kb for each PWM. However, our method results in an expected density of approximately 40 sites per 1,000-base upstream region, which may be a more reasonable number for TFBS-enriched regions such as 1,000-base upstream promoters.
Table 4.
Prediction of TFBSs by multiTF with default settings
| MultiTF | Optimized for function | Optimized for function with high-specificity matrices | Predefined (0.85) | Predefined (0.85) with high-specificity matrices |
|---|---|---|---|---|
| Known sites | 16 | 16 | 16 | 16 |
| Predicted known sites | 5 | 4 | 6 | 5 |
| Missed known sites | 11 | 12 | 10 | 11 |
| Total predicted sites | 68 | 58 | 161 | 76 |
| Overlapped predicted sites | 25 | 24 | 21 | 14 |
| Sensitivity | 31.3% | 25.0% | 37.5% | 31.3% |
| Specificity | 36.8% | 41.4% | 13.0% | 18.4% |
It is important to keep in mind that the thresholds for our approach were only chosen based on the available known sites for the PCK1 promoter. Aside from PCK1, several other promoters, such as the promoters for the actin, casein and insulin genes, are currently being investigated in detail. It is possible that some of our TFBS predictions are false positives due to the fact that a universal threshold for a PWM may not be practical for whole genome analysis. Instead, it might be necessary to fine tune thresholds not only for each PWM, but even for each individual gene. Furthermore, predictions must be verified experimentally, which raises particular challenges for binding sites in a gene promoter that are only functional in certain cells, tissues or developmental stages. We believe that the development of better computational predictions will provide better candidates for further experimental verification. An integration of experimental and computational studies has the potential to greatly advance research on the control of gene transcription.
One setback to the PWM model is that it does not take insertions or deletions (indels) into consideration. Some TFs are flexible in their DNA interactions and can tolerate binding sites of different lengths. In the future, we hope to incorporate indels into our methodologies to better reflect the possibility of such interactions. Additionally, neighboring TFBSs are often clustered into closely located groups to allow for protein interactions between the TFs that bind them. Recently, several methods have been proposed 39., 40. to analyze composite regulatory elements, that is, modules of multiple binding sites. Integration of these approaches should further uncover the interactions of neighboring TFs.
Comparative genomics methods provide tools for studying the potential function of gene promoters that have not been well characterized. Such approaches have the potential to greatly simplify the functional analysis of gene promoters by predicting both the presence and arrangement of specific TFBSs. In this work, we chose to identify conserved TFBSs through a phylogenetic footprinting method using HMR alignments. The genes for both PCK1 and its isoform, PCK2 (the mitochondrial form of the enzyme), are present in every eukaryotic species for which the gene sequence is available. Despite this wide species distribution, its transcriptional control has been primarily studied in rat under the assumption that the pattern of regulation noted in one species is applicable to others. This generalization has more than academic significance, as drugs are being developed for human diseases. When viewed from this perspective, a systematic analysis of the pattern of TFBSs in a broad variety of mammalian species should reveal conserved regulatory elements and suggest candidate regions upon which to focus efforts to control the rate of transcription of a specific gene. It is also likely that additional regulatory elements will be identified, which were not apparent from previous analyses of promoter function. Finally, any analysis of transcriptional regulation of a gene promoter that has not been previously studied would most likely benefit from an initial screening of putative regulatory elements using approaches such as the one outlined in this work. For example, our method was able to successfully recover almost all known elements in the PCK1 promoter.
The phylogentic shadowing method (41) uses more closely related species such as human, chimpanzee and rhesus monkey, or cattle, pig and sheep to identify lineage-specific TFBSs. Applying our approach to alignments generated from additional more closely related species should provide additional insights. Differences in the organization of regulatory elements within a subset of species could indicate physiologically critical differences in the responsiveness of a gene to environmentally induced regulatory signals such as diet and hormones.
Conclusion
In this study, we presented a systematic analysis of conserved TFBSs within the upstream regions of all available RefSeq genes using HMR alignments. Using the PCK1 promoter as an example, our analysis produced a reasonable sensitivity of 75% and a specificity of 32%. In addition to recovering known sites, we also predicted novel candidates, one of which was confirmed by functional assays. It is our hope that the conserved TFBS dataset (available at http://bfgl.anri.barc.usda.gov/tfbsConsSites) will provide a useful resource for the development of further transcriptional regulation hypotheses.
