Abstract
Phytocystatins constitute a multigene family that regulates the activity of endogenous and/or exogenous cysteine proteinases. Cereal crops like wheat are continuously threatened by a multitude of pathogens, therefore cystatins offer to play a pivotal role in deciding the plant response. In order to study the need of having diverse specificities and activities of various cystatins, we conducted comparative analysis of six wheat cystatins (WCs) with twelve rice, seven barley, one sorghum and ten corn cystatin sequences employing different bioinformatics tools. The obtained results identified highly conserved signature sequences in all the cystatins considered. Several other motifs were also identified, based on which the sequences could be categorized into groups in congruence with the phylogenetic clustering. Homology modeling of WCs revealed 3D structural topology so well shared by other cystatins. Protein-protein interaction of WCs with papain supported the notion that functional diversity is a consequence of existing differences in amino acid residues in highly conserved as well as relatively less conserved motifs. Thus there is a significant conservation at the sequential and structural levels; however, concomitant variations maintain the functional diversity in this protein family, which constantly modulates itself to reciprocate the diversity while counteracting the cysteine proteinases.
Key words: wheat cystatins, structural diversity, functional diversity, comparative analysis
Introduction
The explosion taken place in genome sequencing so far has outpaced its structural and functional aspects. With the sequence-structure gap widening further, elucidating functions of proteins from available structural information is a viable approach. However, only 45,000 protein structures are currently available in Protein Data Bank (PDB). Resolving structure of every protein experimentally seems to be an uphill task. Therefore, recoursing to computational model prediction tools has become inevitable. Proteins that belong to the same family or superfamily are suitable candidates for computational analysis as they have conserved sequences, structures and functions to varying degrees depending on their evolutionary patterns.
Cystatins are proteinaceous reversible competitive and non-competitive inhibitors of papain-like cysteine proteinases 1, 2, 3, which constitute an evolutionary superfamily with representatives from all major kingdoms of life (4). Cystatin superfamily has been subdivided into families based on their sequence homology, molecular weights as well as presence and absence of disulphide bonds 5, 6. The earlier classification consisted of only three families: stefins, cystatins and kininogens. Stefins contain ~100 residues with molecular weights of ~11 kDa and no disulphide bonds nor glycosylation. Cystatins are ~115 amino acid residues long with four conserved cysteine residues forming two disulphide bonds and may be glycosylated or non-glycosylated. Kininogens are the longest cystatins whose molecular weights range from 60 to 120 kDa with several cystatin domain repeats and are glycosylated (7). The latest entry into this superfamily includes cystatins found in plants, called phytocystatins (PhyCys). PhyCys share sequence homology with other cystatins but they lack disulphide bonds and have a molecular weight of 12-16 kDa. However, several PhyCys with a molecular mass of ≈23 kDa have a carboxy-terminal extension, which has been involved in the inhibition of a second family of cysteine peptidases, the legumain peptidases (8). Besides possessing the three signature sequences of cystatin superfamily, that is, a conserved N-terminal G residue, a highly conserved QXVXG motif in the central loop region and a P/AW near the C-terminal end, phytocystatins have an exclusive N-terminal conserved motif [LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-X-[EDQV]-[HYFQ]-N that corresponds to the α-helix in the cystatin structure. Phylogenetic grouping also demonstrated PhyCys as a separate cluster, thus qualifying them as an independent family (9).
The diverse roles that PhyCys play in plant physiology have developed this area into a promising research field with high output potentiality. Primarily, they have been implicated in regulating plant protein turnover and defense responses 10, 11. Their expression patterns coincide with major seed storage proteins 12, 13, 14, signifying their role in preventing storage protein degradation by endogenous proteinases during endosperm development. Their role in defense is inferred from their ability to counteract with exogenous proteases secreted by insects, nematodes and fungi 15, 16, 17, 18. This is further supported by their induction with wounding or methyl jasmonate 10, 11, 19. The expression of PhyCys in transgenic plants has conferred enhanced resistance against insects, nematodes and viruses 20, 21, 22, 23, thus making them apt candidates for integrated pest/pathogen management programs. Some PhyCys have also been shown to respond to various abiotic stresses like heat, cold, salinity and anaerobiosis 1, 10, 24. Lately, their involvement in regulating programmed cell death and senescence has also been investigated 25, 26, 27.
In wheat (Triticum aestivum), multiple cystatins have been discovered. WC1, WC2, WC3 and WC4 are expressed both at seed maturation and germination with differential expressions (28). Another wheat cystatin (WC5) has been identified to be expressed only during early embryogenesis and maturation stage of grain development (29). A multidomain cystatin of molecular weight 23 kDa containing a long C-terminal extension region showed elevated expression during cold acclimation. It also exhibits strong antifungal activity against the mycelial growth of the snow mold fungus Microdochium nivale (30). Gliadain, a gibberellin-inducible cysteine protease occurring in germinating seeds of wheat, is regulated by intrinsic cystatins (31). Thus like other PhyCys, WCs also seem to inhibit both endogenous and exogenous cysteine proteinases. These functions can be further consolidated by the structural analysis of these molecules and their possible interactions with papain, a hallmark of all cysteine proteinases.
Structurally, only a single plant cystatin from rice, oryzacystatin (OC1), has been solved so far (32). The oryzacystatin molecule consists of a central helix and a five-stranded antiparallel β-sheet. Its architecture was observed to be similar to other animal cystatins like human stefin A, stefin B and chicken egg white cystatin, confirming their evolutionary relatedness (32). Analyses of different cystatin–papain complexes imply a common mode of inhibition that is accomplished by a high affinity fitting of the “tripartite wedge” of cystatin molecules into a complementary active site groove present in the papain molecule 33, 34. In principle, this bimolecular interaction of CP-CPI keeps the protease inactive for long time, thus preventing the proteolysis of actual substrates. The wedge is formed by the N-terminal glycyl-containing trunk, the highly conserved QXVXG in the first hairpin loop, and the second hairpin loop containing P/AW motif of the cystatin molecule 35, 36.
In the present study, the sequence-structure-function relationships for the wheat cystatin family members are elucidated. All the available sequences of cystatin proteins isolated from wheat were retrieved from NCBI and comparative protein sequence analysis with rice, barley, sorghum and maize cystatins was performed by using different bioinformatics analysis packages. Structures for all the WCs were modeled along with their interactions with papain in order to explore the structural variability and its manifestation at the functional level. This study helped to relate the already known functions of these proteins with their sequences as well as the predicted structures. It also served to better understand the various mechanisms operational in developing this protein family and their implication in plant defense.
