Abstract
In the post-genomic era, various computational methods that predict protein-protein interactions at the genome level are available; however, each method has its own advantages and disadvantages, resulting in false predictions. Here we developed a unique integrated approach to identify interacting partner(s) of Semaphorin 5A (SEMA5A), beginning with seven proteins sharing similar ligand interacting residues as putative binding partners. The methods include Dwyer and Root-Bernstein/Dillon theories of protein evolution, hydropathic complementarity of protein structure, pattern of protein functions among molecules, information on domain-domain interactions, co-expression of genes and protein evolution. Among the set of seven proteins selected as putative SEMA5A interacting partners, we found the functions of Plexin B3 and Neuropilin-2 to be associated with SEMA5A. We modeled the semaphorin domain structure of Plexin B3 and found that it shares similarity with SEMA5A. Moreover, a virtual expression database search and RT-PCR analysis showed co-expression of SEMA5A and Plexin B3 and these proteins were found to have co-evolved. In addition, we confirmed the interaction of SEMA5A with Plexin B3 in co-immunoprecipitation studies. Overall, these studies demonstrate that an integrated method of prediction can be used at the genome level for discovering many unknown protein binding partners with known ligand binding domains.
Key words: domain-domain interaction, semaphorin, plexin, protein interaction prediction
Introduction
Protein-protein interactions play a major role during normal cellular functions or pathological diseases. These protein-protein interactions are physically mediated by 4–7 amino acid residues (ligand binding regions or binding signatures) at the interface of the binding domain, while the other residues are involved in bringing ligand binding residues structurally close 1., 2., 3., 4., 5., 6., 7., 8., 9.. Binding signatures of interacting proteins can be identified from their known structural profile 10., 11. or using phage-display peptide library selection 1., 12., 13.. Since the structure of most of the proteins is unknown, identifying binding signatures using a structural profile is not easy. However, using in vivo phage-display peptide library screening, random seven amino acid peptides, expressed as part of a coat protein gene on the surface of bacteriophages, can be affinity selected against the receptor on the membrane surface of organ-specific endothelial cells by intravenous injection of the phages into mice. These affinity selected peptides are the binding signatures of proteins that interact with the receptor(s) on endothelial cells (1). Therefore, we can predict the protein-protein interactions underlying these binding signatures or peptides.
Recently, we identified Semaphorin 5A (SEMA5A) as a metastasis-associated protein by bioinformatics analysis using binding signatures from phage-display assay and in vivo selection (1). In order to understand the biological significance of SEMA5A expression, it is essential to identify its binding partners or receptor. There are various methods available that are being applied individually to predict protein-protein interactions at the genome level, showing their relevance to predictions (14., 15., 16.). However, each method has its own disadvantages leading to incorrect predictions. Among the experimental procedures, it is already known that the yeast two-hybrid system gives a significant number of false positives, while mass spectrometry is expensive and time consuming 17., 18.. On the other hand, computational methods have their own disadvantages when used alone and only a few methods provide a platform for biologists to predict single protein-protein interactions. In this study, we applied computational methods to predict the interacting partners of SEMA5A using ligand binding signatures screened from an in vivo phage-display peptide library assay from our previous report (1).
To our knowledge, for the first time we have applied ligand binding peptides combined with other protein interaction prediction methods to identify binding partners of SEMA5A. Our approach involved prediction of SEMA5A binding partners from a set of proteins sharing the binding signature, NAFTPDY, identified using phage-display screening combined with bioinformatics analysis (1). The procedure utilizes integration of various useful methodologies of protein-protein interactions already elucidated 14., 15., 16., 19., 20.. Currently, we have used six different theoretical methods for predicting the interacting partners of SEMA5A. At each step, proteins that fail to satisfy the criteria were dropped. This integrative approach for the prediction of protein-protein interactions can be expanded to identify binding partners for more proteins whose binding signatures are already known.
Results
Peptide complementarity predicts protein interactions
Root-Bernstein and Dillon (RBD) theory suggests that protein receptors will contain ligand-like sequences within the binding (extracellular) region, if the peptide is self-complementary (21). A well-known example is glucagon and glucagon receptor sharing 80% similar residues in their ligand binding regions. Similarly, insulin and insulin receptor and gastringastrin binding protein interactions have also been shown (21). The ligand binding sites of proteins can be identified by phage-display peptide library screening (1). In our previous publication, we predicted Plexin B3, Neuropilin-2 (NRP2), Integrin alpha-X and -E, NETO1, Desmocolin 2 and Ephrin B2 along with SEMA5A to contain NAFTPDY ligand binding peptide in their extracellular binding regions through a combination of phage-display peptide library and in silico analyses (1). Since these proteins are predicted to share the same putative ligand binding peptide and the Semaphorin family of proteins is known to undergo homophilic inteaction (self-aggregation), we selected these proteins to identify the binding partner of SEMA5A based on RBD theory of protein evolution. For instance, it is interesting to note that the ligand binding regions of both SEMA5A and Plexin B3 are found in the semaphorin (sema) domain of the proteins. It is already known that semaphorins and plexins interact with each other through the sema domain 22., 23., 24., 25.. This suggests that any of the seven proteins may interact with SEMA5A at their putative ligand binding regions. Hence, these proteins served as a pool of proteins for further study to identify SEMA5A interacting partners.
