Abstract
The life-threatening infections caused by Leptospira serovars demand the need for designing anti-leptospirosis drugs. The present study encompasses exploring inhibitors against phosphoheptose isomerase (GmhA) of Leptospira, which is vital for lipopolysaccharide (LPS) biosynthesis and is identified as a common drug target through the subtractive genomic approach. GmhA model was built in Modeller 9v7. Structural refinement and energy minimization of the predicted model was carried out using Maestro 9.0. The refined model reliability was assessed through Procheck, ProSA, ProQ and Profile 3D. The substrate-based virtual high-throughput screening (VHTS) in Ligand.Info Meta-Database tool generated an in-house library of 354 substrate structural analogs. Furthermore, structure-based VHTS from the in-house library with different conformations of each ligand provided 14 novel competitive inhibitors. The model together with insight gained from the VHTS would be a promising starting point for developing anti-leptospirosis competitive inhibitors targeting LPS biosynthesis pathway.
Key words: LPS biosynthesis, Leptospira, homology modeling, virtual high-throughput screening, GmhA inhibitors
Introduction
Leptospirosis is a widespread zoonotic disease caused by the spirochete Leptospira that is pathogenic for human (1). More than 500,000 cases of severe leptospirosis occur annually in the world (2). It is predominantly an occupational disease that affects persons in frequent contact with infected rodents, pet animals or polluted water, and the spirochete penetrates the human body through skin or mucous membranes 3, 4, 5, 6. The symptoms of leptospirosis are extremely broad from meningitis, pneumonitis, hepatitis, nephritis, pancreatitis, erythema nodosum and death (7). Progression to multi-organ system complications occurs in 5% to 15% of cases, with mortality rates of 5% to 40% (8).
Leptospira has over 200 pathogenic serovars divided into 25 serogroups and many different strains with small antigenic differences in some serovars (7). The complete genomic sequences of four pathogenic Leptospira (L. interrogans serovar Lai str. 56601, L. interrogans serovar Copenhageni str. Fiocruz L1-130, L. borgpetersenii serovar Hardjo-bovis JB197 and L. borgpetersenii serovar Hardjo-bovis L550) were sequenced and released 9, 10, 11. Extensive variation in the number and the distribution of insertion sequences, genomic islands and other genomic contents was observed during genomic comparison of serovars Lai and Copenhageni (2), which eventually determines the unique phenotypes of each strain. The whole genome sequencing of pathogenic and non-pathogenic strains extended the vaccine development process significantly along with development of a vaccine ontology database 7, 12. However, the heterogeneous group of pathogenic and saprophytic leptospires always remained a challenge for immunologists to develop an effective and safe leptospirosis vaccine (7). Although early treatment for leptospirosis is important for ensuring a favorable clinical outcome, it is often difficult to achieve, as symptoms during the early stages of infection resemble those of several other systematic diseases (7). Specific antibiotic treatment using doxycycline or penicillin has shown mixed results for mild or sub-clinical infections, nevertheless treatment of severe leptospirosis (organism localized in tissues) is still unclear 4, 13. Identification of potential drug targets for L. interrogans through the substractive genomic approach 13, 14 has laid an attempt for exploring drug targets for screening novel lead compounds to improve leptospirosis treatments.
