Abstract
It is well established that the vast majority of proteins of all taxonomical groups and species are initiated by an AUG codon, translated into the amino acid methionine (Met). Many attempts were made to evaluate the importance of the sequences surrounding the initiation codon, mostly focusing on the RNA sequence. However, the role and importance of the amino acids following the initiating Met residue were rarely investigated, mostly in bacteria and fungi. Herein, we computationally examined the protein sequences of all major taxonomical groups represented in the Swiss-Prot database, and evaluated the preference of each group to specific amino acids at the positions directly following the initial Met. The results indicate that there is a species-specific preference for the second amino acid of the majority of protein sequences. Interestingly, the preference for a certain amino acid at the second position changes throughout evolution from lysine in prokaryotes, through serine in lower eukaryotes, to alanine in higher plants and animals.
Key words: amino acid, initial methionine, translation ribosome, computational biology, bioinformatics
Introduction
In recent years it has become clear that the initiation of translation is affected by many factors 1, 2, 3. These factors might be embedded in the mRNA sequence surrounding the initiation codon [usually AUG, coding for methionine (Met)], especially upstream 4, 5, or following the initiating codon 6, 7 and even further downstream 8, 9. Many of these studies have shown that the sequence directly following the initiation codon might be an important factor in both initiation of translation 10, 11 and efficiency of translation in prokaryotes 12, 13, 14, 15, 16 and eukaryotes 17, 18, 19. Although directly affecting the protein sequence and function, as well as its translation rate, the encoded amino acids directly following the most abundant Met residue were rarely studied. Yet, it was established that the amino acid sequence as well as the related coding RNA sequence and codon usage are important for control of translation and stability of proteins (20). More specifically, the amino acid following the initial Met is important in determining the rates of methionine aminopeptidase (MAP) function in proteins where the initial Met is cleaved 21, 22. Some researchers even attribute the functional features of the amino acid following the Met to the size of the side-chain (23). It was also found that there is a direct relation between the metabolic stability of a protein and the identity of its N-terminal residue, mediated by different degradation mechanisms (24).
Analysis of the sequence surrounding the initial AUG codon in different organisms revealed a surprising diversity. In Escherichia coli, the most common second amino acid following the Met (AUG) is lysine (Lys), mostly with its AAA codon (25). This specific codon is also the most expression promoting. In contradiction, the presence of XGG codons in several positions in the open reading frame sequence, including the second position, considerably reduces the level of expression (26). In the yeast Saccharomyces cerevisiae, the most abundant codon following the AUG was found to be UC U/C 8, 27, giving rise to serine (Ser) as the second amino acid. However, a recent publication (28) indicates that when examining highly expressed proteins in a set of bacteria (9), archeae and unicellular eukaryotes (3), the most abundant amino acid following the initial Met was alanine (Ala). This finding is further supported by a study in which replacement of the three downstream codons following the initial AUG of tobacco (bases +4 to +12) with the sequence GCU UCC UCC (coding for Ala, Ser, Ser) increased the expression level of a reporter gene up to 40 fold (19).
The importance and potential functionality of the amino acids directly following the initial Met, as well as the apparent difference between species, raises the question of the evolutionary conservation of the second amino acid and the mechanisms by which the control of translation and protein expression co-evolved with other elements sharing the same system or mechanism. It is therefore interesting to look at the abundance of the second amino acid throughout evolution and try to make sense of the probable mechanisms by which this feature controls the expression of proteins. In this work, we attempted to examine the abundance of the second amino acid in a large dataset of known/predicted proteins from prokaryotes and lower eukaryotes, such as bacteria and yeast, throughout different evolutionary steps, up to higher eukaryotes, such as mammals and higher plants. By doing this we wish to explore the importance of the second amino acid in determining protein characteristics like expression, stability and function, as well as establish a tool for functional and evolutionary predictions.