Results
Sequence analysis
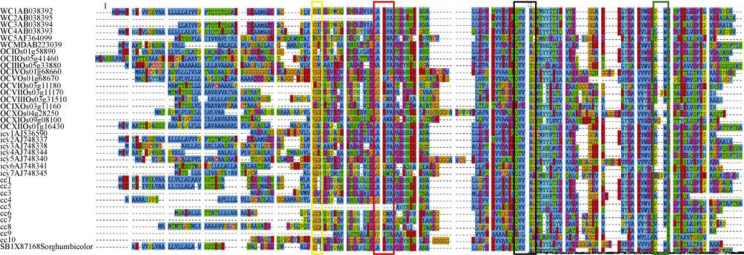
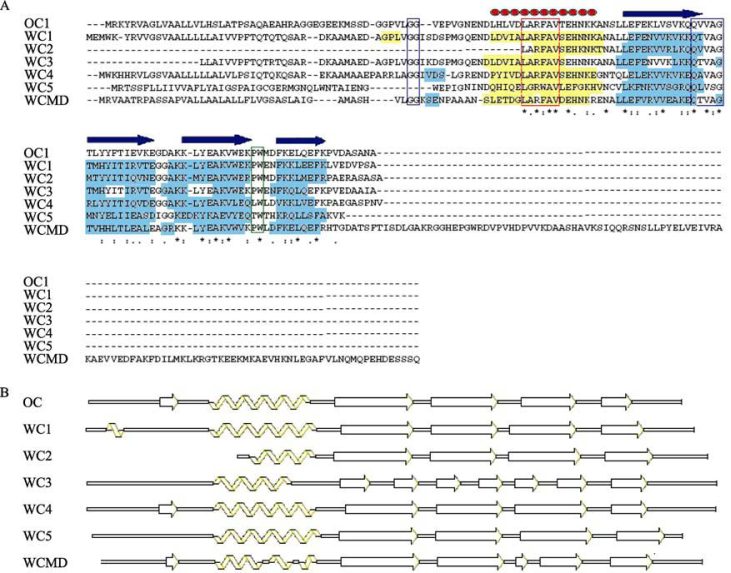
All the sequences were aligned using ClustalW to find out the extent of similarity present among the sequences of the same family, which enjoy a common phylogeny. The four signature motifs of PhyCys are essentially conserved through the sequences with minor alterations (Figure 1). The plant cystatin exclusive, N-terminal motif LARFAV, is fairly conserved, although larger deviations from the consensus are not exceptions and the motif shows a complete absence from cc9. Presence of N-terminal G/GG is also observed to be highly conserved. The functionally indispensable pentapeptide sequence of QXVXG is also observed in all cystatins, although in icy7 it is disrupted by an E (QIVAEG) and in OCIX and cc9 the consensus is almost replaced with a sequence of RFEAG and EHLQE. The fourth conserved motif of P/AW is also present in all cystatins, but in icy7, OCVII, OCIX, OCXI, cc8 and cc9 where interestingly no W is found at the C-terminal end. SignalP predicted the presence of signal peptides in all PhyCys except WC2, WC3, icy1, cc3, cc5, cc7 and cc10. We could not confirm the absence of signal peptide in WC2 owing to the non-availability of its N-terminal end information. Cleavage site prediction ran parallel by TargetP, thus confirming the results. It also predicted most of the cystatins as secretory, except WC3, OCIII and icy1, for which a mitochondrial destination was suggested, but the results were not reliable owing to very low confidence levels of the outputs (Table S1). WC3, like WC2, perhaps does not represent the complete preprotein sequence. ClustalW alignment shows much similarity between N-terminal sequence of WC1, WC3 and WC4. Assignment of WC3 as a mitochondrial protein by TargetP and absence of signal peptide by SignalP were probably due to partially known N-terminal sequence of WC3. Accordingly, WC3, as multiple sequence alignment suggests, could be a secretory protein as well.
Figure 1.
ClustalW alignment of all the cystatin sequences from wheat, rice, barley, sorghum and corn. Few N-terminal and C-terminal amino acid residues are not shown for the clarity of the picture. The conserved signature sequences of the PhyCys are highlighted by enclosing in colored rectangles (Yellow: N-terminal G; Red: LARFAV; Black: QXVXG; Green: P/AW).
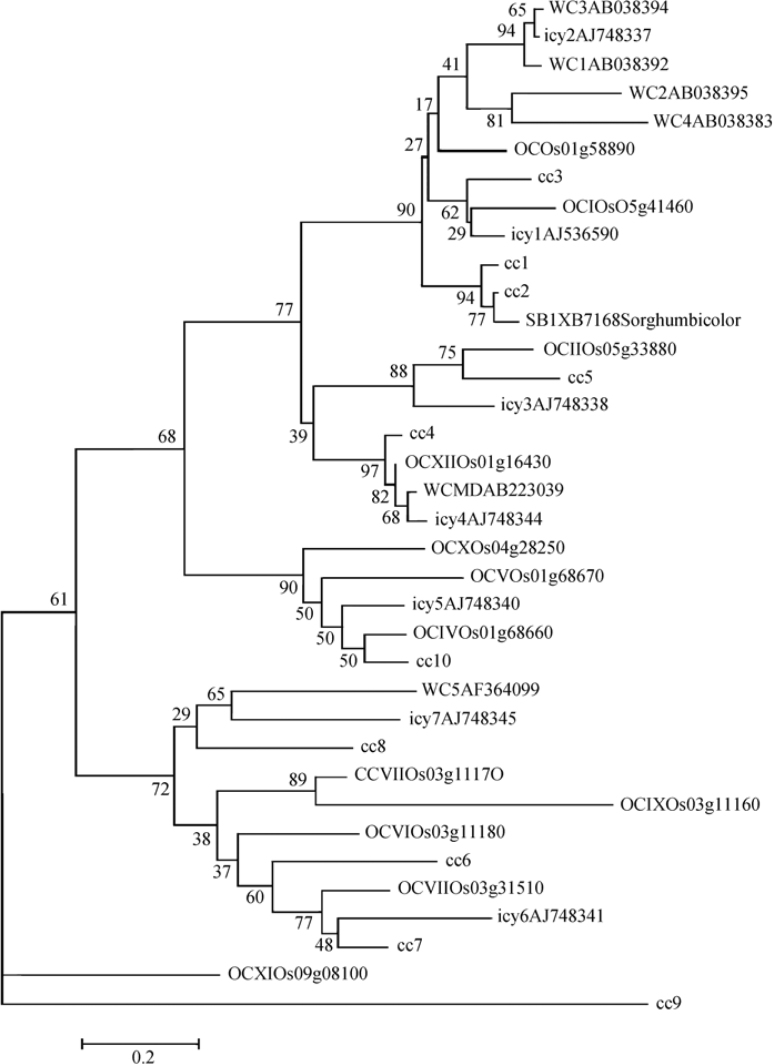
For evaluating the phylogenetic relationships of WCs with other cystatins, an unrooted phylogenetic tree was constructed by neighbor-joining method keeping bootstrap replication size at 1,000 (Figure 2). All the major clusters gave bootstrap values higher than 60. WC1 and WC3 are orthologues of icy2 (bootstrap value 94), while WC5 seems to be an orthologue of icy7 (bootstrap value 65; 57% pairwise similarity). WCMD, OCXII, cc4 and icy4 conspicuously showed orthologous evolution (bootstrap value 97) as is also apparent from their motif analysis and Pfam results (data not shown). WC2 and WC4 are grouped together with 69% pairwise similarity and are thus presumed to be paralogues (bootstrap value 81).