Materials and Methods
Resources and tools
Multiz alignment files of 1,000-base upstream promoter regions of RefSeq genes for hg18 (build 36) (42) were downloaded from the UCSC Genome Browser (29) at http://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz17way/. These files contain multiple alignments of the following 16 vertebrate genomes to the human genome (hg18, Mar. 2006): chimpanzee (panTro1, Nov. 2003), macaque (rheMac2, Jan. 2006), mouse (mm8, Feb. 2006), rat (rn4, Nov. 2004), rabbit (oryCun1, May 2005), cow (bosTau2, Mar. 2005), dog (canFam2, May 2005), armadillo (dasNov1, May 2005), elephant (loxAfr1, May 2005), tenrec (echTel1, Jul. 2005), opossum (monDom4, Jun. 2006), chicken (galGal2, Feb. 2004), frog (xenTro1, Oct. 2004), zebrafish (danRer3, May 2005), tetraodon (tetNig1, Feb. 2004), and fugu (fr1, Aug. 2002). The multiz and TBA alignment programs were downloaded from the Miller Lab at Pennsylvania State University (http://www.bx.psu.edu/miller_lab/). The maf_project.pl script was used to extract 3-way alignments (human, mouse and rat) from the 17-way alignments using each species as the reference.
A total of 23,688 17-way alignments were downloaded from the UCSC Genome Browser. An initial filter removed 2,817 alignments in which mouse, rat or both did not have corresponding orthologous sequences. Another filter removed 502 genes with multiple promoter alignments, probably due to alternative splicing or gene family isoforms. The final dataset included a total of 20,369 HMR alignments, containing 20.4 Mb of human sequences, 13.7 Mb of mouse sequences and 13.4 Mb of rat sequences. Throughout this report, chromosome positions in human genome assembly hg18 (build 36) were used unless otherwise noted.
Known cis-regulatory elements were either annotated manually or with the help of MatInspector (19) running on a single sequence within alignments. Version 2 of the JASPAR TFBS matrix database was downloaded from its website (http://mordor.cgb.ki.se/jaspar2005/download/). The JASPAR PWMs were transformed into TRANSFAC format for use in TFLOC. The consensus sequences of matrices were deduced from frequency matrices. Separate sets of matrices were created for each JASPAR collection (CORE: MA; FAM: MF; phyloFACTS: PF).
Identification of matrix hits with TFLOC
We used a modified version of TFLOC, a program developed to identify conserved TFBSs in Human/Mouse/Rat alignments for display in the UCSC Genome Browser (29). Briefly, TFLOC identifies matches to a PWM of length n that are conserved across ns sequences. Denote the multispecies alignment s, such that sji is the nucleotide at position i of species j. Meanwhile, define an ns×4 background matrix (back) giving the background frequencies of each nucleotide (A, C, G, T) in each species. A sliding window (of length n) calculated the “species score” for each species at each position:
| (1) |
A log-odds score (log_scorei) was calculated for each species normalized by the length of the matrix n and the number of species ns in the alignment:
| (2) |
These scores were then summed for all species, yielding a final log-odds score for the window starting at position i. The log-odds score of each species must exceed the threshold (thresi) for the current position to be considered a match (see Results and Table S2):
| (3) |
Next, the maximum and minimum possible log-odds scores were computed and summed across all species for the given PWM:
| (4) |
| (5) |
These scores were then used to normalize the final, raw log-odds score so that its range was between 0 and 1:
| (6) |
We determined the best threshold for each PWM using all raw scores within the 1,000-base upstream regions of all available RefSeq genes (taken from the RefGene table for hg18). The distribution histograms (ranging from 0 to 1, with bin size 0.001) of raw scores were then created (Figure S1). Using the PCK1 promoter, individualized thresholds for each PWM were determined as the top 0.03% to maximize predictive power (see Results and Discussion and Table 2). TFLOC was then run with the individualized threshold for each PWM as the threshold for the 3-species multiz alignments.
After all PWM hits were recorded, one additional merging/filtering step was performed. In the event that multiple sites bind the same factors, only those sites overlapping each other greater than 80% were merged by keeping the site with the highest raw score. Upon determining the overlap relationship between known and predicted sites, a positive call (Table 2) was made only when the mutual overlap coverage was greater than 50%.