Results
Distribution of second amino acid changes between different taxa
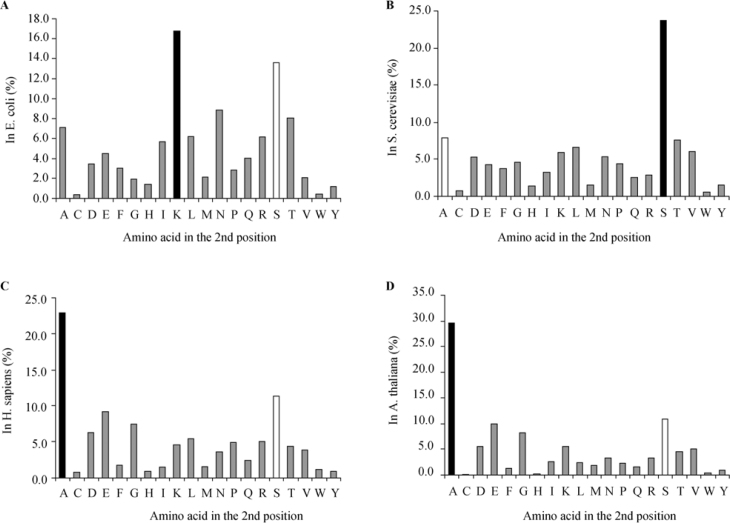
We have conducted a survey on all available Swiss-Prot protein sequences (see Materials and Methods). The results presented in Table S1 show vast differences between phylogenic taxa. Figure 1 shows the differences in amino acid distribution at the second position through three major groups. In bacteria, represented by E. coli (Figure 1A), the most abundant amino acid in the second position is Lys (16.7%), followed by Ser (13.6%). In unicellular eukaryotes, represented by S. cerevisiae (Figure 1B), the most common second amino acid is Ser (23.7%), with Ala in second place (7.9%). However, in mammals, represented by Homo sapiens (Figure 1C), the order is reversed as the most common second amino acid is Ala (22.9%) followed by Ser (11.4%); the effect is even more apparent in plants, represented by Arabidopsis thaliana (Figure 1D), in which the most common second amino acid is Ala (29.6%) followed by Ser (10.8%). The results presented clearly indicate that there are considerable differences in the distribution and selection of the amino acid at the second position of protein sequences. These differences might be related to evolutionary and developmental stages, or might indicate different translation-related mechanisms.
Figure 1.
Prevalence of amino acids in the second positions of proteins initiated by a Met residue in selected species. Panels A-D illustrate the distribution of second position amino acids for selected representatives of each of the evolutionary groups. A. Bacteria, represented by E. coli (n=4,474). B. Unicellular eukaryotes, represented by S. cerevisiae (n=4,819). C. Animals, represented by H. sapiens (n=10,790). D. Plants, represented by A. thaliana (n=3,056). Representative groups were selected based on the largest amount of protein sequences present in the Swiss-Prot database. Black columns are the most abundant second position amino acids. White columns are the next most abundant amino acids.
Distribution of the second amino acid gradually changes throughout evolution
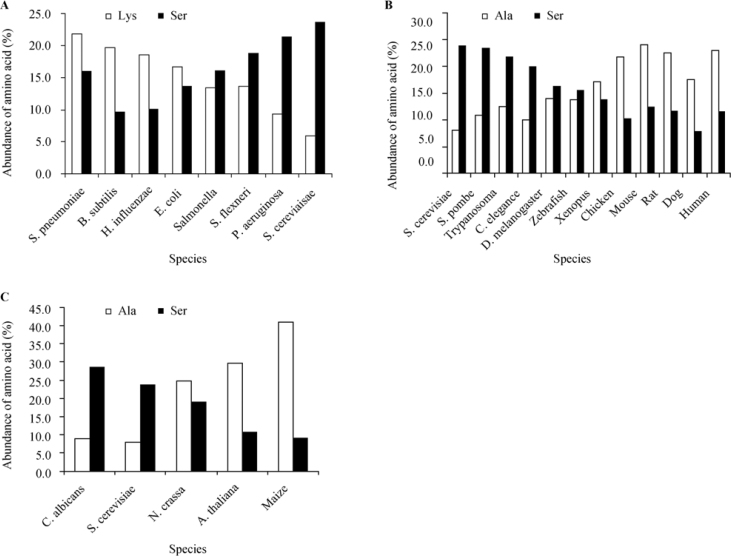
Further analysis of the data presented in Table S1 indicates that the pattern is conserved in related species. For example, some bacteria species (Bacillus subtilis, Haemophilus influenzae or Streptococcus pneumoniae) keep the same preference for the amino acid at the second position. However, a closer examination reveals that there are other bacteria species (Salmonella sp., Shigella flexneri and Pseudomonas aeruginosa) that show a different pattern in which the most preferred amino acid in the second position is Ser (16.1%, 18.8% and 21.3%, respectively) with Lys (or Ala in P. aeruginosa) only at the second place. This deviation from the common bacterial pattern shows a tendency for evolutionary development (illustrated in Figure 2A) from prokaryotes to simple eukaryotes.
