Abstract
The direct negative impact of the transcriptional activity of one component on the second one in cis is referred to as transcriptional interference (TI). U6 is a type III RNA polymerase III promoter commonly used for driving small hairpin RNA (shRNA) expression in vector-based RNAi. In the design and construction of viral vectors, multiple transcription units may be arranged in close proximity in a space-limited vector. Determining if U6 promoter activity can be affected by TI is critical for the expression of target shRNA in gene therapy or loss-of-function studies. In this research, we designed and implemented a modified retroviral system where shRNA and exogenous gene expressions were driven by two independent transcriptional units. We arranged U6 promoter driving shRNA expression and UbiC promoter in two promoter arrangements. In primary macrophages, we found U6 promoter activity was inhibited by UbiC promoter when in the divergent arrangement but not in tandem. In contrast, PKG promoter had no such negative impact. Instead of enhancing U6 promoter activity, CMV enhancer had significant negative impact on U6 promoter activity in the presence of UbiC promoter. Our results indicate that U6 promoter activity can be affected by TI in a proximal promoter-specific and arrangement-dependent manner.
Key words: transcriptional interference, U6 promoter, shRNA, retrovirus
Introduction
RNA interference (RNAi) is an evolutionarily conserved, sequence-specific gene silencing mechanism triggered by two types of RNA in animals: small interfering RNA (siRNA) and microRNA (miRNA). siRNAs are usually generated from precursors including transpons, viruses, endogenously expressed long double-stranded RNA (dsRNA), or experimentally introduced synthetic dsRNA 1, 2, 3, while miRNAs are processed products from transcripts of endogenous non-coding genes that form small stem loops 4, 5, 6, 7, 8, 9. RNAi technology provides not only a powerful tool for loss-of-function genetic research in a variety of systems, but also potentially for in vivo gene therapy by depleting disease-related transcripts while rescuing with the wild-type counterpart. The development of vectors encoding small hairpin RNA (shRNA) allows for the depletion of disease-related transcripts, and has provided potential tools for gene therapy 10, 11, 12, 13, 14. For these purposes, targeting the transcripts with potent and persistent vector-based RNAi is the key to the outcome of the treatment.
So far, several strategies have been tested to enhance the efficacy of vector-based RNAi. These include the design of more efficient RNAi target sequence based on different siRNA selection criteria (15), the promoter choice of type III RNA polymerase III (U6, H1) (16), polymerase II 17, 18, 19, shRNA stem length (20), loop structure (21), mimicking miRNA-based shRNA (22), and addition of U6 leader sequence (12). Among these strategies, increasing the shRNA expression in the cells is one of the most important ways to effectively deplete the target transcripts. Recently, U6 promoter has been proved to be stronger than H1 promoter in vitro and in vivo (16). Enhancing U6 or H1 promoter activity using CMV enhancer to increase the shRNA expression in the target cells leads to strengthened gene silencing effects 23, 24. Thus, developing strategies to enhance or maintain shRNA potency and dose loading in cells is one of the concerns for the design of vector-based RNAi.
The proximal arrangement of two independent transcriptional units may affect each other’s activity by promoter cross-talk (25). Direct negative impact of one transcriptional activity on another in cis is referred to as transcriptional interference (TI). TI is usually asymmetric and results from the existence of two promoters, the stronger promoter reduces the expression of the weaker one. Different promoter arrangements can lead to TI (26). In a viral vector system where space is often limited, it is common to have multiple transgenes in close proximity to drive the expression of a therapeutic transcript or fluorescent marker for research purposes (27), thus maintaining intact U6 promoter activity is critical for successful gene silencing. In this research, we study the U6 promoter activity regulation by TI within the viral vector. We found that U6 promoter activity is inhibited if the shRNA expression cassette is in a divergent arrangement with respect to the UbiC promoter, but not in the tandem, while PKG promoter has no inhibitory effect. The CMV enhancer adjacent to U6 promoter has significant negative effect on U6 promoter activity in the presence of UbiC promoter. Our results suggest that U6 promoter activity can be affected by TI, and this effect is specific to both the proximal promoter as well as its arrangement.