Functional association of proteins with SEMA5A
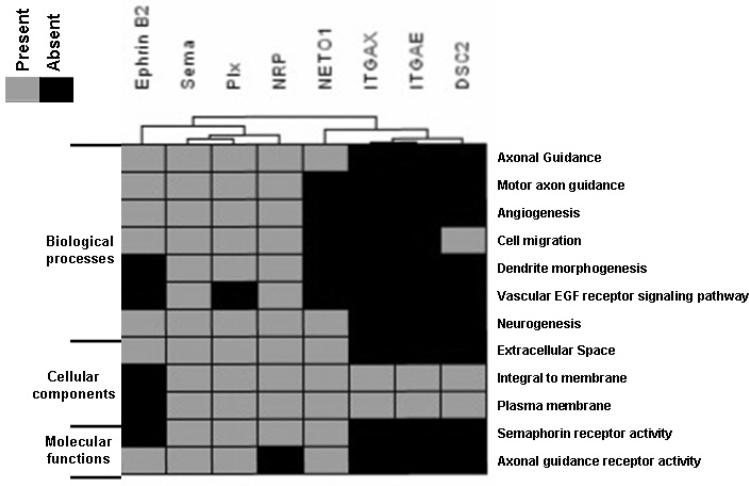
Proteins associated with the same functions and biological processes may interact with each other (26). We tried to identify potential relationships of SEMA5A with proteins sharing the putative binding signature, NAFTPDY, as a function of their co-occurrence in the Gene Ontology (GO) database. Therefore, we searched the functions of the proteins associated with GO terms for semaphorins using the GO database and Medline. The pattern of functional co-occurrence was generated as described in the Methods section. The most significant biological processes, cellular components and molecular functions for the co-occurrence of these proteins are shown in Figure 1. From the GO database, we have selected twelve terms associated with the Semaphorin family of proteins. Among these terms, seven are from biological processes, three are from cellular components and two are from molecular functions. This is a representational analysis of GO terms for each protein for their known functions and biological processes associated with SEMA5A. This way of identifying relationships is critical to predict the shared functions of these proteins with SEMA5A. The frequency of co-occurrence of the other proteins and their association rules in relation to semaphorins were calculated. As per the association rule, the minimum confidence at which Plexin B3 and NRP2 co-occur with semaphorins is 91.67% (Table 1), respectively, whereas other proteins occur at a confidence lower than the 75% arbitrary cut-off. Similarly, modeling of known interacting partners SEMA3C and NRP2 identified using the NAFTPDY peptide can be validated from this functional association. Furthermore, the hierarchical clustering shows that except for Plexin B3 and NRP2, all the other proteins are clustered away from SEMA5A. These results suggest that Plexin B3 and NRP2 may be interacting partners for SEMA5A. Therefore, Plexin B3 and NRP2 were considered for further analyses.
Fig. 1.
Similar pattern of protein functions with SEMA5A. Functional association of proteins with SEMA5A was modeled using GO terms associated with SEMA5A. If a protein shares the GO terms with SEMA5A, it is scored as 1 or else 0. The proteins are clustered using hierarchical clustering based on the score.
Table 1.
Prediction of functional association between SEMA5A and listed proteins using functional terms from GO database*
| Proteins (X) | Freq(Sema U X) | Freq(X) | Conf(Sema→X) | Conf(X→Sema) | Min. Confidence |
|---|---|---|---|---|---|
| Plexin B3 | 91.67 | 91.67 | 91.67 | 100 | 91.67 |
| NRP2 | 91.67 | 91.67 | 91.67 | 100 | 91.67 |
| Ephrin B2 | 58.33 | 58.33 | 58.33 | 100 | 58.33 |
| NETO1 | 58.33 | 58.33 | 58.33 | 100 | 58.33 |
| DSC2 | 25 | 25 | 25 | 100 | 25 |
| ITGAE | 16.67 | 16.67 | 16.67 | 100 | 16.67 |
| ITGAX | 16.67 | 16.67 | 16.67 | 100 | 16.67 |
All the values represented are in percent. Freq(Sema U X) is the frequency of co-occurrence of all the functional terms between SEMA5A and given proteins (X). Conf(Sema→X) or Conf(X→Sema) represents the confidence of the functional association between SEMA5A and other proteins or vice versa. “Min. Confidence” is the minimal confidence valued between Conf(Sema→X) and Conf(X→Sema). NRP2, neuropilin-2; NETO1, neuropilin and tolloid-like 1; DSC2, desmocolin 2; ITGAE, integrin alpha-E, ITGAX, integrin alpha-X.
Association of protein function based on literature
Proteins cited in the same literature or text may be involved in the same function. One suitable technique for screening protein-protein interactions is text mining (26). We constructed a similarity score using a cosine similarity measure that quantitatively measures the co-occurrences of selected proteins with SEMA5A in searches using Google Scholar and PubMed. It gives an index calculated between normalized vector pairs (SEMA5A and selected proteins), which implies whether or not similarity exists between SEMA5A and a given protein but not the magnitude of the similarity. However, the magnitude of interaction has already been demonstrated using hierarchical clustering as shown in Figure 1. Even though cosine similarity could be used for protein interaction studies, it becomes difficult to fit the measure to suit proteins containing few citations. Therefore, we have eliminated proteins having less than 500 hits from Google Scholar or PubMed and are clustered away from SEMA5A. The results show that Plexin B3 and NRP2 have cosine similarity of 0.98, which is close to 1. The closer the cosine similarity measure to 1, the more often the proteins coexist in the literature. This in turn suggests that Plexin B3 and NRP2 may interact with SEMA5A.