Figure 2.
Evolutionary development associated with changes of preference for amino acids on the second position. The most and the next most abundant second position amino acids on the second position for all species were analyzed. The different species examined were divided into groups representing different evolutionary processes. A. The development from prokaryotes, represented by a few bacteria species (ordered according to the predicted phylogenetic tree based on 16S rRNA), to one unicellular eukaryote represented by yeast. B. The development from unicellular eukaryotes represented by yeasts and trypanosome to higher animals. C. The development from lower eukaryotes represented by yeasts and fungi to higher plants.
When examining the preference of the amino acid at the second position in other lower eukaryotes (unicellular eukaryotes such as Schyzosaccharomyces pombe, Candida albicans and Trypanosoma sp.), the same pattern as in S. cerevisiae was revealed (Figure 2B). However, when examining higher multicellular eukaryotes (such as Caenorhabditis elegans, Drosophila melanogaster and Danio rerio), one can figure out that although Ser is the most preferred amino acid at the second position of proteins, followed by Ala, the difference in frequency between Ser and Ala is gradually reduced while moving up the evolutionary tree (starting with Xenopus through chicken, mouse, rat, dog and human). It seems that Ala takes over as the preferred second position choice, and Ser is pushed to second place with an increasing difference in frequency values (Figure 2B).
When taking a different evolutionary path from the lower eukaryotes (unicellular eukaryotes) to multicellular fungi (Neurospora crassa) and plants (Arabidopsis and maize), one can illustrate the same pattern, to some extent in a more pronounced way, in which the most preferred amino acid at the second position changes from Ser in the lower organisms and fungi to Ala in higher plants (Figure 2C).
The distribution of the amino acid at the second position is specific to this position
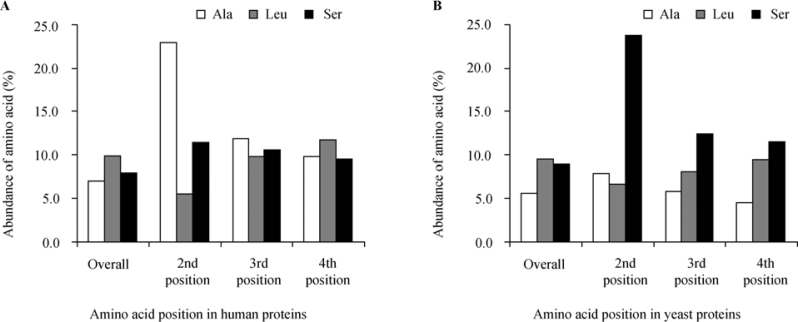
In order to rule out the possibility that the results presented above were just coincidental (i.e., based on an artifact involving biased positions within the protein sequences), we have tested both the abundance of each amino acid at the second position and its overall abundance in the whole examined proteome of a selected taxon. The results presented in Table 1 and Figure 3 clearly indicate that both for human and yeast, there is little correlation. It is clear that the most abundant amino acid in both species proteomes is leucine (Leu), whereas Ala and Ser, which are the most frequently represented at the second position in human and yeast, respectively, are not over represented on the whole proteomes.
Table 1.