Results
A modified retroviral system capable of gene knockdown and expression
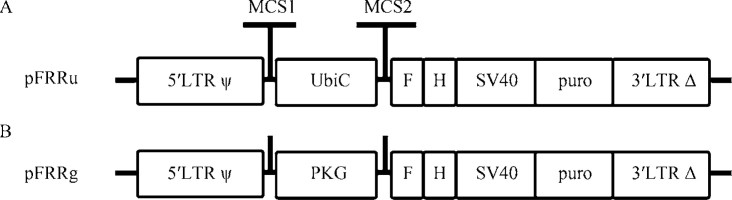
Mouse primary macrophages are hard to be transfected or nucleofected. However, they can be infected with retroviruses. We compared several commercialized retroviral vectors and found that pSuper-Retro-puro from Oligoengine worked best for virus production. It has at least two confirmed features, one is the shRNA expression driven by H1 promoter, the other is the expression of puromycin drug resistance marker driven by PKG promoter. However, H1 promoter may not function with the same strength as a U6 promoter for shRNA expression (16), and this vector lacks the cloning sites for exogenous gene expression. We therefore modified this vector into a new retroviral vector, capable of gene knockdown and expression while reserving the puromycin resistance expression function. As illustrated in Figure 1A, we removed the H1 promoter in pSuperRetro-puro and reserved the multiple cloning sites (MCS1) for cloning of U6 promoter driving shRNA expression. Human UbiC promoter has been shown to be constitutively active in a variety of cells and tissues (27), and was selected to drive the expression of an exogenous gene and puromycin resistance gene separated by SV40 promoter. MCS2 between UbiC and SV40 allows the cloning of exogenous gene. We derived vectors with C-terminal Flag-His6 (FH) to allow for immunoblot or immunoprecipitation of the expressed protein, and named this new vector as pFRRu. The other vector pFRRg has the same design strategy, but UbiC promoter was replaced with PKG promoter (Figure 1B). With these new retroviral vectors we have transduced a variety of mouse cell types (e.g., epithelial cells, keratinocytes, fibroblasts, and hematopoietic cells) as well as primary cells [e.g., mouse embryonic fibroblasts and bone marrow-derived macrophages (BMDMs)]. The efficiency of transduction varies with cell type and the virus titer applied. However, following puromycin selection, all the residual cells were virally transduced. In this study, the viral titer we applied led to 30%-60% transduction efficiency for macrophages, avoiding multiple entries of viral transcripts into one cell.
Figure 1.
A modified retroviral system with gene knockdown and exogenous gene expression functions. A. Map of the modified retroviral vector with UbiC promoter. pFRRu was generated as described in Materials and Methods. There are two multiple cloning sites: 5′-end of UbiC has the multiple cloning sites for shRNA expression cassette (MCS1), and 3′-end of UbiC has the multiple cloning sites for exogenous gene expression (MCS2) containing an in-frame Flag-His6 (FH) tag. Puromycin (puro) resistance expression was reserved with SV40 promoter. B. The modified retroviral vector with PKG promoter. UbiC promoter as described above was replaced with PKG promoter.