Hydropathic profile predicts SEMA5A interacting proteins
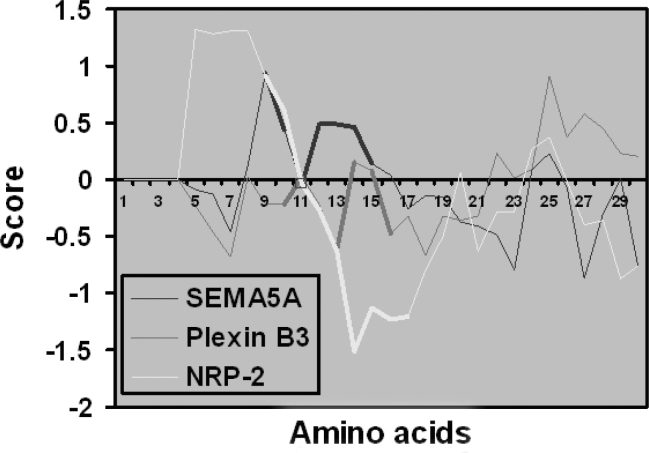
According to molecular recognition theory, the ligand binding residues of two interacting proteins may have complementary hydropathic profiles 27., 28.. We compared the hydropathic profile of SEMA5A with Plexin B3 and NRP2 as shown in Figure 2. The results demonstrate that the putative ligand binding regions of Plexin B3 and NRP2 have complementary hydropathic indices compared to that of SEMA5A as shown in the thicker lines (Figure 2). This further predicts the interaction of SEMA5A with Plexin B3 and NRP2.
Fig. 2.
Hydropathic complementarity of proteins with SEMA5A. Kyte and Doolittle hydropathic profiles of SEMA5A, Plexin B3 and NRP2 were calculated using ProtScale. The hydropathic scores were plotted using Microsoft Excel™. Thick regions in the graph represent the regions corresponding to NAFTPDY residues in each protein. The binding peptide corresponding to Plexin B3 and NRP2 have negative hydropathic index (complementary hydropathic values) compared with that of SEMA5A.
SEMA5A binding partners are co-expressed in tissues
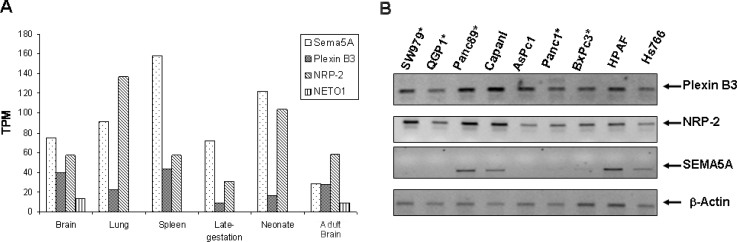
Interacting proteins are co-expressed in many tissues (29). Therefore, we tested the expression of SEMA5A, Plexin B3 and NRP2 using Gene Expression Atlas, UniGene expression profile and reverse transcription polymerase chain reaction (RT-PCR). Gene Expression Atlas showed that SEMA5A, Plexin B3 and NRP2 are co-expressed in spinal cord, brain and prostate tumors (data not shown). The UniGene expression profile shown in Figure 3A demonstrates the co-expression of SEMA5A, Plexin B3 and NRP2 in different tissues (brain, lung and spleen) and embryonic stages (late gestational and neonatal mice). As a control, NETO1 was tested and was not co-expressed in any of the SEMA5A expressing tissues except brain tissue (Figure 3A). Similarly, RT-PCR using pancreatic cancer cell lines shows that SEMA5A, Plexin B3 and NRP2 are co-expressed in pancreatic cancer cell lines established from metastases (Figure 3B). The RT-PCR data suggest that SEMA5A interaction with Plexin B3 and NRP2 may be involved in pancreatic tumor progression and metastasis. Overall, these results suggest that SEMA5A may interact with Plexin B3 or NRP2.
Fig. 3.
Co-expression of SEMA5A with its putative binding partners. A. UniGene expression data for SEMA5A, Plexin B3, NRP2 and NETO1 in different adult and fetal tissues. Except NETO1, the other genes are co-expressed with SEMA5A. TPM, tags per million. B. RT-PCR analysis showing the co-expression of Plexin B3 and NRP2 with SEMA5A in pancreatic cancer cell lines established from metastasis. Note that there is no expression of SEMA5A in cell lines established from primary tumor.
Modeling of Plexin B3 structure
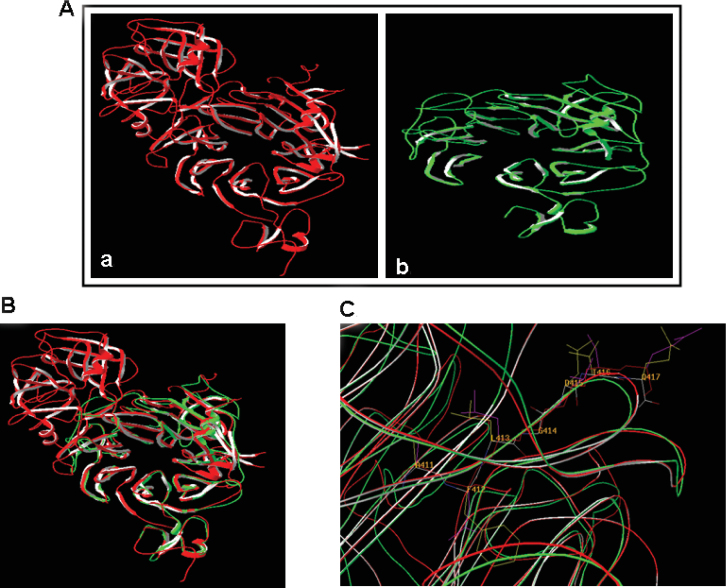
Homology modeling using the remote protein structure prediction server CPHmodels was performed to design the structure of the sema domain of Plexin B3, which has been predicted using the sema domain of Met receptor. The structure of the sema domain of Met receptor and Plexin B3 is shown in Figure 4A and their overlapping structure is shown in Figure 4B. Furthermore, Figure 4C shows that the predicted ligand binding region from NAFTPDY peptide has structural similarity to Met receptor.