Analysis of amino acid representation in the analyzed protein sequences of human and yeast
| Human |
Yeast |
|||||||
|---|---|---|---|---|---|---|---|---|
| Amino acid | Overall (%) | Second position (%) | Third position (%) | Fourth position (%) | Overall (%) | Second position (%) | Third position (%) | Fourth position (%) |
| A | 7.02 | 22.9 | 11.9 | 9.8 | 5.60 | 7.93 | 5.8 | 4.5 |
| C | 2.33 | 0.8 | 1.8 | 1.9 | 1.29 | 0.75 | 0.9 | 0.9 |
| D | 4.84 | 6.2 | 4.3 | 3.0 | 5.83 | 5.25 | 5.1 | 5.0 |
| E | 6.95 | 9.1 | 6.1 | 6.1 | 6.55 | 4.30 | 6.1 | 6.5 |
| F | 3.76 | 1.8 | 2.9 | 3.4 | 4.49 | 3.71 | 4.8 | 5.0 |
| G | 6.72 | 7.5 | 7.0 | 6.6 | 5.06 | 4.59 | 4.4 | 3.0 |
| H | 2.57 | 0.9 | 1.9 | 1.7 | 2.13 | 1.41 | 2.0 | 1.8 |
| I | 4.49 | 1.5 | 2.2 | 3.0 | 6.54 | 3.24 | 5.8 | 6.0 |
| K | 5.67 | 4.6 | 4.0 | 4.7 | 7.30 | 5.93 | 8.3 | 7.4 |
| L | 9.86 | 5.5 | 9.8 | 11.7 | 9.53 | 6.62 | 8.1 | 9.4 |
| M | 2.19 | 1.6 | 1.7 | 2.1 | 2.08 | 1.58 | 1.7 | 1.3 |
| N | 3.69 | 3.6 | 3.0 | 3.1 | 6.08 | 5.42 | 5.5 | 6.3 |
| P | 6.14 | 4.9 | 8.1 | 8.1 | 4.33 | 4.42 | 3.5 | 4.8 |
| Q | 4.69 | 2.4 | 3.5 | 4.3 | 3.91 | 2.55 | 4.2 | 4.6 |
| R | 5.55 | 5.0 | 7.1 | 7.1 | 4.43 | 2.88 | 6.0 | 5.8 |
| S | 8.03 | 11.4 | 10.5 | 9.5 | 8.93 | 23.74 | 12.5 | 11.5 |
| T | 5.35 | 4.4 | 6.3 | 5.4 | 5.86 | 7.55 | 6.1 | 6.9 |
| V | 6.11 | 3.8 | 5.0 | 5.2 | 5.64 | 6.04 | 5.5 | 5.3 |
| W | 1.22 | 1.2 | 1.5 | 1.5 | 1.05 | 0.58 | 0.7 | 0.7 |
| Y | 2.81 | 0.9 | 1.3 | 1.8 | 3.35 | 1.51 | 3.1 | 3.1 |
Note: The most abundant amino acids (L, A and S) are presented in bold.
Figure 3.
Overall abundance of specified amino acids and distribution around the N-terminus of protein sequences of human and yeast. The abundance of all amino acids was examined on entire protein sequences, as well as on the second, third and fourth positions following the initial Met. A. The abundance in human (H. sapiens) proteins (n=10,790). B. The abundance in yeast (S. cerevisiae) proteins (n=4,819). In both cases Leu was the most abundant amino acid in the overall calculation (although not significantly higher than other amino acids). However, Ala and Ser was the most abundant amino acid on the second position in human (22.9%) and in yeast (23.7%), respectively. In both of human and yeast, no significant deviation was found for either Ala or Ser from the overall abundance of these amino acids in the proteome.
To further search for possible bias in the distribution of the relevant amino acids in both species, we compared the frequency of all amino acids in the third and fourth positions following the initial Met to that of the second position. As indicated in Figure 3 and Table 1, the distributions of amino acids, especially Ser, Ala and Leu in the third and fourth positions, are correlated with their overall frequencies in the proteome rather than with their abundances at the second position, indicating that the observation of species-specific amino acid selectivity at the second position is real, and not based on an artifact. These results support the initial assumption that there is a special functional importance to the amino acid at the second position, which is much stronger than that of the following two residues.