Inhibition of U6 promoter activity by divergent promoter arrangement of U6 and UbiC
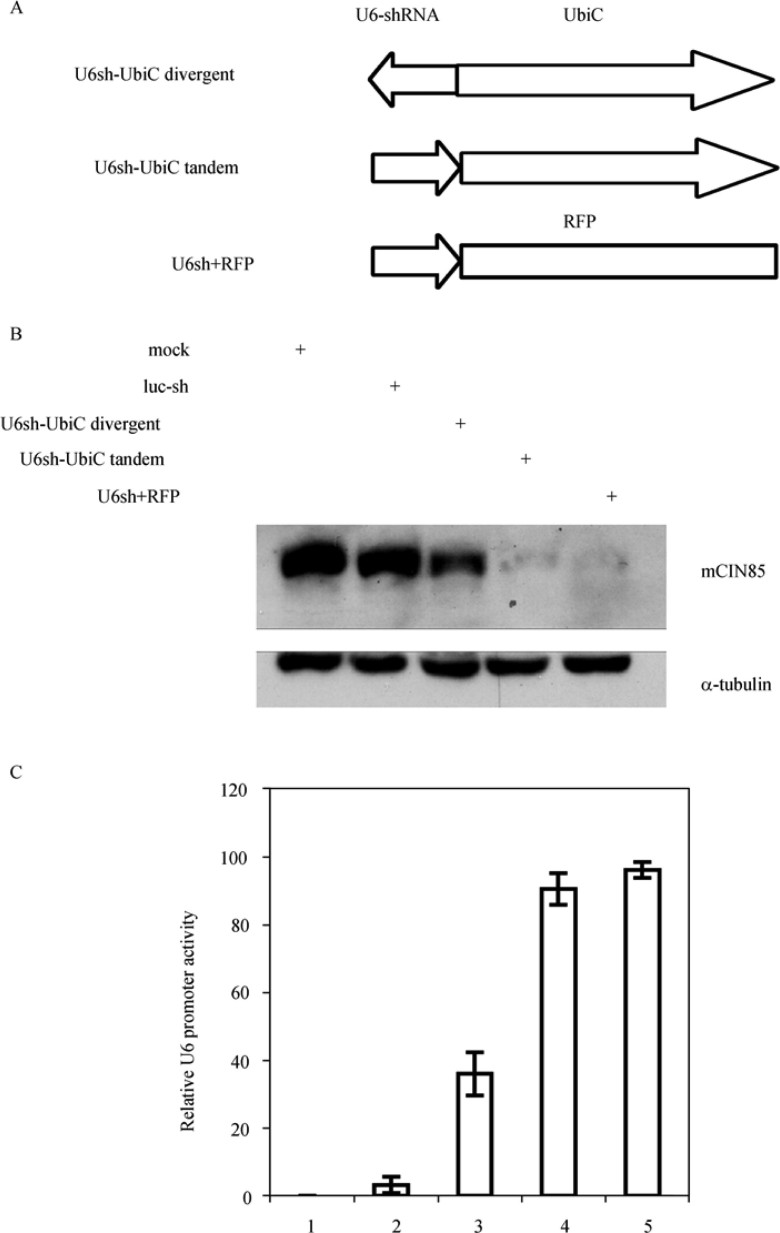
Usually three promoter arrangements may lead to TI, namely convergent promoters, tandem promoters and overlapping (divergent) promoters (26). Since convergent promoter arrangement for U6 and other promoters is rare in viral vectors, we focused on tandem and divergent promoter arrangements to explore which arrangements led to significant impairment to U6 promoter activity. We constructed U6 promoter driving mCIN85 shRNA expression cassette and inserted this expression cassette in MCS1 of pFRRu in different orientations. One was divergent promoter arrangement in which U6 and UbiC were in opposite direction (pU6sh-pUbiC divergent), and the other was tandem in the same direction as UbiC (pU6sh-pUbiC-tandem). We also generated a luciferase shRNA expression vector as a non-specific shRNA control and a pFRRrfp-U6-mCIN85 shRNA as a control to minimize promoter interaction by replacing UbiC promoter with a non-promoter DNA fragment (Figure 2A). These vectors were introduced into Plat E cells to produce viruses. BMDMs were transduced with respective virues and subsequently selected with puromycin to remove the non-transduced cells. After four days of selection the remaining cells were lysed for protein extraction. Western blots were carried out using rabbit anti-CIN85 antibody (Figure 2B). Relative U6 promoter activity was determined using endogenous mCIN85 protein level normalized against the α-tubulin level as the loading control. As shown in Figure 2C, the arrangement of U6 promoter relative to UbiC promoter in the vector had a significant impact on U6 promoter activity. Tandem arrangement maintained much higher U6 promoter activity than divergent arrangement. This suggested that U6 promoter activity can be regulated by TI in a promoter arrangement-dependent manner.