Fig. 4.
Molecular modeling of Plexin B3 protein. A. Molecular modeling of the sema domain of Plexin B3 was performed using the web-based homology modeling server CPHmodels 2.0 utilizing Met receptor as a template. All modeling was carried out using Swiss Model and was viewed using Swiss-PdbViewer. Note the structure of the sema domain of Met (a) and Plexin B3 (b). B. The structure of the sema domain from Plexin B3 overlapped with that of Met receptor. C. The amino acids corresponding to the binding peptide NAFTPDY are conserved and surface exposed in Plexin B3 similar to Met receptor.
Presence of similar domains infers protein interactions
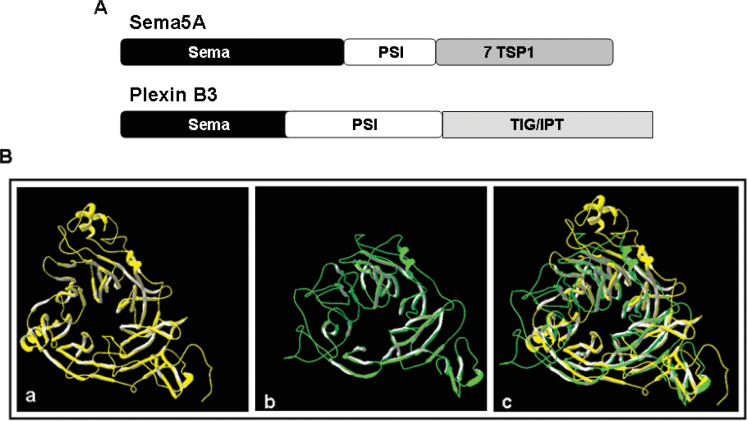
As a derivation of the Rosetta Stone model, proteins with similar domains may interact with each other (26). PSI-BLAST and CDD search results showed that Plexin B3 and SEMA5A have similar domains. We have already predicted the structure of SEMA5A in our previous publication (1). Here we compared the structure of the sema domain of SEMA5A and Plexin B3. The extracellular domains of Plexin B3 and SEMA5A shown in Figure 5A demonstrate that they share the sema domain and PSI domain. Furthermore, the structure of the sema domain of both SEMA5A and Plexin B3 are similar as shown in Figure 5B. This suggests that SEMA5A and Plexin B3 may interact with each other through the sema domain.
Fig. 5.
Comparison of domains between SEMA5A and Plexin B3. A. Different domains in the extracellular regions of SEMA5A and Plexin B3. Sema, semaphorin domain; PSI, photosystem I; TSP-1, thrombospondin specific repeats; TIG/IPT, immunoglobulin-like fold shared by plexin and transcription factors. B. Structure of the sema domain of SEMA5A (a), Plexin B3 (b) and their overlapping image (c).
SEMA5A and Plexin B3 co-evolved
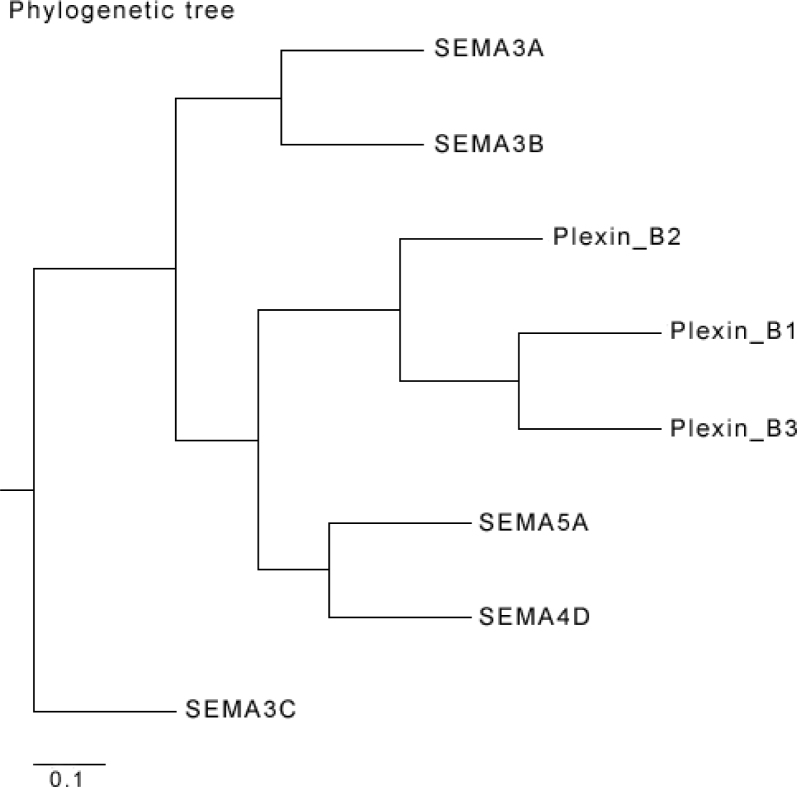
Proteins clustered together phylogenetically tend to function together and hence, interact with each other 30., 31.. To test this, we generated a phylogenetic tree using semaphorins and plexins. The result in Figure 6 shows a close evolutionary relationship between SEMA5A and Plexin B3 together with less distance within the Plexin family of proteins. This predicts that SEMA5A and Plexin B3 may have co-evolved as binding partners.