Figure 2.
Unrooted phylogenetic tree of wheat, rice, maize, sorghum and barley cystatins constructed by the neighbor-joining method. Bootstrap values are indicated against each branch. Bootstrap similarity is >50% and the tree was built after 1,000 cycles.
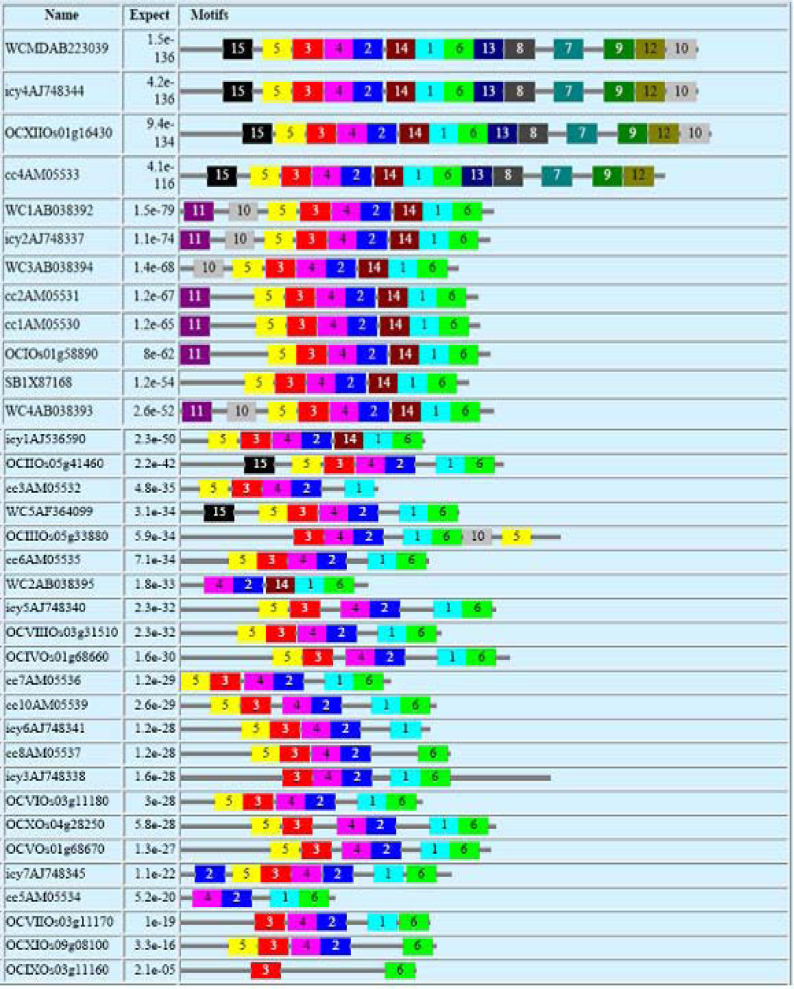
An extensive search of the motifs and their positions was executed by MEME software, which identified several conserved motifs in the cystatin sequences (Figure 3). WC1, WC3, WC4 and icy2 contain similar motifs arranged in identical order. WC2, which is phylogenetically related to WC4, possesses only five motifs present in the same order. The software could not detect any other motif in this cystatin owing to the availability of only the truncated protein in the database. Nevertheless, it can be implied that the full-length protein of WC2 would possess other motifs in the same order. Similarly, WC5 and icy2 also share identical motifs in the same array; however, they did not show any close relationship in phylogenetic analysis. WCMD, OCXII, cc4 and icy4 can also be clearly demarcated by their motif characterization, which is distinct from the rest of the cystatins both in terms of number and order. Motifs numbered 7, 8, 9, 12 and 13 are unique to the four cystatins and thus can be used to identify related proteins in other plant systems as well. Detailed sequence information of the motifs identified by MEME software is provided in Table S2.
Figure 3.
Block diagram representation and distribution of different non-overlapping motifs predicted by MEME software in cystatins of wheat, rice and barley. The sequences of the motifs are given in Table S3.
A close enquiry of the motifs revealed some interesting features. For instance, motif 10, which is present at the C-terminal end of WCMD, OCXII, cc4 and icy4, is placed upstream of motif 5 in cystatins like WC1 and icy4. This might be the consequence of a duplication event of cystatin domain within an ancestral gene, which possibly existed before diversification of rice, wheat and barley. After duplication it probably underwent divergence and lost the sequences that were critical for enzymatic performance. Pfam also identified the extended C-terminal ends of these proteins as cystatin domains, albeit they lack the signature motifs of the family (data not shown). Both motifs 10 and 5 are present at the C-terminal end of OCIII, although these motifs are present upstream of motif 3 in most other sequences. Motif 14, present between the first and second motifs in WC1, WC3, WC4, WCMD, icy2 and several other cystatins (all closely grouped in the phylogenetic tree), is not identified in some other cystatins (e.g., WC5, OCIII, IV, V) where a different stretch of amino acid residues is present. This might have taken place by accumulation of mutations, which does not seem to affect the structure of the final product (discussed below). In some sequences like OCXI and icy6, motif 1 is also absent, and a different stretch of amino acids is present. Such small but significant differences in motif arrangements indicate occurrence of microevolution that might have taken place in the development of this protein family and contributed to broaden its range of specificities.
To further expand our knowledge about the presence of other related motifs involved in regulation of PhyCys, we examined the upstream sequences of the rice and barley cystatin genes. Based on the phylogenetic study of the PhyCys, we conducted promoter analysis of the rice cystatins that were depicted as orthologues of different WCs, as they would give the nearest approximation of the existence of possible motifs and their functions, which might be present in the related WC promoters. Wheat promoter analysis can only be done by isolating and sequencing the promoters, which is not a prerequisite in rice where the whole genome sequence is known and such studies can be carried out by using any annotated or predicted gene. We chose OCI and OCII, the nearest neighbors of WC1-WC4, and OCXII, the orthologue of WCMD. The sequence of barley icy1 gene promoter was also studied, as barley is evolutionarily the closest neighbor of wheat among grasses. Results showed the presence of a number of different motifs that are known to regulate/participate in various phytophysiological processes. Table S3 enlists the major motifs discovered in the positive strand of the promoters of OCI, OCII, OCXII and icy1. Some of the motifs have been excluded due to functional redundancy, unknown function or their presence on the negative strand. Some motifs like CAAT box, TATA box, Skn-1 and Sp1 are present in every promoter, while some exist in only one, including motif IIb in OCI, TGA in OCII, box W1, CAT box and TCA box in OCXII, as well as GCN4 and TC-rich repeats in icy1. Light responsive elements were structurally and functionally redundant and therefore most of them have not been referred to in this paper. Several motifs appear to be responsive to different phytohormones like auxin, methyl jasmonate (a methyl derivative of jasmonic acid), salicylic acid and abscisic acid; the later phytohormones are known to play major roles in the intricately controlled defense responses of the plants. Some motifs can reciprocate to external stress stimuli, such as box W1 to fungal invasion, LTR to low temperatures, MBS to water deficiency, and TC-rich repeats to defense and stress responsiveness. Others like CAT box, GCN4 and Skn-1 are possibly involved in the temporal expression regulation of different cystatins.