The distribution of the amino acid at the second position is similar for proteins in which the initial Met is removed by MAP
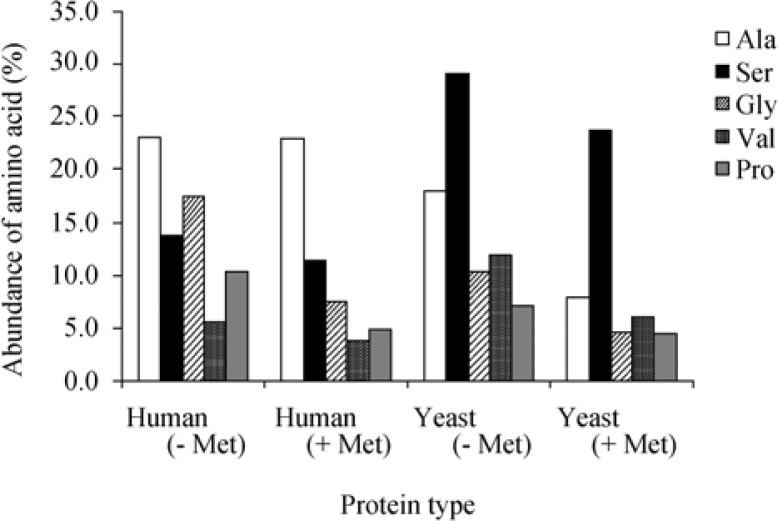
The analysis presented above was performed only on proteins containing an initial Met residue at the N-terminus of their predicted protein sequences. It was proposed that one option for the functional importance of the second position amino acid is related to the efficiency of MAP function in removing the initial Met residue from the precursor to mature the protein. Indeed, both Ala and Ser are among the amino acids that are most suitable substrates for MAP when following the initial Met, together with glycine (Gly), valine (Val) and proline (Pro). To evaluate this assumption, we examined the frequency of the above amino acids at the first position of a subset of Swiss-Prot protein sequences lacking either Met or Leu at their N-terminal end. The results (Figure 4) indicate that even though there is a slight increase in the abundance of Ala and Gly in non-Met yeast genes, the overall effect is not significant enough to establish a direct correlation between the amino acid preference at the second position and MAP activity.
Figure 4.
Abundance of amino acids on the second position in proteins potentially cleaved by MAP in comparison with proteins containing the initial Met in the mature protein. The abundance of the most common amino acids at the N-terminus of proteins undergoing cleavage by MAP (Ala, Ser, Gly, Val and Pro) was compared between the first position of possible MAP cleaved proteins (−Met) in human (n=1,034) and yeast (S. cerevisiae, n=268), and the second position of Met initiated proteins (+Met). The analysis revealed no significant difference between abundance of Ala as second amino acid in human and Ser as second amino acid in yeast between MAP cleaved and non-cleaved proteins. However, a more apparent difference was shown for Gly, Val and Pro in both human and yeast.
Evolutionary conserved proteins tend to have an even higher preference for specific amino acids at the second position
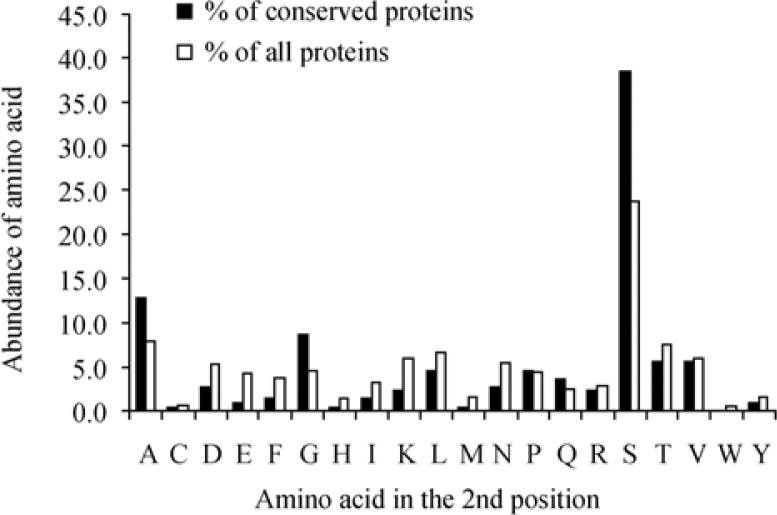
As natural selection and evolution play an important role in conserving or changing the preference for a certain amino acid at the second position, we decided to examine the abundance of the second amino acid in a subset of highly conserved proteins both in yeast and in human. As an indicator for conservation of yeast proteins, we used the UCSC-BLAT [release May 2004 (hg17)] analysis. A yeast protein for which we got a significant (sequence similarity covering over 80% of a gene’s protein-coding sequences) hit in the human genome was designated as conserved. As a result, a set of 218 such proteins was selected and examined. The results indicated a higher abundance of both Ser and Ala at the second position as compared to the overall yeast proteome (Figure 5), with the increase more pronounced for Ser (23.7% to 38.5%), suggesting that proteins containing these amino acids at the second position are more evolutionarily stable.