Fig. 6.
Co-evolution of SEMA5A with Plexin B3. Phylogenetic tree with distance is shown for Semaphorin and Plexin families of proteins. The phylogenetic tree shows a close evolutionary relationship between SEMA5A and Plexin B3, representing the co-evolution of the two proteins.
SEMA5A interacts with Plexin B3
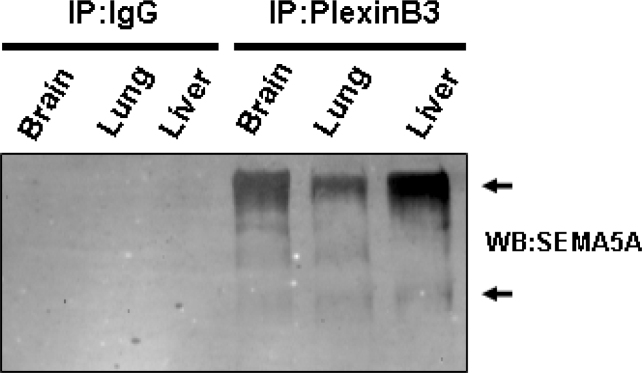
Next, we confirmed the interaction of SEMA5A with Plexin B3 in normal tissues co-expressing SEMA5A and Plexin B3. We performed co-immunoprecipitation using anti-PlexinB3 antibody followed by immunobloting using anti-SEMA5A antibody. Normal brain, lung and liver tissues from rats were used. Results shown in Figure 7 demonstrate that SEMA5A and Plexin B3 interact with each other as they co-immunoprecipitated in brain, lung and liver lysates.
Fig. 7.
Co-immunoprecipitation of SEMA5A and PlexinB3. Tissue lysates were immunoprecipitated using anti-PlexinB3 and IgG control antibodies and were then immunoblotted using anti-SEMA5A antibody.
Discussion
Recently, our laboratory has implicated the expression of SEMA5A in metastatic pancreatic cancer cell lines but not in primary cancer cell lines (1). In addition, the report that mice knockouted for SEMA5A died due to defects in the cranial vascular system (32) demonstrates the importance of SEMA5A in the development of the vascular system in embryos. These studies show the significance of SEMA5A in cancer and developmental biology, suggesting the importance of identifying SEMA5A interacting proteins. Identification of a SEMA5A binding partner would add great value to its functional study. In this report, we have shown Plexin B3 as a primary binding partner for SEMA5A using a novel sequence of computational protein interaction prediction method. We are illustrating for the first time the application of peptide and hydropathic complementarity along with a unique sequence of methods to predict protein-protein interactions. We have also modeled the 3D structure of the sema domain of Plexin B3 and identified the putative ligand binding peptide that may regulate the binding of SEMA5A with Plexin B3. However, this method of predicting protein-protein interactions is not a genome level analysis and there could be other binding partners for SEMA5A. Interestingly, along with our prediction, Artigiani et al. identified Plexin B3 as a functional receptor for SEMA5A using transfection experiments in Cos-7 cells (29). Nevertheless, to our knowledge, our report is the first one to show the interaction in the normal physiology of any type of cells and in cancer cell lines.
The present study is a small scale prediction of protein-protein interactions involving proteins sharing similar ligand binding regions. The ligand binding regions of interacting proteins can be identified by computational prediction. However, we previously used in vivo phage-display peptide library screening to identify natural ligand binding regions (1). Furthermore, in the same study we identified seven putative proteins sharing ligand binding regions with SEMA5A using bioinformatics strategies. In the present study, we have tried to identify the binding partner for SEMA5A within the seven putative proteins. Root-Bernstein and Dillon have demonstrated that glucagon and glucagon receptor have peptide complementarity in their ligand binding regions (21), as well as many other proteins and their receptors. In this study, we have observed similar protein complementarity in SEMA5A and putative proteins screened from phage-display peptide library. Therefore, we chose these proteins for further analysis as putative SEMA5A binding proteins.
The verification of a SEMA5A binding partner involved many analytical challenges. Careful consideration and trustworthiness of the partners selected were done to avoid the identification of false positives. Furthermore, we collected well-known functions of SEMA5A and modeled other proteins to those functions using the association rule. This increases the likelihood of finding SEMA5A interacting partners and reducing false positives. Previously, the functions of proteins using the GO database have been used to predict protein interactions (16). In the present study, we have modeled a combination of GO functional terms with the association rule to predict the binding partners of SEMA5A. This analysis helped to narrow down Plexin B3 and NRP2 as the most possible interacting partners of SEMA5A. In addition, small scale text mining using cosine similarity demonstrated the co-occurrence of Plexin B3 and NRP2 in the literature. The co-occurrence of proteins in the literature is an indication of functional relevance between proteins 16., 33.. Cosine similarity may not give the right magnitude of distance between two proteins that have been compensated by hierarchical clustering of proteins using the association rule calculation. However, many proteins of unknown function with few citations in the literature were chosen. Consequently, cosine similarity for those proteins was not considered as they were not significantly clustered near SEMA5A. Furthermore, we did not consider interactions within Semaphorin family of proteins for simplicity. Therefore, we removed SEMA3C from the list of SEMA5A interacting partners tested.