Structure analysis
Structures of six known WCs were homology modeled using MOE software taking the NMR structure of oryzacystatin (OC1, PDB accession No. 1eqk) as a template. For each molecule, eleven structures were generated in the database, out of which the minimized average model with maximum score was selected. The energies of the designed structures were minimized using the energy minimization tool of MOE. The following parameters were used for energy minimization: Forcefield = MMFF94X; Gradient = 0.01; Cutoff: On = 8, Off = 10; Solvation: Dielectric = 1, Exterior = 80. The minimized structures were finally saved as .pdb files, which were validated online by PROCHECK software (data not shown).
The NMR structure available for OC1 consists of 102 residues. MOE-designed structures contained 104, 78, 104, 104, 98 and 102 residues for WC1, 2, 3, 4, 5 and WCMD, respectively. The C-terminal half of WCMD could not be modeled because OC1 did not provide any template for it.
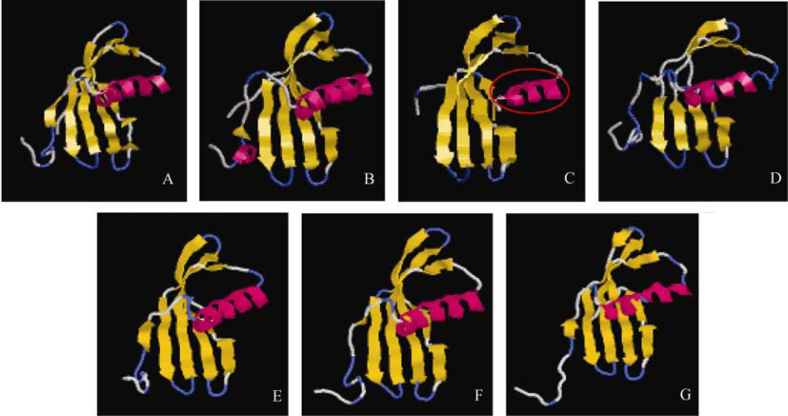
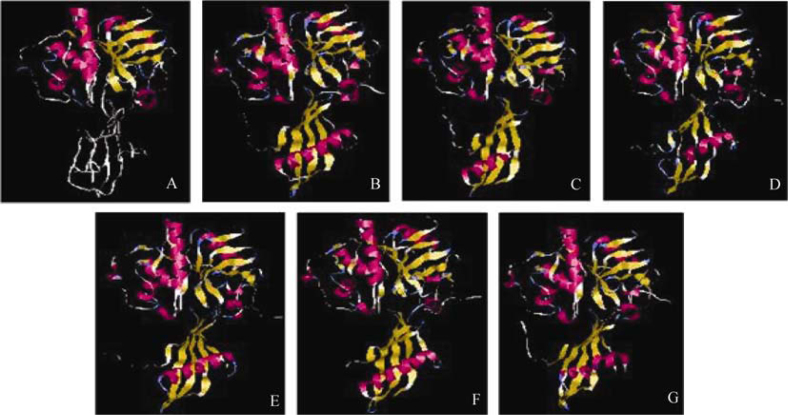
The core structure of the OC1 involving an α-helix and four β-strands is well preserved in all the WCs, with slight differences that do not bring any major changes in the 3D structures (Figure 4). Structurally, WC1, 2, 4 and 5 are almost identical to OC1, with an α-helix that is wrapped around from one side by a somewhat coiled β sheet comprising of four antiparallel β strands. Strands β2 and β5 contain bulges that help in wrapping the helix better. WCMD contains a bulge in β4 strand as well. WC1 has an additional short α-helix at the N-terminal end, while the helix in WCMD contains two kinks, although the helix remains straight. In WC2 the α-helix is shorter as compared to that in OC1 and other WCs (Figure 4C). The structure of WC3 contains two antiparallel β sheets that are formed because of the profound bulges present in the β strands. Thus, the helix in WC3 is more extensively wrapped around by the β sheets. Figure 5A highlights the above-mentioned similarities and differences among the WCs, with respect to the secondary structure of OC1. The hairpin loops L1 and L2, which are known to play an important role in interaction with the cysteine proteases and contain the highly conserved residues of QXVXG and P/AW, possess the same number of residues in WCs as in OC1. As a result, the length of the loops is identical and the amino acid content is also almost alike. Figure 5B gives a diagrammatic representation of the secondary structures of WCs and OC1.
Figure 4.
3D structures of oryzacystatin (A), WC1 (B), WC2 (C), WC3 (D), WC4 (E), WC5 (F) and WCMD (G). Variation in helix is marked with a red circle. 3D structures were constructed by MOE software using oryzacystatin as the template.
Figure 5.
A. Amino acid sequence alignment of oryzacystatin (OC1) and wheat cystatins (WCs). The positions of secondary structures of OC1 are indicated except the first β-strand, which is not shared by most of the WCs. An array of red ovals indicates an α-helix and block arrows indicate β-strands. α-helices of WCs are shown in yellow boxes and β-strands are in cyan boxes. The four signature sequences of PhyCys are shown enclosed in differently colored rectangles. B. Diagrammatic representation of secondary structures of different wheat cystatins. Structure of OC1 is shown for comparison. Structures are not up to scale.
To investigate the interactions of different WCs with cysteine proteinases, stefin B–papain complex (PDB accession No. 1stf) was used as a template to predict the interactions of papain with the six WCs (Figure 6). In this complex, the two hairpin loops form a wedge, which suitably fits into the active site groove of papain due to structural complementarity, by making extensive interactions at the interface that are predominantly hydrophobic. In WC–papain complexes too, the two hairpin loops form a wedge that fits into the groove of papain and make contacts with residues in papain in a fashion quite akin to stefin B. The amino terminal end of stefin B interacts with the unprimed sites of papain running along the surface of the enzyme like a trunk. This prevents the amino terminal residues, particularly the highly conserved Gly present in an improper orientation, to be acted upon by the Cys25 of papain, which is the catalytically active amino acid (37). Although the N-termini of WCs (except WC2) are longer than those of stefin B, they present an analogous structural topology thus rendering them unfit for the proteolytic activity of papain. Table S4 compares the pairwise interactions (<4Å) at the protein–protein interface of 1stf to complexes of WC1 and WC5 with papain. In WC–papain interactions, the major contribution comes from loop 1 harboring the QXVXG pentapeptide. On the other hand, loop 2 makes few or no contacts.