Statistical tests
The properties of the raw score distributions, including mean, standard deviation, KS distance, skewness and kurtosis, were determined using SAS Proc Univariate program. All histograms of raw scores were also manually inspected. Distribution curves with means more than one standard deviation to the left of 0.5 were denoted as “shifted to the left”. Deviations from the Gaussian distribution were assessed using KS distance with a cutoff of 0.1. Distribution asymmetry was measured by skewness. A distribution with an asymmetric tail extending to the left was referred to as “skewed to the left” or “negatively skewed”, while a distribution with an asymmetric tail extending to the right was referred to as “skewed to the right” or “positively skewed”. A cutoff of 0.2 was used for skewness. Based on these three properties, all 308 histograms of JASPAR TFBS matrices were assigned into one of eight histogram categories (Figures 1 and S1, Tables 1 and S1).
Cell culture, transfections and luciferase assay
HepG2 hepatoma cells were cultured at 37°C in an atmosphere of 95% air and 5% CO2. The medium was a half-half mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F-12, supplemented with 5% fetal bovine serum, 5% calf bovine serum, 50 units/mL of penicillin and 50 μg/mL of streptomycin (Invitrogen). Wild-type (WT) cells, that is, mouse primary hepatocytes transformed with a temperature-sensitive SV40 large T antigen, were propagated at 33°C and assayed at 37°C in ɑ-minimal essential medium (AMEM), supplemented with 4% fetal calf serum, 2 mM glutamine, 22 nM dexamethasone, 50 units/mL of penicillin and 50 μg/mL of streptomycin (Invitrogen). Transient transfection was performed in triplicates using 24-well plates as described previously (43). Briefly, a quarter million of HepG2 cells were cultured in 1 well of 24-well plates for 24 h. Then 0.05 mg plasmids that over-express SP1 or SP3 were co-transfected with 0.2 μg PCK1 luciferase reporter plasmid p490-Luc to cells, using FuGENE6 (Roche) according to the manufacture’s protocol. At the end of 24-h transfection, cells were washed once with ice-cold 1X phosphate-buffered saline (PBS). Proteins were extracted using cell culture lysis reagent (Promega) and luciferase activity was measured using a luminometer (Molecular Devices). The protein content of the extracts was determined using a Bio-Rad protein assay kit (Bio-Rad Laboratories). Final data were expressed as relative luciferase units per μg of extract protein.
Nuclear extracts and fluorescent EMSA
Nuclear extracts were isolated according to a protocol described previously (43) with minor modifications. Essentially, WT cells were collected from a 150-mm plate using a plastic scraper and washed once with ice-cold 1X PBS. Cell pellets were then resuspended in 1,200 μL of ice-cold buffer A. After 10 min of incubation on ice, the cells were lysed via adding Nonidet P-40 to a final concentration of 0.25%, followed by 10 pulses of mixing using a vortex. The solution was centrifuged at 5,000 rpm for 1 min to obtain nuclei (pellet). This pellet was then resuspended in 100 μL of ice-cold buffer B and incubated in ice for 5 min. Buffer C was added dropwise (about 50 μL) to achieve a final concentration of 0.3 M KCl. The mixture was placed on ice for 30 min with occasional gentle shaking and then centrifuged at 12,000 rpm for 15 min at 4°C to obtain the nuclear extract (supernatant). This extract was then dialyzed against 100 volumes of buffer D for 2 h at 4°C, using Slide-A-Lyzer Dialysis Cassette (Pierce). The nuclear extract was quantified and stored at −70°C. Fluorescent EMSA was performed with a modified protocol of Yang et al. (43). DNA fragments were generated via annealing two complementary oligouncleotides, one of which was labeled with 6-FAM at the 5′-end (IDT Inc.). The labeled oligonucleotides were 5′-/6-FAM/-TCCAGCTGAGGGGCAGGGCTGTCCTCC-3′ for the wild-type sequence of SP1 binding site in rat PCK1 promoter (−61/−47), and 5′-/6-FAM/-TCCAGCTGAttttCAtttCTGTCCTCC-3′ for its mutant sequence (small letters are mutated nucleotides). The EMSA reaction was carried out and the product was analyzed as described previously (43). After electrophoresis, the gel was scanned using a Typhoon 9200 PhosphorImager® scanner (GE Healthcare). The image was analyzed using ImageQuant® software (GE Healthcare).