Molecular evidence indicates that the interaction between proteins takes place through 4–7 amino acids (1) at the ligand binding regions, and these ligand binding regions of interacting proteins display inverse hydropathic profiles leading to hydropathic complementarity 27., 28.. Furthermore, molecular recognition theory states that amino acids from complementary strands of DNA will have hydropathic complementarity and can bind to each other 27., 28., 34.. More accumulating evidence indicates that this theory has been applied to design biologically active synthetic analogs of receptor binding sites and to map epitopes for antibodies 28., 34., 35., 36.. Previously, hydropathic complementarity has not been applied to identify protein-proteins interactions. In this report, Plexin B3 and NRP2 were demonstrated to have hydropathic complementarity with SEMA5A at the putative ligand binding region, suggesting their interaction. To our knowledge, for the first time we have used hydropathic complementarity to verify the interactions between putative binding partners.
Proteins sharing structural similarity and ligand binding regions are already known to interact with each other (21). We have already identified the structure of the sema domain of SEMA5A (1). Here we have predicted the structure of the sema domain of Plexin B3 by homology prediction method using the structure of Met receptor as a template and showed the similarity of Plexin B3 to SEMA5A. This is another clue that these proteins may interact with each other. These steps further suggest that Plexin B3 could be the binding partner for SEMA5A.
Evolutionarily interacting proteins are found to be co-expressed in the same tissues and previously, this co-expression was used to identify protein-protein interactions 16., 37., 38., 39.. In the present study, we verified the interaction of the identified proteins by examining their co-expression in the same tissues using Unigene expression and GNF microarray expression databases as well as RT-PCR analysis. Expression was further confirmed by performing RT-PCR analysis using pancreatic cancer cell lines. We observed expression of Plexin B3 and NRP2 in SEMA5A-expressing and aggressive pancreatic cancer cell lines, indicating the co-expression of these proteins. Meanwhile, SEMA5A and Plexin B3 co-evolved together. Previously, interacting proteins were reported to co-evolve 16., 20.. We showed that Plexin B3 is closely related to SEMA5A phylogenetically.
Among the set of proteins analyzed, Plexin B3 is a promising candidate as a binding partner for SEMA5A. We have confirmed our prediction using immunoprecipitation experiments in rat tissues. Independently, our prediction was also confirmed by a different group (29). By predicting the interacting partners for SEMA5A, we are not only predicting the functional receptor for SEMA5A, but also applying a new approach to predict protein-protein interactions through combinations of many different concepts. We are aware that ligand binding regions of most proteins are not known. However, it will be significant to develop this analysis for genome-wide prediction of protein binding partners by developing a database of ligand binding sites of proteins identified either using phage-display peptide library or structure determination methods. We believe this is a confirmation for the interaction of SEMA5A with Plexin B3, and we predict that interaction is critical for cellular responses associated with pancreatic cancer progression and metastasis.
Materials and Methods
Peptide complementarity
Proteins containing a sema domain are already known to self-aggregate (or undergo homophilic interactions) 40., 41.. SEMA5A may self-aggregate due to the presence of a sema domain (unpublished data). Therefore, the putative proteins that share the binding signature NAFTPDY with SEMA5A were grouped together based on RBD theories that peptide receptors will contain ligand-like sequences within the binding (extracellular) region, if the peptide is self-complementary (21).
Hydropathic complementarity
Kyte and Doolittle hydropathic profile for proteins was generated using ProtScale 42., 43.. The values were plotted using Microsoft Excel™. The complementary regions on the graph were shaded in dark solid lines (Figure 2).
Patterns of protein functions
To identify proteins that perform the same function, biological processes, molecular functions and cytoplasmic components (in general, called terms) associated with SEMA5A were listed from the GO database (44). Functional association of proteins was done using the frequency and confidence of co-occurrence of each protein with SEMA5A. Let P = (p1, p2,…,pn) be a set of proteins and F = (f1, f2,…,fm) be the set of functional terms such that each protein may or may not perform the function. Hence, Fi ⊆ P. Depending on the presence (or absence) of Fi for P, a value of 1 (or 0) was assigned. The proteins were later clustered using a hierarchical clustering method with the Genesis software (45). Later, the fraction of proteins containing the functional terms with pattern X was calculated as Frac(X) = X/N, where X is the number of Fi qualified as 1 for a protein and N is the number of functional terms. The frequency of co-occurrence of all the functional terms for proteins with patterns X and Y for two different proteins was calculated as Freq(X U Y). The assumption is that for two proteins to have the same function, the presence of pattern X for one protein implies the presence of pattern Y for the second protein with same functional terms, Fi. Therefore, the confidence of the functional association for two proteins was calculated as Conf(X) = [Freq(X U Y)/Frac(X)]×100% (26). Those associations greater than an arbitrary minimal confidence of 75% were considered valid.
Analysis of biological literature for functional relationship
To find the functional relationship between two proteins, the co-occurrence of the proteins in the biological literature was analyzed. Two bibliographic search databases, Google Scholar and PubMed were queried. The number of entries under each search is listed in Table 2, and cosine similarity based distance metric was used to find the relationship. Cosine similarity is the measure of the cosine of the angle between two vectors. The value is 1 when the two vectors are identical and is 0 when the vectors are completely orthogonal. The protein that has maximum cosine similarity to the given protein is the one that is most similar to it. Given two protein vectors X and Y, the cosine co-efficient is: Sim(X, Y) = X · Y/|X||Y| (46). The cosine co-efficient measured the similarity between protein vectors.