Figure 5.
Comparison between distribution of the second position amino acid in all yeast proteins and conserved yeast proteins that have orthologs in human. The prevalence of the second amino acid in a subset of yeast (S. cerevisiae) proteins that have human orthologs (“conserved”, n=218) was compared with the prevalence in the total number of yeast proteins examined (“all”, n=4,819). The figure indicates a significant increase in the fraction of proteins containing Ser at the second position in the subset of “conserved” yeast proteins. An increase is also apparent for Ala and Gly, but to a lesser extent.
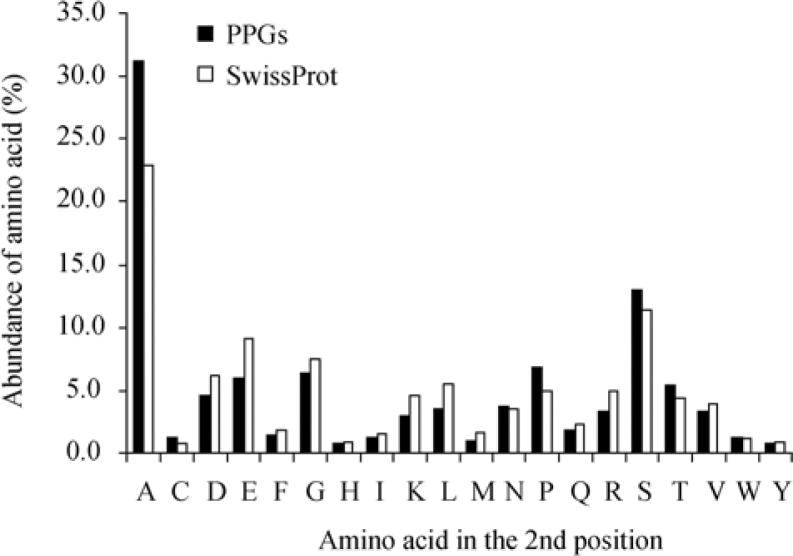
It was previously established that processed pseudogenes (PPGs) (29) are highly expressed genes that can represent relics of ancient genes (30). In order to select for a subset of most ancient and evolutionary conserved human proteins, we have selected proteins derived from genes that gave rise to PPGs in the database (www.pseudogene.org) created at the Yale University (30). We randomly selected 500 PPG represented genes and analyzed their annotated protein sequences for the abundance of the second position amino acid (Figure 6). Much like the evolutionary conserved yeast proteins, the human ancient and conserved proteins showed an increase in frequencies of both Ala and Ser with the increase more prominent for Ala (22.9% to 31.2%).
Figure 6.
Comparison between distribution of the second position amino acid in human proteins translated from genes that have PPGs. Since PPGs are more commonly derived from highly expressed and evolutionary conserved genes, the prevalence of the second amino acid in a randomly picked subset of human proteins that have a PPG (“PPGs”, n=500) from www.pseudogene.org was compared with the prevalence in the total number of human proteins examined (“SwissProt”, n=10,790). The figure indicates a significant increase in the fraction of proteins containing Ala at the second position in the subset of “PPGs” human proteins. An increase is also apparent for Ser and Pro, but to a lesser extent.
The fact that the prevalence of the preferred amino acid is even higher in evolutionary stable proteins might indicate a role in expression or translation, as there is a good correlation between levels of expression and conservation.