Figure 2.
Divergent promoter arrangement of U6 and UbiC inhibits U6 promoter activity. A. Diagram of U6 and UbiC promoter arrangements. mCIN85 shRNA expression cassette was constructed as described in Materials and Methods, and was inserted into pFRRu to form divergent and tandem promoter arrangements. Minimal interfered U6 promoter activity control was set by replacing the UbiC promoter with a non-promoter cDNA fragment of RFP. B. Residual endogenous mCIN85 protein level to reflect U6 promoter activity. Mouse BMDMs were transduced with retroviruses produced with different vectors as indicated. Four days after viral transduction and drug selection, residual cells were collected and lysed. Western blots were performed using purified rabbit anti-CIN85 as primary antibody. C. Normalized mCIN85 knocking down efficiency reflecting relative U6 promoter activity. Sample sequences are as indicted in Panel B. Values are statistics from three independent experiments.
CMV enhancer negatively impacts U6 promoter activity in the presence of UbiC promoter
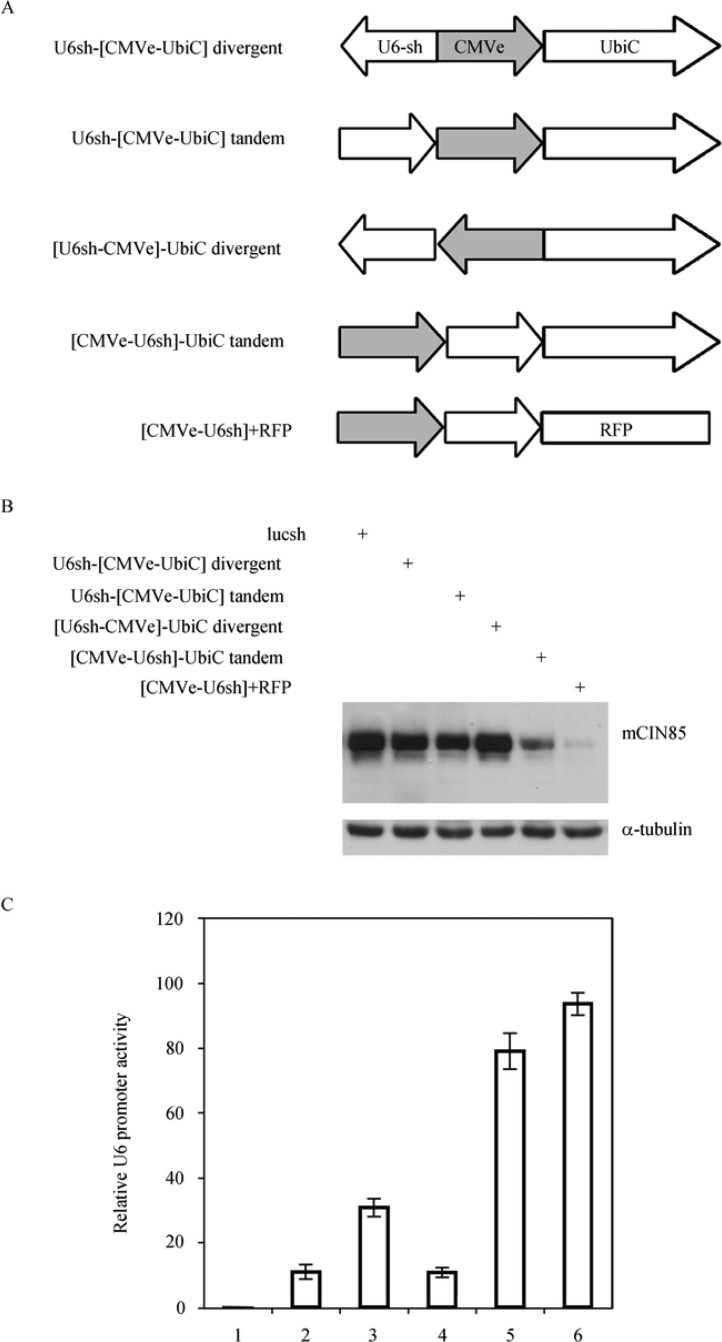
Previous studies have shown that the CMV enhancer has a positive effect on U6 promoter activity (23). However, the effect of the CMV enhancer on U6 promoter activity in the presence of TI is unknown. To answer this question, we placed a CMV enhancer between U6 and UbiC promoters in both promoter arrangements by fusing the enhancer to the upstream of UbiC or U6 promoter (Figure 3A). We then tested the U6 promoter activity. To our surprise, instead of enhancing U6 promoter activity, CMV enhancer in all four configurations significantly strengthened the UbiC inhibitory effect on U6 promoter activity in both promoter arrangements (Figure 3B and C). However, the level of inhibition varied with the promoter arrangements. Fusing the CMV enhancer upstream of U6 and keeping tandem arrangement of the promoters gave relatively less inhibition, while the divergent configuration gave the highest inhibition. This result indicates that the CMV enhancer can boost TI and significantly inhibit U6 promoter activity in the presence of UbiC promoter in either promoter arrangements.