Table 2.
The number of entries for each bibliographic search using Google Scholar and PubMed*
| Search key word | Google scholar | PubMed |
|---|---|---|
| Semaphorin + plexin + neuropilin | 983 | 77 |
| Semaphorin + plexin | 1,930 | 202 |
| Plexin + neuropilin | 1,080 | 85 |
| Semaphorin + neuropilin | 2,710 | 305 |
| Semaphorin | 7,900 | 948 |
| Neuropilin | 7,000 | 698 |
| Plexin | 2,860 | 259 |
Searched on December 31, 2007.
Co-expression of genes and protein interaction
To find the co-expression of genes, Gene Expression Atlas (http://expression.gnf.org) text query was used. The result for each gene was compared manually with that of the other genes. Furthermore, the number of expressed sequence tags per million tags for each gene in different organs was listed from Unigene expression database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). The values were plotted in a graph and compared with that of the other genes. The co-expression of SEMA5A, Plexin B3 and NRP2 in pancreatic cancer was examined by RT-PCR using respective primers. Total cellular RNA was isolated from nine pancreatic cancer cell lines of different origin using Trizol reagent (Invitrogen, Carlsbad, CA, USA) (47) and RT-PCR was performed as described (48). First-strand cDNA was synthesized using total RNA (2 μg), oligo dT18 primer and superscript II RT (Invitrogen); 2 μL of first-strand cDNA (1:10 dilutions) was amplified using PCR primer sets and a thermal cycler (MJ Research, Waltham, MA, USA). PCR fragments were separated on a 2% agarose gel containing ethidium bromide (0.25 μg/mL), visualized and analyzed using the Alpha Imager gel documentation system (Alpha Innotech, San Leandro, CA, USA).
Domain-domain interactions
To elucidate the presence of similar domains in SEMA5A and candidate proteins, SEMA5A was used as a query for PSI-BLAST (49) with default parameters and the complete non-redundant database of Homo sapiens. Furthermore, a BLAST-conserved domain search was performed for all three proteins with default parameters and conserved domain database (CDD) version 2.05 50., 51..
Protein modeling
Modeling of Plexin B3 was performed using the web-based homology modeling server CPHmodels 2.0 (52) and further analysis was performed using Swiss-PdbViewer as described earlier 1., 53..
Co-immunoprecipitaion of SEMA5A and Plexin B3
Normal brain, lung and liver tissues were lysed in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100 and protease inhibitors cocktail (Pierce, Rockford, IL, USA). For immunoprecipitaion, tissue lysates were incubated for 2 h with anti-PlexinB3 antibody (pAbB3-A, generous gift from Dr. Ulrich Finckh, Hamburg University, Germany) or IgG control antibodies (1 μg/mL) followed by overnight incubation with Protein A/G agarose (GE Healthcare, Piscataway, NJ, USA). Immune complexes were washed three times with 1% Triton X-100 containing PBS (PBS-T), resolved on 8% SDS-PAGE and immunoblotted with indicated antibodies. Following protein electrotransfer, nylon membranes (Millepore, Billerica, MA, USA) were probed with anti-SEMA5A antibody. The blots were developed using an ECL plus enhanced chemiluminescence kit (GE Helthcare).
Phylogenetic profile and protein interactions
The sequences of members belonging to the Semaphorin and Plexin families were selected from NCBI Entrez and aligned using Clustal W (54) (http://workbench.sdsc.edu). A distance matrix was generated using ProtDist in Phylip program 55., 56. with Felsenstein’s categories, bootstrap options and all other default parameters for the Clustal W alignment file (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). Furthermore, the neighborjoining tree was generated using the Phylip neighbor program with the bootstrap options for the distance matrix obtained from ProtDist. Finally, Phylodendron software (D.G. Gilbert version 0.8d) was used to generate trees (http://www.es.embnet.org/Doc/phylodendron/).
Authors’ contributions
AS performed most of the studies, analyzed the data and drafted the manuscript. MLV performed the immunoprecipitation studies and helped in RT-PCR analysis. AS and RKS conceived the study. RKS participated in its design and coordination and helped in preparation of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was partly supported by Molecular Therapeutics Program, Nebraska Department of Health and Human Services and by Grant CA72781 (to RKS) and Cancer Center Support Grant (P30CA036727) from National Cancer Institute, National Institutes of Health, USA.