Figure 6.
Protein–protein interactions of different WCs with papain molecule by taking the stefin–papain complex (A) as template, modeled in Swiss-Pdb viewer. B. WC1–papain complex. C. WC2–papain complex. D. WC3–papain complex. E. WC4–papain complex. F. WC5–papain complex. G. WCMD–papain complex. Structural graphics are produced by using Rasmol software version 2.7.3.1.
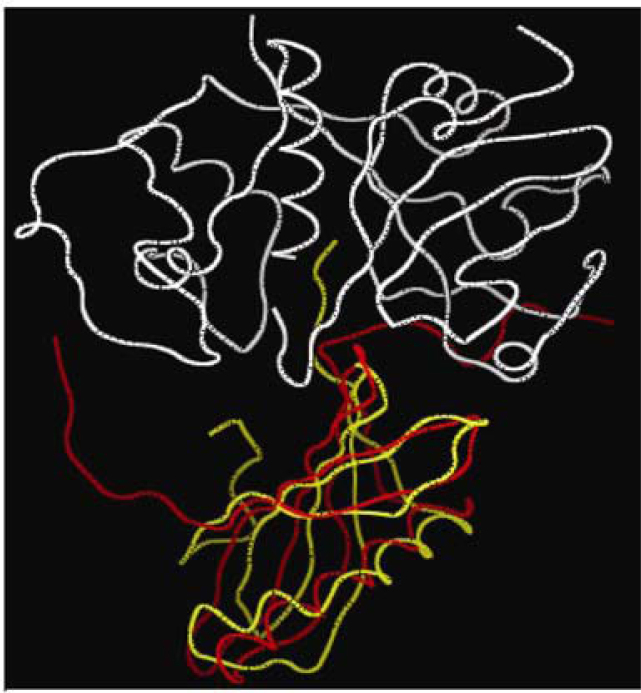
Figure 7 presents the superimposition of WC1–papain complex on 1stf. As is clearly visible, main bodies of the two cystatins are very much alike in structure, although shifts in the backbone are quite distinct. Both amino and carboxy termini of WC1 are longer than that of stefin B. The next level of difference is at the amino acid residue sequence (data not shown). The third level of variation that contributes to the differences in interactions is at the conformational level of the interacting amino acids and their side chains. Table S5 shows the differences of conformations of interacting amino acid residues with respect to their phi, psi and omega values. Thus, WCs interact with papain in an overall similar manner but with minor changes, which can affect their interactions with different cysteine proteinases.
Figure 7.
Superimposition of WC1–papain complex on stefin B–papain complex (1stf). Graphics were generated by MOE software. Color scheme: papain (white), WC1 (red), stefin B (yellow).
Discussion
PhyCys comprise an interesting family of proteins, members of which play critical roles in diverse biological processes occurring in plants. The diversity of roles played by PhyCys can be traced back to the ancestral sequence from which they originated, incorporating various modifications on their way to attain the present stage. In wheat, six different cystatins have been experimentally identified and characterized. Each of them display characteristic features of cystatins but with variations depending on spatial, temporal and conditional stimuli regulating their expressions. The purpose of this work was to find out a correlation between sequence, structure and function of wheat cystatins by a detailed study of cystatin sequences of five important poaceae members, including wheat, rice, barley, sorghum and maize, and creat the structures of WCs by homology modeling.
Multiple sequence alignment highlighted the sequence conservation of amino acid residues among different members of cystatin families in these species. This conservation, however, is concomitant with differences that are sufficient enough to support the variations subsequently reflected at the structural and functional levels. For example, motif LARFAV, which forms the hydrophobic core of the cystatin molecules, and QXVXG and P/AW motifs, which are directly involved in protein-protein interactions, are essentially conserved in all wheat sequences with the exception of WCMD, where LARFAV is replaced by LGRWAV. However, this deviation does not seem to affect either structure or function, as the residues substituted share similar physiochemical properties. In addition to the conservation of signature motifs, residues flanking these motifs also find a considerable similarity that is expected from the members of the same gene family. Yet these motifs may be present or absent in a particular cystatin, or show different patterns of arrangements in different cystatins. Considering the number of cystatin genes annotated in the rice genome (38), one would expect the existence of several more in wheat, whose genome is comprised of three distinct genomes and is roughly 40 times bigger than that of rice. In that case, proteins with much more sequential, structural and functional diversity would be expected. However, this hypothesis needs experimental verification along with a genome-wide search for wheat cystatins requiring whole genome sequence information, determination of which still remains an uphill task given the huge size and ploidy of the wheat genome.
Phylogenetic tree results outline the development of cystatins in wheat, rice, barley, maize and sorghum; many of them exhibited orthologous and paralogous relations with each other. This observation indicates that this gene family of proteins is strictly conserved and has evolved from some ancestral grass species undergoing speciation-ensuing duplication and diversification events. Constant exposure to attacks by various pests and diseases could be the reason for the existing sequence and structure variability of the proteins (38). Motif analysis also communicates the same fundamental necessity for the development of this gene family. Motifs containing the signature sequences are either well conserved or are having substitutions that do not change their activity, while the ones that do not have a direct impact on the active site contain altered residues and are clearly the outcome of accumulation of mutations or have been subjected to rearrangements. Most of the cystatins are recognized as secretory proteins as they contain signal peptides that direct them to endoplasmic reticulum from where they are finally targeted to extracellular locations. Here they can neutralize their cognate cysteine proteinases, which may be endogenous or exogenous, and thus participate in developmental or defense responses. Different corn cystatins showed distinct spatiotemporal patterns as well as kinetics. They were not only developmentally regulated, but also affected by cold (cc8 and cc9) and water deficiency (cc2, cc3, cc4, cc5 and cc9) (39).
The structural analysis of WCs also delivers the same message as is received from their sequence study. The 3D structures of the cystatins are well conserved throughout the species, but different cystatins display different specificities and inhibitory functions for different proteinases. For instance, the Ki value of OCI for cathepsinH is higher than for papain. Thus OCI is a papain inhibitory type of cystatin as against the cathepsinH inhibitory type of OCII, for which Ki value for papain is higher than for cathepsinH (13). Seven barley cystatins show discrete activities toward different cysteine proteases (40). Thus cystatins from the same species exhibit different inhibitory spectra. The bottom line is: different cystatins possess different inhibitory capacities (both in terms of specificity and activity). This is reflected in their sequences as well as structures. However, whatever variations are found at the structural and functional levels, cystatins are found to conserve the basic 3D structural fold that so clearly distinguishes them as cystatins. Such pattern of conservation is also found in the sequences of PhyCys in the form of signature motifs.