Authors’ contributions
GEL, MTW, RWH and JY conceived and designed the experiments. MTW wrote TFLOC. JY performed gel shift and reporter assays. GEL and JY performed the predictions. GEL, CPVT, RWL, TSS, LKM and EEC analyzed the data. GEL, MTW, RWH and JY wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank M. Hou, O. Couronne, and L.C. Gasbarre for helpful comments in the preparation of this manuscript, and also thank G. Wei and T. Brown for technical support. This work was supported in part by CRIS Project (No. 1265-31000-090-00D and 1265-31000-081-00D) from US Department of Agriculture and by NIH Grant DK-25541 (to RWH). JY was supported by the NIH Metabolism Training Program (DK-07139). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by US Department of Agriculture.
Contributor Information
George E. Liu, Email: george.liu@ars.usda.gov.
Jianqi Yang, Email: jianqi.yang@case.edu.
Supporting Online Materials
Figure S1 and Tables S1-S5
References
- 1.Collins F.S. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R.H. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Matys V. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlieghe D. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GuhaThakurta D. Cis-regulatory variations: a study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics. 2006;7:235. doi: 10.1186/1471-2164-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 8.Sandelin A. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandelin A., Wasserman W.W. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 10.Xie X. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer T. Evaluating phylogenetic footprinting for human-rodent comparisons. Bioinformatics. 2006;22:430–437. doi: 10.1093/bioinformatics/bti819. [DOI] [PubMed] [Google Scholar]
- 12.Wasserman W.W. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 13.Cooper G.M. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulyk M.L. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X. Reliable prediction of transcription factor binding sites by phylogenetic verification. Proc. Natl. Acad. Sci. USA. 2005;102:16945–16950. doi: 10.1073/pnas.0504201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siepel A. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanchette M., Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenhard B., Wasserman W.W. TFBS: computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 19.Cartharius K. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 20.Sandelin A. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loots G.G., Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ovcharenko I. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tompa M. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 24.Hanson R.W., Reshef L. Glyceroneogenesis revisited. Biochimie. 2003;85:1199–1205. doi: 10.1016/j.biochi.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Hanson R.W. Metabolism in the era of molecular biology. J. Biol. Chem. 2005;280:1705–1715. doi: 10.1074/jbc.X400009200. [DOI] [PubMed] [Google Scholar]
- 26.Pilkis S.J., Granner D.K. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu. Rev. Physiol. 1992;54:885–909. doi: 10.1146/annurev.ph.54.030192.004321. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland C. New connections in the regulation of PEPCK gene expression by insulin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996;351:191–199. doi: 10.1098/rstb.1996.0016. [DOI] [PubMed] [Google Scholar]
- 28.Chakravarty K. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit. Rev. Biochem. Mol. Biol. 2005;40:129–154. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 29.Hinrichs A.S. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor J. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolbe D. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leclerc G.M., Boockfor F.R. Pulses of prolactin promoter activity depend on a noncanonical E-box that can bind the circadian proteins CLOCK and BMAL1. Endocrinology. 2005;146:2782–2790. doi: 10.1210/en.2005-0100. [DOI] [PubMed] [Google Scholar]
- 33.Jacob K.K., Stanley F.M. Elk-1, C/EBPalpha, and Pit-1 confer an insulin-responsive phenotype on prolactin promoter expression in Chinese hamster ovary cells and define the factors required for insulin-increased transcription. J. Biol. Chem. 2001;276:24931–24936. doi: 10.1074/jbc.M102826200. [DOI] [PubMed] [Google Scholar]
- 34.Carninci P. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 35.Carninci P. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 36.Giardine B. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margulies E.H. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kel A.E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferretti V. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–D126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanchette M. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boffelli D. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 42.Pruitt K.D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J. Sodium butyrate induces transcription from the G alpha(i2) gene promoter through multiple Sp1 sites in the promoter and by activating the MEK-ERK signal transduction pathway. J. Biol. Chem. 2001;276:25742–25752. doi: 10.1074/jbc.M102821200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 and Tables S1-S5