Figure 3.
Negative impact of UbiC on U6 promoter activity enhanced in the presence of CMV enhancer. Legends in Figure 2 was followed except that CMV enhancer was placed between U6 and UbiC promoter or fused to the upstream of U6 as plotted. A. Diagram of U6 promoter, CMV enhancer, and UbiC promoter arrangements. CMV enhancer was fused to the upstream of either U6 promoter or UbiC promoter forming divergent or tandem arrangements as indicated. B. Western blot of mCIN85 to reflect the residual mCIN85 left in the cells. Samples were generated using different virus transduction as indicated. C. Relative U6 promoter activity after normalization against α-tubulin. Values are statistics from three independent experiments.
Regulation of U6 promoter activity by TI is promoter-specific
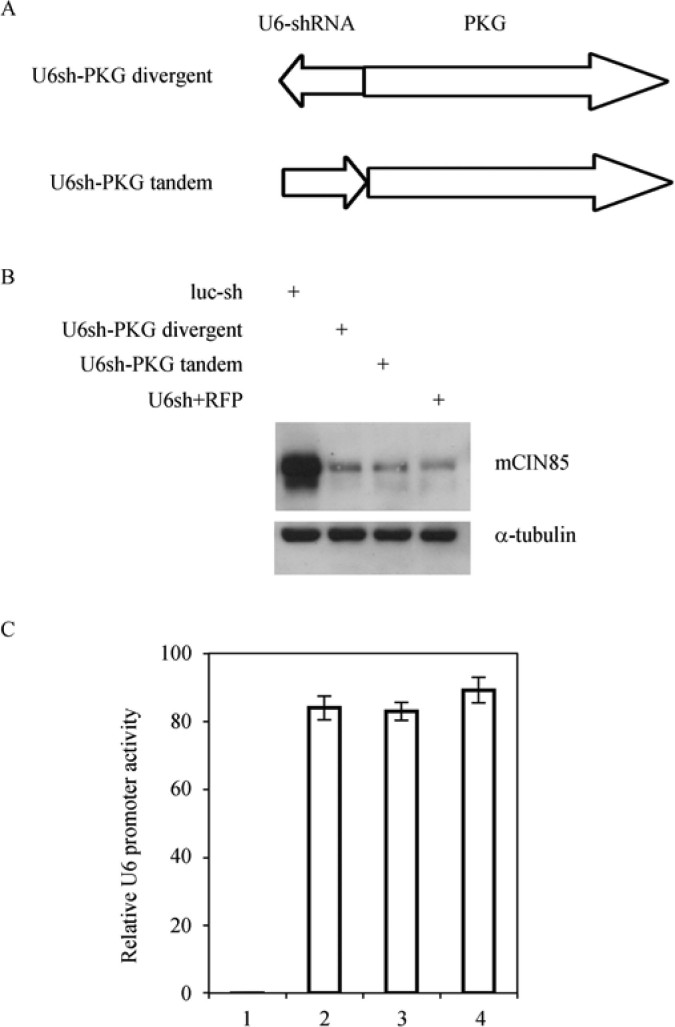
TI is often originated from asymmetric strength of two closely arranged promoters, the stronger promoter reduces the activity of the weaker one (26). UbiC promoter is ubiquitously active in a variety of cells and is a relatively strong promoter (27). We next asked whether U6 promoter activity can be maintained if we replace the UbiC promoter with a weaker PKG promoter to balance the previously asymmetric strength. We constructed similar viral vectors in both U6 promoter arrangements with PKG promoter as shown in Figure 4A and tested U6 promoter activity in transduced BMDMs. As expected, no significant inhibition of U6 promoter activity was observed in either arrangement (Figure 4B and C). This result suggests that regulation of U6 promoter activity by TI is promoter-specific. Balancing the strength of the two adjacent promoters is the key. PKG, a weaker promoter, has minimum TI effect on U6 promoter activity.