In the cystatin–cysteine proteinase interaction, the two hairpin loops and the amino terminus are the major players. Yet the role of N-terminus remains ambiguous. N-terminal 21 residues of OC1 are not essential for its papain inhibitory function (41). Contradictorily, involvement of NH2-terminal region of OC1 was found to be involved in cysteine proteinase inhibition (42). Modeling studies using cowpea cystatin revealed presence of five amino terminal residues responsible for the stabilization of enzyme–inhibitor complex by providing a substantial fraction of hydrophobic interaction at the interface (43). A pineapple cystatin, AcCYS1, contains an extended N-terminal trunk (NTT) of 63 residues rich in Ala and Glu. A signal peptide preceding the NTT is processed in vitro by microsomal membranes giving rise to a 27 kDa species. The proteolytic removal of the NTT results in the decrease of the inhibitory potency of Ac-CYS1 against fruit bromelain during fruit ripening to increase tissue proteolysis, softening and degradation (44). WC–papain complexes also envisage significant contribution of N-terminal residues, particularly the highly conserved Gly. Unlike the N-terminus, the role of the first binding loop in the inhibition of papain-like proteinases is well established. Randomly selected mutants of soyacystatins binding to papain from a library by phage display invariantly had the QVVAG sequence in the first binding loop (45). However, using the same technique, two variants, DVVSA and NTSSA, were found with low affinity for papain (46). In addition, mutations at the central Val residue of the QXVXG region have moderate effects on activity (41). In fact, the Val→Gly mutant was as active as wild oryzacystatin. Several hypervariable sites have been located at strategic positions on the protein: on each side of the conserved glycine residues in the N-terminal trunk, within the first and second inhibitory loops entering the active site of target enzymes, and surrounding the LARFAV motif. These hypervariable sites have been assumed to be positively selected and thus implicated in fuctional diversity (47). Icy7 contains two aberrations in its sequence: an E in the QXVX(E)G region and absence of W in the second loop. It shows no inhibitory activity against papain, cathepsinB or cathepsinH. It also fails to inhibit the fungal growth of two phytopathogenic fungi, Botrytis cineria and Fusarium oxysporum (40). Presence of E perhaps disrupts the hydrophobic interactions at the interface of icy7 and papain. The QXVXG sequence in WCs is highly conserved and plays an important part during interaction with papain. The importance of the W residue in the second loop has also been confirmed by phage display experiments with soyacystatin (45). The second hairpin contributes ~13% of the total energy of complex formation as compared to the 40%-60% done by the first loop between chicken cystatin and papain (48). In 1stf, the second binding loop is also of minor importance. In WCs, the loop is placed wider with respect to the papain active site crevice and form very few <4Å contacts.
The functional diversity of cystatins can be explained by the sequence-structure variations, and is also controlled at the expression levels. Kuroda et al. (28) discovered distinct spatial and temporal expressions for WC1, 2, 3 and 4. While spatiotemporal profiles were similar for two different cystatins, it varied in the amount expressed. WC5 expression has been found to be restricted to the maturation stage of grain development (29). WCMD is an inducible multidomain cystatin that inhibits the growth of the snow mold fungus M. nivale (30). Interestingly, the promoter of OCXII, which is an orthologue of WCMD, contains a fungal elicitor responsive element. This implies the presence of similar motif(s) in WCMD. However, no cold responsive element was found in OCXII promoter, albeit it is present in OCI and icy1. The icy1 mRNA expression in vegetative tissues increases in response to anerobiosis, dark and cold shock (1). Indeed, the motifs that can respond to low temperatures and anoxia were identified in icy1 by the program. Given the evolutionary proximity of barley to wheat, one can expect the presence of similar motifs in wheat cystatin orthologues (WC1-4). Conclusively, the impetus that drives the evolution of a protein family like cystatin functions at two levels. Firstly, it is regulated at the gene expression level, which controls the spatiotemporal expression as well as the amounts to be expressed. Secondly, it is at the level of the minor deviations in the structural components of the genes, which governs the specificities and activities of the cystatins.
Conclusion
PhyCys are members of a multigene family and contain a number of different cystatins in the grass species we studied (wheat, barley, rice, maize and sorghum). Presence and maintenance of many cystatin genes explains the wide variety of roles they play in different processes ranging from counteracting biotic and abiotic stresses to regulating endogenous protein turnover. PhyCys are inhibitors of cysteine proteinases that are used as potent weapons by phytopathogens and pests for invasion and colonization. In such a system of interacting proteins, each partner coevolves in response to the changes occurring in the cognate molecule (49). In order to post an effective attack, pathogens select suitable mutations that can augment their pathogenicity. Plants need to counterbalance it by evolving their own genes and this leads to the ever diversification of this family of proteins. Cystatins in each species have conserved residues particularly the ones involved in maintaining structure and related biochemical function. Despite the conservation of the sequences, sufficient variations are also present, which do not bring gross alterations in structure and function but do introduce changes in specificity and inhibitory activity. This is desirable for improving the repertoire of cysteine proteinase inhibitors in plants against offenders. The overall structure is similar for different cystatins. However, variations can be explained by occurrence of slight shifts in the backbone, changes in amino acid residues and their conformations. Activity of the cystatins is not merely controlled at the sequence and structure levels, but also affected by the regulatory elements (e.g., promoters) of the genes, which ultimately decide when and where to express.
Materials and Methods
Sequence analysis
Cystatin sequences for wheat, sorghum, maize and barley were obtained from the publicly available NCBI database (http://www.ncbi.nlm.nih.gov/). Sequences for rice (Oryza sativa L. ssp. japonica) were extracted from the International Rice Genome Sequencing Project (IRGSP) at The Institute for Genomic Research (TIGR; http://www.tigr.org). Gen-Bank accession numbers of all the phytocystatins are given in Table 1. All protein sequences were aligned by employing EBI tools ClustalW (http://www.ebi.ac.uk/clustalw) (50) with default parameters. Analysis of conserved motifs was performed by MEME (Multiple Em for Motif Elicitation) software version 3.5.4 (http://meme.sdsc.edu) (51) using minimum and maximum motif width of 8 and 15 residues respectively, and a maximum number of 15 motifs, keeping the rest of the parameters at default. Phylogenetic analysis of the sequences was done by MEGA (Molecular Evolutionary Genetic Analysis) software version 4.0 (52), using neighbor-joining method with complete deletion and Poisson correction settings. Signal peptide analysis was executed by using the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/signalP) (53). Cellular localizations for the various cystatins were predicted by TargetP (http://www.cbs.dtu.dk/services/TargetP/) (54), which was also programmed for cleavage site prediction simultaneously. Rice and barley promoter sequences were examined using plantCARE database (http://bioinformatics.pbs.ugent.be/webtools/plantcare/html/) (55). A stretch of 1,000 bases (961 bases in icy1) upstream of the start site was considered for analysis.
Table 1.