Examining the conservation of the second amino acid in yeast-human conserved proteins revealed that from a subset of 84 yeast proteins containing a Ser at the second position, only 22 proteins (25%) retained the same amino acid at the same position in human orthologs, whereas 37 proteins (44%) showed replacement of Ser with Ala. The rest showed a non-specific replacement of the amino acid at the second position (data not shown).
Discussion
It is widely accepted that the vast majority of the proteins are translated from an AUG codon interpreted by the ribosome as a Met. In many cases Met is also the first amino acid of mature proteins. However, maturation of some proteins causes removal of this Met residue by a designated aminopeptidase enzyme, leaving the protein with an amino terminal residue that was originally the second amino acid of the translated protein precursor 21, 23. The importance of the second amino acid in proteins is not only implied in such cases, but might rather be interpreted in more than one way. The evidence for correlation between taxa, evolutionary stage and preference of certain amino acids at the second position of proteins may serve as a powerful tool for many aspects of studying the proteome and functionality of proteins.
In this work we have shown that the preference for specific amino acids at the second position of proteins has changed through evolution, and yet can be determined as still being conserved between related taxonomical classes. The gradual changes in preference for the amino acid at the second position may be attributed to co-evolution with translational mechanisms such as ribosomal structure and function, initiation/elongation factors, and other related components of the translation machinery. When examined, the data presented in Figure 2A and Table S1 show strong correlation between second amino acid preferences in different prokaryote species and their taxonomic or developmental status based on 16S rRNA sequences, but to a lesser extent on the sequence resemblance of house keeping genes such as arsC (31). Evolutionary developed transitions from one preferred amino acid to another may also indicate changes in ribosomal structure and function throughout the evolution of eukaryotes from unicellular yeasts, through multicellular fungi, to higher plants and animals.
The fact that we could detect no apparent correlation between the abundance of amino acids in the general proteome of yeast and human (represented by the Swiss-Prot database) with the abundance of the second position amino acids, whereas a good correlation exists for the third and fourth positions examined, indicates that there is a functional selection for the amino acid occupying the second position, much like the Met at the first position but to a lesser extent. The function of the second amino acid is to be evaluated further. However, we are able to attribute it partially to the availability of the N-terminus of a protein to cleavage by MAP. Further analysis of the MAP mechanism, as well as of other translational and post-translational mechanisms, regarding the sequence preference of the first two amino acids at the N-terminus of proteins, is needed to better understand the nature of this mechanism.
The species-specific preference for the amino acid in the second position may thus help us in predicting the evolutionary stage of a given new taxon. According to the results presented, assuming that the transition from Ser to Ala as the most abundant second amino acid indicates an evolutionary stage within eukaryotes, we may speculate that higher plants (such as Arabidopsis and maize) are more evolutionarily advanced than higher mammals such as human, at least in the complexity of protein production mechanism. This might also indicate that the rate of evolution is more rapid for plants than for animals. It may also enable us in sorting the proteome of a species with regards to each protein’s translation efficiency, evolutionary conservation, potential subcellular localization and function.
In cases where a predicted gene/transcript contains more than one potential initiation site, such as multiple clustered Met residues, alternatively spliced variants with a variety of potential initiation sites, or alternative promoters, the finding presented in this work might help in predicting the most probable initiation site. Furthermore, it might also help in predicting the most efficient site when multiple sites are known or predicted.
We believe that the data presented in this work will point a spotlight to a somewhat previously neglected aspect of the proteome and protein sequence analysis. The importance of the second amino acid, especially in highly expressed proteins may prove to be a bioinformatic tool, not only in evolution and proteome research, but rather in protein engineering and production. We suggest that understanding the basis of the amino acid selection differences between taxa can help enhance mammalian protein production in bacteria or fungi much to the same extent as understanding the importance of other controlling elements.
Materials and Methods
Analysis of the second amino acid in proteins of different species
Protein sequences were downloaded in FASTA format from Swiss-Prot (UniProt Release 4.0 with Swiss-Prot Release 46.0 of 01-Feb-2005) (32). Only protein sequences starting with methionine (M), the vast majority of all sequences, were selected and copied into a different subset. The initial M was removed and all the protein sequences were arranged and sorted by their new initial letter. A simple counter command was applied on all sequences determining how many sequences belong to each group.