Figure 4.
U6 promoter activity response to TI is promoter-specific. Legends in Figure 2 were followed except that UbiC promoter was replaced with PKG promoter. A. Diagram of U6 and PKG promoter arrangements. B. Residual endogenous mCIN85 protein level to reflect U6 promoter activity. C. Relative U6 promoter activity after normalization against α-tubulin. Values are statistics from three independent experiments.
Discussion
In this research, we have designed and implemented a retroviral system where shRNA and exogenous gene expressions are allowed by two independent transcriptional units. Our results indicate that U6 promoter activity is inhibited when in the divergent arrangement with respect to UbiC promoter, but in tandem arrangement the activity remains unchanged. However, PKG promoter has no such impact. Instead of enhancing the U6 promoter activity, CMV enhancer has significant negative impact on U6 promoter activity by potentiating TI in the presence of UbiC promoter in both tandem and divergent promoter arrangements. We demonstrate that U6 promoter activity can be affected by TI in a promoter arrangement-dependent and proximal promoter-specific manner.
Maintaining U6 promoter activity is crucial for expression of shRNA in the cells and subsequent depletion of target transcripts in vector-based strategy. In designing of vectors for gene therapy or loss-of-function research, promoters with different strengths may be arranged in a space-limited viral vector. TI occurs between two promoters of varying strength, where the stronger one suppresses the activity of the weaker 26, 28. Since U6 promoter may not be strong enough to balance the asymmetry of promoter strength, therefore, it is suppressed by the adjacent stronger promoter. Our results indicate that promoter arrangement is the key for maintaining U6 promoter activity, providing guidelines for maintaining U6 promoter activity in designing viral vectors.
Enhancing U6 promoter activity by CMV enhancer in target cells has been proved to be an effective way to obtain satisfactory gene silencing (23). U6 promoter is constitutively active in a variety of cell types and maintains relatively high activity by providing approximately 4×105 transcripts per cell (29). However, under some circumstances, unmodified Pol III promoter is not sufficient to provide satisfactory depletion of target transcripts even without reported TI 12, 23, 24, 30, 31, 32. The addition of a CMV enhancer adjacent to U6 promoter or hybrid CMV-H1 promoter has been reported to improve the efficiency of RNAi 23, 24 or shRNA delivery in vivo (33). Our results indicate that in a relative more complex vector niche, addition of CMV enhancer adjacent to U6 promoter may even enhance the asymmetry of the two independent transcriptional units and potentiate suppression to U6 promoter activity by enhancing TI in either divergent or tandem promoter arrangement.
Materials and Methods
Cell culture
Human embryonic kidney 293T cells and retroviral packaging cell line Plat E cells (30) were maintained in DMEM-10 (DMEM containing 10% FBS, Glutamine and pen/str). Mouse primary BMDMs were isolated and cultured using alpha-10 (alpha MEM containing 10% FBS, Glutamine and pen/str) and a conditional medium containing M-CSF as described (31).
Antibodies
Mouse monoclonal α-tubulin antibody and rabbit anti-CIN85 antibody were purchased from Sigma.