Accession numbers of different cystatins studied
| Species | Cystatin | Accession No. | No. of amino acids |
|---|---|---|---|
| Wheat (Triticum aestivum) |
WC1 | AB038392 | 142 |
| WC2 | AB038395 | 78 | |
| WC3 | AB038394 | 125 | |
| WC4 | AB038393 | 142 | |
| WC5 | AF364099 | 128 | |
| WCMD | AB223039 | 243 | |
| Rice (Oryza sativa) |
OCI | Os01g58890 | 139 |
| OCII | Os05g41460 | 156 | |
| OCIII | Os05g33880 | 150 | |
| OCIV | Os01g68660 | 158 | |
| OCV | Os01g68670 | 147 | |
| OCVI | Os03g11180 | 113 | |
| OCVII | Os03g11170 | 117 | |
| OCVIII | Os03g31510 | 123 | |
| OCIX | Os03g11160 | 114 | |
| OCX | Os04g28250 | 156 | |
| OCXI | Os09g08100 | 120 | |
| OCXII | Os01g16430 | 250 | |
| Barley (Hordeum vulgare) |
icy1 | AJ536590 | 107 |
| icy2 | AJ748337 | 140 | |
| icy3 | AJ748338 | 145 | |
| icy4 | AJ748344 | 169 | |
| icy5 | AJ748340 | 147 | |
| icy6 | AJ748341 | 123 | |
| icy7 | AJ748345 | 124 | |
| Maize (Zea mays) |
cc1 | AM05530 | 135 |
| cc2 | AM05531 | 134 | |
| cc3 | AM05532 | 97 | |
| cc4 | AM05533 | 226 | |
| cc5 | AM05534 | 68 | |
| cc6 | AM05535 | 116 | |
| cc7 | AM05536 | 97 | |
| cc8 | AM05537 | 127 | |
| cc9 | AM05538 | 78 | |
| cc10 | AM05539 | 120 | |
| Sorghum (Sorghum bicolor) |
SB1 | X87168 | 130 |
Structure modeling and analysis
For constructing the structures of WCs, a template for homology modeling was searched with BLAST program on the Protein Data Bank (www.rcsb.org/pdb/) (56). Template structure was selected with a cutoff sequence identity of <40%. The 3D structures of WCs were modeled using MOE (Molecular Operating Environment) software version 2006.08 (Chemical Computing Group, Inc.). Secondary structure components of the cystatin sequences were analyzed using SWISS-PDB viewer (http://spdbv.vital-it.ch) and Geno3D (http://geno3d-pbil.ibcp.fr) tools. Model consistency and viability were appraised by PROCHECK software available online (http://www.ebi.ac.uk./Thorton/software.html) for protein structure verification (57). The protein–protein interactions of different WCs with papain were predicted by superimposing their structures onto the 3D structure of the stefin B–papain complex (PDB accession No. 1stf) using the magic fit option available in the program of SWISS-PDB viewer (58). Analysis of the modeled structures was performed using the RasMol version 2.7.3.1 (59).
Authors’ contributions
SD designed the study, collected the datasets, carried out sequence and structural analyses, interpreted the results to find out the structure-function relationships and drafted the manuscript. VKS contributed in software tool usage and in applying various parameters in model building and protein-protein interactions. SSM participated in the design and coordination of the study, helped in structure-function relation analysis and revised the manuscript. AK conceived the study, participated in its design and coordination and revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Supplementary Material
Tables S1-S5
References
- 1.Gaddour K. A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol. Biol. 2001;45:599–608. doi: 10.1023/a:1010697204686. [DOI] [PubMed] [Google Scholar]
- 2.Barrett A.J. Cysteine proteinase inhibitors of the cystatin superfamily. In: Barrett A.J., Salvesen G., editors. Proteinase Inhibitors. Elsevier Science Publishers; Amsterdam, Netherlands: 1986. pp. 515–569. [Google Scholar]
- 3.Abe M. Corn cystatin I expressed in Escherichia coli: investigation of its inhibitory profile and occurrence in corn kernels. J. Biochem. 1994;116:488–492. doi: 10.1093/oxfordjournals.jbchem.a124551. [DOI] [PubMed] [Google Scholar]
- 4.Brown W.N., Dziegielewska K.M. Friends and relations of the cystatin superfamily—new members and their evolution. Protein Sci. 1997;6:5–12. doi: 10.1002/pro.5560060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turk V., Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 6.Barrett A.J. Nomenclature and classification of the proteins homologous with the cysteine proteinase inhibitor chicken cystatin. Biochem. J. 1986;236:312. doi: 10.1042/bj2360312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett A.J. The cystatins: a new class of peptidase inhibitors. Trends Biochem. Sci. 1987;12:193–196. [Google Scholar]
- 8.Martinez M. Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteases. FEBS Lett. 2007;581:2914–2918. doi: 10.1016/j.febslet.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 9.Margis R. Structural and phylogenetic relationships among plant and animal cystatins. Arch. Biochem. Biophys. 1998;359:24–30. doi: 10.1006/abbi.1998.0875. [DOI] [PubMed] [Google Scholar]
- 10.Pernas M. Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett. 2000;467:206–210. doi: 10.1016/s0014-5793(00)01157-1. [DOI] [PubMed] [Google Scholar]
- 11.Botella M.A. Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol. 1996;112:1201–1210. doi: 10.1104/pp.112.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe M. Corn kernel cysteine proteinase inhibitor as a novel cystatin superfamily member of plant origin. Molecular cloning and expression studies. Eur. J. Biochem. 1992;209:933–937. doi: 10.1111/j.1432-1033.1992.tb17365.x. [DOI] [PubMed] [Google Scholar]
- 13.Kondo H. Two distinct cystatin species in rice seeds with different specificities against cysteine proteinases. Molecular cloning, expression, and biochemical studies on oryzacystatin-II. J. Biol. Chem. 1990;265:15832–15837. [PubMed] [Google Scholar]
- 14.Abe M. Molecular cloning of a cysteine proteinase inhibitor of rice (oryzacystatin). Homology with animal cystatins and transient expression in the ripening process of rice seeds. J. Biol. Chem. 1987;262:16793–16797. [PubMed] [Google Scholar]
- 15.Yang A.H., Yeh K.W. Molecular cloning, recombinant gene expression, and antifungal activity of cystatin from taro (Colocasia esculenta cv. Kaosiung no.1) Planta. 2005;221:493–501. doi: 10.1007/s00425-004-1462-8. [DOI] [PubMed] [Google Scholar]
- 16.Martinez M. The strawberry gene Cyf1 encodes a phytocystatin with antifungal properties. J. Exp. Bot. 2005;56:1821–1829. doi: 10.1093/jxb/eri172. [DOI] [PubMed] [Google Scholar]
- 17.Telang M. Bitter gourd proteinase inhibitors: potential growth inhibitors of Helicoverpa armigera and Spodoptera litura. Phytochemistry. 2003;63:643–652. doi: 10.1016/s0031-9422(03)00239-5. [DOI] [PubMed] [Google Scholar]
- 18.Hines M.E. Isolation and partial characterization of soybean cystatin cysteine proteinase inhibitor of coleopteran digestive proteolytic activity. J. Agric. Food Chem. 1991;39:1515–1520. [Google Scholar]