Analysis of the third and fourth amino acid in human and yeast proteins
We have utilized the above method, only removed either the first two or three amino acids from the protein sequences.
Analysis of the second amino acid in human and yeast proteins potentially cleaved by MAP
Protein sequences were downloaded in FASTA format from Swiss-Prot (UniProt Release 4.0 with Swiss-Prot Release 46.0 of 01-Feb-2005) (32). Only protein sequences not initiating with either methionine (M) or leucine (L) were selected and copied into a different subset (leucine is the second common initiating amino acid). The initial M or L was removed and all the protein sequences were arranged and ordered by their new initial letter. A simple counter command was applied on all sequences determining how many sequences belong to each group.
Authors’ contributions
RS and AN collected data and conducted analysis. RS conceived the idea and prepared the manuscript. YC approved and supervised the study. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing intersts exist.
Supplementary Material
Table S1
References
- 1.Boni I.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komarova A.V. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komarova A.V. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005;187:1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shine J., Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stormo G.D. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyasaka H. The positive relationship between codon usage bias and translation initiation AUG context in Saccharomyces cerevisiae. Yeast. 1999;15:633–637. doi: 10.1002/(SICI)1097-0061(19990615)15:8<633::AID-YEA407>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 9.Rocha E.P. Translation in Bacillus subtilis: roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 1999;27:3567–3576. doi: 10.1093/nar/27.17.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor M. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl. Acad. Sci. USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito D. In vivo evidence for the prokaryotic model of extended codon-anticodon interaction in translation initiation. EMBO J. 2003;22:651–656. doi: 10.1093/emboj/cdg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez G. Preference for guanosine at first codon position in highly expressed Escherichia coli genes. A relationship with translational efficiency. Nucleic Acids Res. 1996;24:2525–2527. doi: 10.1093/nar/24.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohno H. Preferential usage of some minor codons in bacteria. Gene. 2001;276:107–115. doi: 10.1016/s0378-1119(01)00670-9. [DOI] [PubMed] [Google Scholar]
- 14.Stenstrom C.M., Isaksson L.A. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 15.Stenstrom C.M. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 16.Fuglsang A. Nucleotides downstream of start codons show marked non-randomness in Escherichia coli but not in Bacillus subtilis. Antonie Van Leeuwenhoek. 2004;86:149–158. doi: 10.1023/B:ANTO.0000036147.66445.7a. [DOI] [PubMed] [Google Scholar]
- 17.Joshi C.P. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 18.Sawant S.V. Sequence architecture downstream of the initiator codon enhances gene expression and protein stability in plants. Plant Physiol. 2001;126:1630–1636. doi: 10.1104/pp.126.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuglsang A. Bioinformatic analysis of the link between gene composition and expressivity in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Antonie Van Leeuwenhoek. 2004;86:135–147. doi: 10.1023/B:ANTO.0000036119.00001.3b. [DOI] [PubMed] [Google Scholar]
- 20.Brooks D.J. Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code. Mol. Biol. Evol. 2002;19:1645–1655. doi: 10.1093/oxfordjournals.molbev.a003988. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Bassat A. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kendall R.L., Bradshaw R.A. Isolation and characterization of the methionine aminopeptidase from porcine liver responsible for the co-translational processing of proteins. J. Biol. Chem. 1992;267:20667–20673. [PubMed] [Google Scholar]
- 23.Hirel P.H. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenstrom C.M. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez de Valdivia E.I., Isaksson L.A. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton R. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tats A. Highly expressed proteins have an increased frequency of alanine in the second amino acid position. BMC Genomics. 2006;7:28. doi: 10.1186/1471-2164-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shemesh R. Genomic fossils as a snapshot of the human transcriptome. Proc. Natl. Acad. Sci. USA. 2006;103:1364–1369. doi: 10.1073/pnas.0509330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson C.R., Dugas S.L. Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 2003;3:18. doi: 10.1186/1471-2148-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bairoch A. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1