References
- 1.Sadanandam A. Identification of functional cell adhesion molecules with a potential role in metastasis by a combination of in vivo phage display and in silico analysis. OMICS. 2007;11:41–57. doi: 10.1089/omi.2006.0004. [DOI] [PubMed] [Google Scholar]
- 2.Bogan A.A., Thorn K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti P., Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 4.Jones S., Thornton J.M. Prediction of protein-protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 5.Guharoy M., Chakrabarti P. Conservation and relative importance of residues across protein-protein interfaces. Proc. Natl. Acad. Sci. USA. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLano W.L. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 7.Keskin O. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Phys. Biol. 2005;2:S24–S35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 8.Res I., Lichtarge O. Character and evolution of protein-protein interfaces. Phys. Biol. 2005;2:S36–S43. doi: 10.1088/1478-3975/2/2/S04. [DOI] [PubMed] [Google Scholar]
- 9.Barbas C.F., III . Cold Spring Harbor Laboratory Press, Cold Spring Harbor; USA: 2001. Phage Display: A Laboratory Manual. [Google Scholar]
- 10.Ma B. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ofran Y., Rost B. Predicted protein-protein interaction sites from local sequence information. FEBS Lett. 2003;544:236–239. doi: 10.1016/s0014-5793(03)00456-3. [DOI] [PubMed] [Google Scholar]
- 12.Pasqualini R., Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualini R. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 14.Deng M. Inferring domain-domain interactions from protein-protein interactions. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcotte E.M. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes D.R. Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 17.Shoemaker B.A., Panchenko A.R. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker B.A., Panchenko A.R. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLoS Comput. Biol. 2007;3:e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsh E., Sharan R. Identification of conserved protein complexes based on a model of protein network evolution. Bioinformatics. 2007;23:E170–E176. doi: 10.1093/bioinformatics/btl295. [DOI] [PubMed] [Google Scholar]
- 20.Pazos F., Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 21.Root-Bernstein R.S., Dillon P.F. Molecular complementarity I: the complementarity theory of the origin and evolution of life. J. Theor. Biol. 1997;188:447–479. doi: 10.1006/jtbi.1997.0476. [DOI] [PubMed] [Google Scholar]
- 22.Suto F. Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron. 2007;53:535–547. doi: 10.1016/j.neuron.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99:59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 24.Tamagnone L. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 25.Winberg M.L. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95:903–916. doi: 10.1016/s0092-8674(00)81715-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q., Chen Y.P. Mining frequent patterns for AMP-activated protein kinase regulation on skeletal muscle. BMC Bioinformatics. 2006;7:394. [Google Scholar]
- 27.Blalock J.E. Genetic origins of protein shape and interaction rules. Nat. Med. 1995;1:876–878. doi: 10.1038/nm0995-876. [DOI] [PubMed] [Google Scholar]
- 28.Gomez I. Hydropathic complementarity determines interaction of epitope 869HITDTNNK876 in Manduca sexta Bt-R1 receptor with loop 2 of domain II of Bacillus thuringiensis Cry1A toxins. J. Biol. Chem. 2002;277:30137–30143. doi: 10.1074/jbc.M203121200. [DOI] [PubMed] [Google Scholar]
- 29.Artigiani S. Plexin-B3 is a functional receptor for semaphorin 5A. EMBO Rep. 2004;5:710–714. doi: 10.1038/sj.embor.7400189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser H.B. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 31.Waddell P.J. Phylogenetic methodology for detecting protein interactions. Mol. Biol. Evol. 2007;24:650–659. doi: 10.1093/molbev/msl193. [DOI] [PubMed] [Google Scholar]
- 32.Fiore R. Inactivation of the Sema5a gene results in embryonic lethality and defective remodeling of the cranial vascular system. Mol. Cell. Biol. 2005;25:2310–2319. doi: 10.1128/MCB.25.6.2310-2319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberts B. Fourth edition. Garland Publishing, Inc.; New York, USA: 2002. Molecular Biology of the Cell. [Google Scholar]
- 34.Bost K.L. Similarity between the corticotropin (ACTH) receptor and a peptide encoded by an RNA that is complementary to ACTH mRNA. Proc. Natl. Acad. Sci. USA. 1985;82:1372–1375. doi: 10.1073/pnas.82.5.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillon P.F. Augmentation of aortic ring contractions by angiotensin II antisense peptide. Hypertension. 1998;31:854–860. doi: 10.1161/01.hyp.31.3.854. [DOI] [PubMed] [Google Scholar]
- 36.Heal J.R. A search within the IL-1 type I receptor reveals a peptide with hydropathic complementarity to the IL-1beta trigger loop which binds to IL-1 and inhibits in vitro responses. Mol. Immunol. 1999;36:1141–1148. doi: 10.1016/s0161-5890(99)00129-7. [DOI] [PubMed] [Google Scholar]
- 37.Ge H. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 38.Tirosh I., Barkai N. Computational verification of protein-protein interactions by orthologous co-expression. BMC Bioinformatics. 2005;6:40. doi: 10.1186/1471-2105-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj N., Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 40.Follenzi A. Cross-talk between the protooncogenes Met and Ron. Oncogene. 2000;19:3041–3049. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 41.Hartwig C. Plexin B3 promotes neurite outgrowth, interacts homophilically, and interacts with Rin. BMC Neurosci. 2005;6:53. doi: 10.1186/1471-2202-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasteiger E. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasteiger E. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 44.Ashburner M. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturn A. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 46.Lewis J. Text similarity: an alternative way to search MEDLINE. Bioinformatics. 2006;22:2298–2304. doi: 10.1093/bioinformatics/btl388. [DOI] [PubMed] [Google Scholar]
- 47.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 48.Singh R.K. Immune dysfunction despite high levels of immunoregulatory cytokine gene expression in autologous peripheral blood stem cell transplanted non-Hodgkin’s lymphoma patients. Exp. Hematol. 2000;28:499–507. doi: 10.1016/s0301-472x(00)00145-4. [DOI] [PubMed] [Google Scholar]
- 49.Schaffer A.A. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchler-Bauer A. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul S.F. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Lund, O., et al. 2002. CPHmodels 2.0: X3M a computer program to extract 3D models. Abstract at the CASP5 conference A102.
- 53.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 54.Thompson J.D. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 56.Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]