- 19.Bolter C.J. Methyl jasmonate induces papain inhibitor(s) in tomato leaves. Plant Physiol. 1993;103:1347–1353. doi: 10.1104/pp.103.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haq S.K. Protein proteinase inhibitor genes in combat against insects, pests and pathogens: natural and engineered phytoprotection. Arch. Biochem. Biophys. 2004;431:145–159. doi: 10.1016/j.abb.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Atkinson H.J. Engineering plants for nematode resistance. Annu. Rev. Phytopathol. 2003;41:615–639. doi: 10.1146/annurev.phyto.41.052002.095737. [DOI] [PubMed] [Google Scholar]
- 22.Delledonne M. Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol. Breed. 2001;7:35–42. [Google Scholar]
- 23.Gutierrez-Campos R. The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 1999;17:1223–1226. doi: 10.1038/70781. [DOI] [PubMed] [Google Scholar]
- 24.Valdés-Rodríguez S. Cloning of a cDNA encoding a cystatin from grain amaranth (Amaranthus hypochondriacus) showing a tissue-specific expression that is modified by germination and abiotic stress. Plant Physiol. Biochem. 2007;45:790–798. doi: 10.1016/j.plaphy.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Belenghi B. AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur. J. Biochem. 2003;270:2593–2604. doi: 10.1046/j.1432-1033.2003.03630.x. [DOI] [PubMed] [Google Scholar]
- 26.Sugawara H. Is a cysteine proteinase inhibitor involved in the regulation of petal wilting in senescing carnation (Dianthus caryophyllus L.) flowers? J. Exp. Bot. 2002;53:407–413. doi: 10.1093/jexbot/53.368.407. [DOI] [PubMed] [Google Scholar]
- 27.Solomon M. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–444. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda M. Molecular cloning, characterization, and expression of wheat cystatins. Biosci. Biotechnol. Biochem. 2001;65:22–28. doi: 10.1271/bbb.65.22. [DOI] [PubMed] [Google Scholar]
- 29.Corr-Menguy F. Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Mol. Biol. 2002;50:687–698. doi: 10.1023/a:1019906031305. [DOI] [PubMed] [Google Scholar]
- 30.Christova P.K. A cold inducible multidomain cystatin from winter wheat inhibits growth of the snow mold fungus Microdochium nivale. Planta. 2006;223:1207–1218. doi: 10.1007/s00425-005-0169-9. [DOI] [PubMed] [Google Scholar]
- 31.Kiyosaki T. Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS J. 2007;274:1908–1917. doi: 10.1111/j.1742-4658.2007.05749.x. [DOI] [PubMed] [Google Scholar]
- 32.Nagata K. Three-dimensional solution of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry. 2000;39:14753–14760. doi: 10.1021/bi0006971. [DOI] [PubMed] [Google Scholar]
- 33.Machleidt W. Mechanism of inhibition of papain by chicken egg white cystatin. Inhibition constants of N-terminally truncated forms and cyanogen bromide fragments of the inhibitor. FEBS Lett. 1989;243:234–238. doi: 10.1016/0014-5793(89)80135-8. [DOI] [PubMed] [Google Scholar]
- 34.Bode W. The 2.0 Å X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 1988;7:2593–2599. doi: 10.1002/j.1460-2075.1988.tb03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nycander M., Bjork I. Evidence by chemical modification that tryptophan-104 of the cysteine proteinase inhibitor chicken cystatin is located in or near the proteinase-binding site. Biochem. J. 1990;271:281–284. doi: 10.1042/bj2710281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrahamson M. Identification of the probable inhibitory reactive sites of the cysteine proteinase inhibitors, human cystatin C and chicken cystatin. J. Biol. Chem. 1987;262:9688–9694. [PubMed] [Google Scholar]
- 37.Stubbs M.T. The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990;9:1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez M. Comparative phylogenetic analysis of cystatin gene families from arabidopsis, rice and barley. Mol. Genet. Genomics. 2005;273:423–432. doi: 10.1007/s00438-005-1147-4. [DOI] [PubMed] [Google Scholar]
- 39.Mossonneau A. Maize cystatins respond to developmental cues, cold stress, and drought. Biochim. Biophys. Acta. 2005;1729:186–199. doi: 10.1016/j.bbaexp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Abraham Z. Structural and functional diversity within the cystatin gene family of Hordeum vulgare. J. Exp. Bot. 2006;57:4245–4255. doi: 10.1093/jxb/erl200. [DOI] [PubMed] [Google Scholar]
- 41.Arai S. Papain-inhibitory activity of oryzacystatin, a rice seed cysteine proteinase inhibitor, depends on the central Gln-Val-Val-Ala-Gly region conserved among cystatin superfamily members. J. Biochem. 1991;109:294–298. [PubMed] [Google Scholar]
- 42.Urwin P.E. Involvement of the NH2-terminal region of oryzacystatin-1 in cysteine proteinase inhibition. Protein Eng. 1995;8:1303–1307. doi: 10.1093/protein/8.12.1303. [DOI] [PubMed] [Google Scholar]
- 43.Aguiar J.M. Molecular modeling and inhibitory activity of cowpea cystatin against bean bruchid pests. Proteins. 2006;63:662–670. doi: 10.1002/prot.20901. [DOI] [PubMed] [Google Scholar]
- 44.Neuteboom L.W. An extended AE-rich N-terminal trunk in secreted pineapple cystatin enhances inhibition of fruit bromelain and is posttranslationally removed during ripening. Plant Physiol. 2009;151:515–527. doi: 10.1104/pp.109.142232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koiwa H. Phage display selection of hairpin loop soyacystatin variants that mediate high affinity inhibition of a cysteine proteinase. Plant J. 2001;27:383–391. doi: 10.1046/j.1365-313x.2001.01104.x. [DOI] [PubMed] [Google Scholar]
- 46.Melo F.R. Use of phage display to select novel cystatins specific for Acanthoscelides obtectus cysteine proteinases. Biochim Biophys. Acta. 2003;1651:146–152. doi: 10.1016/s1570-9639(03)00264-4. [DOI] [PubMed] [Google Scholar]
- 47.Kiggundu A. Modulating the protease inhibitory profile of a plant cystatin by single mutations at positively selected amino acid sites. Plant J. 2006;48:403–413. doi: 10.1111/j.1365-313X.2006.02878.x. [DOI] [PubMed] [Google Scholar]
- 48.Machleidt W. Molecular mechanism of inhibition of cysteine proteinases by their inhibitors: kinetic studies with natural and recombinant variants of cystatins and stefins. Biomed. Biochim. Acta. 1991;50:613–620. [PubMed] [Google Scholar]
- 49.Martinez M., Diaz Z. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 2008;8:198. doi: 10.1186/1471-2148-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higgins D. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey T.L., Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press; Menlo Park, USA: 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- 52.Tamura K. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 53.Bendtsen J.D. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 54.Emanuelsson O. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 55.Lescot M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berman H.M. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laskowski R.A. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 58.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 59.Sayle R., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S5