Construction of multifunctional retroviral vectors
UbiC promoter containing multiple cloning sites was amplified by PCR using pFG12 (16) as template. SV40-puro fragment was obtained by PCR using pMX-puro as template (32). These two fragments were spliced together using joint PCR and were cloned into BglII/SacII sites in pSuper-Retro-puro (Oligoengine), and we named this new vector as pFRRu. The other vector pFRRg was obtained by replacing UbiC promoter in pFRRu with PCR fragment of PKG promoter. CMV enhancer was obtained by PCR using pDsRed2 (Clontech) as template with primer pairs: TCTAGAAGATCTCGCGTTACATAACTTACGGTAAATG (forward), AGTCGGATCCAAAACAAACTCCCATTGACGTCAATG (reverse). BamHI/BglII-cut CMV enhancer fragment was fused to UbiC promoter by a subcloning into BglII site of pFRRu. Modified regions of these vectors were confirmed by sequencing.
mCIN85 shRNA expression cassette
Mouse CIN85 shRNA target sequence was predicted with Rational_siRNA_Design Program (33). We selected target sequence in the 3′-UTR of mCIN85 mRNA: GCTACCATGATTCCAAATAA.
Mouse CIN85 shRNA expression cassette was constructed by joint PCR. Briefly, we obtained the hU6 promoter (f1) by PCR using pBS-hU6-1 as template (16) with primer pairs p1 and p2, and the shRNA fragment (f2) was obtained by PCR using primer pairs p3 and p4 (Table 1).
Table 1.
PCR primer pairs used for mouse CIN85 shRNA expression cassette
| Primer | Sequence | Orientation |
|---|---|---|
| p1 | GCACAGATCTATCTAGAACCCCAGTGGAAAGACGCGCAG | forward |
| p2 | GGTGTTTCGTCCTTTCCACAAG | reverse |
| p3 | GTGGAAAGGACGAAACACCGCTACCATGATTCCAAATAATTCAAGAGATTATTTG | forward |
| p4 | TCCAGCTCGAGAAAAAGCTACCATGATTCCAAATAATCTCTTGAATTATTTG | reverse |
| p5 | AGCTGTCGACAGATCTATCTAGAACCCCAGTGGAAAGACGCGCAG | forward |
| p6 | TCCAGGGATCCCTCGAGAAAAAGCTACCATGATTCCAAATAATCTCTTG | reverse |
Note: Nucleotides in italics are hairpin sequences while in bold face are target siRNA sequences.
After electrophoresis, f1 was purified from a 1% TAE-agrose gel with gel purification kit (Invitrogen) while f2 was purified from a 12% TAE-polyacrylamide gel. Joint PCRs were carried out using hU6 forward primer p1, shRNA reverse primer p4 and mixed template of f1 and f2. The PCR products were purified, cut with BglII/XhoI and subcloned into a pFRRu or pFRRg vector as depicted in the text. To change the orientation of U6-driven shRNA expression cassette in the vectors, we re-amplified the above shRNA expression cassette with PCR primer pairs p5 and p6 (Table 1). This fragment was cut with SalI/BamHI and was cloned into XhoI/BglII-cut pFRRu. All constructs containing shRNA expression cassette sequence were sequenced using hU6 forward primer p1.
Retrovirus production and transduction of mouse BMDMs
Protocols described in Ilves et al. (31) were followed with some modifications. Briefly, a subconfluent culture of Plat E cells in six-well plate was transfected with pFRRu- or pFRRg-derived plasmids. After 24 h, cells were washed once with LPS-free alpha-10, and fresh alpha-10 was applied in each well for another 24 h to accumulate viruses. Viruses were collected and filtered with 0.45-μm syringe filter and mixed with equal volumes of alpha-10, and 10% conditional medium containing M-CSF (equivalent to 10 ng/mL M-CSF final) and protamine (10 μg/mL final, purchased from Sigma). The prepared medium was applied to the BMDMs for overnight infection, and the infected BMDMs were further cultured in fresh alpha-10 containing 10% M-CSF conditional medium and puromycin (3.5 μg/mL final) for four days to remove non-transduced cells. This protocol allowed about 30%-60% viral transduction efficiency. Residual cells after drug selection were considered as viral-transduced. The cells were collected and washed with 1×PBS and lysed with cell lysis buffer [20 mM Hepes pH 7.6, 300 mM NaCl, 0.5 mM EDTA, 5 mM NaF, 1 mM sodium orthovanadate, 5% glycerol, 0.1% NP-40, 1 mM DTT, protease inhibitor cocktail (Sigma)]. Protein concentration was measured with Bio-Rad rapid protein assay kit. For Western blot analysis, 20 μg of cell lysate was loaded to each lane.
Indirect U6 promoter activity assay
In this research, U6 promoter activity was defined as the percentage of endogenous mCIN85 depletion. Residual endogenous mCIN85 and α-tubulin protein level were determined by Western blot. Bar-charts were plotted by measuring the density of mCIN85 bands in the film by ImageJ while using α-tubulin band density as loading control for normalization.
Authors’ contributions
LN performed the experiments. MDT, YW, QS, and YZ prepared the reagents. MDT helped data analysis and figure drawing. YF supervised the project and wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
References
- 1.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Zamore P.D. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir S.M. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau N.C. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 8.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 9.Lim L.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 10.Paddison P.J. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummelkamp T.R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 12.Paul C.P. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 13.Sui G. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J.Y. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taxman D.J. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makinen P.I. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J. Gene Med. 2006;8:433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- 17.Song J. Poly(U) and polyadenylation termination signals are interchangeable for terminating the expression of shRNA from a pol II promoter. Biochem. Biophys. Res. Commun. 2004;323:573–578. doi: 10.1016/j.bbrc.2004.08.128. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J. shRNA transcribed by RNA Pol II promoter induce RNA interference in mammalian cell. Mol. Biol. Rep. 2006;33:43–49. doi: 10.1007/s11033-005-3965-1. [DOI] [PubMed] [Google Scholar]
- 20.Siolas D. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 21.Miyagishi M. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J. Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 22.Chang K. Lessons from Nature: microRNA-based shRNA libraries. Nat. Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 23.Xia X.G. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong S.T. Hybrid cytomegalovirus enhancer-h1 promoter-based plasmid and baculovirus vectors mediate effective RNA interference. Hum. Gene. Ther. 2005;16:1404–1412. doi: 10.1089/hum.2005.16.1404. [DOI] [PubMed] [Google Scholar]
- 25.Hampf M., Gossen M. Promoter crosstalk effects on gene expression. J. Mol. Biol. 2007;365:911–920. doi: 10.1016/j.jmb.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Shearwin K.E. Transcriptional interference-a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lois C. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 28.Eszterhas S.K. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell Biol. 2002;22:469–479. doi: 10.1128/MCB.22.2.469-479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthess Y. Conditional inhibition of cancer cell proliferation by tetracycline-responsive, H1 promoter-driven silencing of PLK1. Oncogene. 2005;24:2973–2980. doi: 10.1038/sj.onc.1208472. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J.D. Improved accumulation and activity of ribozymes expressed from a tRNA-based RNA polymerase III promoter. Nucleic Acids Res. 1995;23:2259–2268. doi: 10.1093/nar/23.12.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilves H. Retroviral vectors designed for targeted expression of RNA polymerase III-driven transcripts: a comparative study. Gene. 1996;171:203–208. doi: 10.1016/0378-1119(96)00075-3. [DOI] [PubMed] [Google Scholar]
- 32.Boden D. Promoter choice affects the potency of HIV-1 specific RNA interference. Nucleic Acids Res. 2003;31:5033–5038. doi: 10.1093/nar/gkg704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassani Z. A hybrid CMV-H1 construct improves efficiency of PEI-delivered shRNA in the mouse brain. Nucleic Acids Res. 2007;35:e65. doi: 10.1093/nar/gkm152. [DOI] [PMC free article] [PubMed] [Google Scholar]