Lipopolysaccharide (LPS) is an essential component of the outer membrane in Gram-negative bacteria 15, 16. LPS not only functions as a protective barrier preventing cell entry of hydrophobic molecules, but also helps maintain the structural integrity of the outer membrane. LPS of Leptospira contributes to the pathology associated with diseases and comprises the major surface component of leptospires 16, 17. Studies with monoclonal antibodies have shown that LPS is a target for agglutinating and osponizing antibodies (18). Thus, LPS plays a key role in immunity to infection. Indeed, anti-LPS monoclonal antibodies provide passive protection against infection, and purified LPS can stimulate active immunity 19, 20. LPS being an agglutinating antigen is also important for serological classification of leptospires. Thus, LPS is vital for leptospiral survival, virulence and antibiotic sensitivity, and enzymes controlling this pathway are of significant interest as molecular targets for new antimicrobial intervention. The structure of LPS is largely unknown. However, chemical analysis revealed that LPS of L. interrogans serovar Cophanegeni and L. borgepetersenii serovars Hardajo is consistent with composition similar to that of the typical Gram-negative bacteria 21, 22. LPS comprises of Lipid A, a core oligosaccharide, and in some bacteria, an O-specific polysaccharide chain. The core oligosaccharide has an inner core region consisting of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) and one or more heptose units, and an outer core consisting of an additional core residue 22, 23, 24, 25. Lipid A and Kdo are highly conserved in Gram-negative bacteria and essential for cell viability. The biosynthesis of these molecules is therefore target for traditional antibiotic discovery efforts (26). Most Gram-negative bacteria also contain one or more L-glycero-D-manno-heptose molecules attached to Kdo. Mutants in heptose metabolism are avirulent and highly susceptible to antibiotics, thus inhibitors of heptose biosynthesis could be used as anti-virulence drugs (27).
Phosphoheptose isomerase (GmhA) was identified as putative common drug target for L. interrogans serovars 13, 14. Heptoses targeted to inner core LPS are synthesized within the cytosol as ADP-activated L-glycero-β-D-manno-heptose molecule 28, 29, 30. The biosynthesis starts with GmhA catalyzing D-sedoheptulose-7-phosphate (S7P) to D-glycero-D-manno-heptose 7-phosphate. The isomerization reaction is the first committed step of LPS biosynthesis (16). Thus, understanding the structure and function of GmhA would be useful to discover novel inhibitors.
In the present study, a 3D structural model of GmhA was predicted using homology modeling technique. Structural refinement and energy minimization of built 3D model was done using Maestro 9.0. The structural quality of the predicted model was verified using Procheck, ProSA, ProQ and discrete optimized protein energy (DOPE) profile. Validity of the model was assessed by docking natural substrate S7P. The purpose of our study was to use virtual high-throughput screening (VHTS) to find novel inhibitors of the modeled GmhA followed by scoring and ranking of the compounds to identify potential leads. The novel competitive inhibitors proposed here would be highly useful for developing antimicrobial drug against leptospirosis.
Results and Discussion
GmhA as drug target
Diverse groups of pathogenic and saprophytic leptospires, and differences in the number and the distribution of insertion sequences, genomic islands and other genomic contents among Leptospira serovars 2, 7 make it difficult to develop effective drug or vaccine against leptospirosis. A common drug target for all pathogenic leptospires would be ideal to discover novel leptospirosis drug candidate. GmhA was identified as a common drug target among four pathogenic leptospires (genome sequencing completed till date) through the subtractive genomic approach. GmhA is a homodimer consisting of chains A, D and B, C. Each chain has 195 amino acid residues. Raw sequence of GmhA, available at the UniProt (ID: Q72RC1), was retrieved (Table 1). The selected drug target protein had 100% sequence identity with both L. interrogans serovars (Lai and Copenhageni) and 92% sequence identity with two L. borgpetersenii serovars (Hardjo-bovis JB197 and Hardjo-bovis L550). The GmhA active site residues were 100% conserved among four pathogenic Leptospira serovars. Current research approach was intended towards proposing GmhA as the molecular target for structure-based drug discovery against leptospirosis.
Table 1.
Raw sequence of GmhA of Leptospira accessed from UniProt (Q72RC1)
| MDIKEIALGQIRDSIATKQKCIDSILEDIIKAGEIVSKILQAGNTIFLCGNGGSSCDASHIAAELVVRYKSGNERKALPALSLS ADSAVLTACSNDYGYEEIFSRQIEAFGRKGDLLIGLSTSGNSKNVLLALEKAKTRGVKTISLLGGDGGKMKNLSDLDVIVP SNVTARIQESHILIGHIICSIVEYNLFKME |
The enzyme binds with S7P to give rise to D-glycero-D-manno-heptose 7-phosphate in an isomerization reaction, which is the starting point of LPS biosynthesis pathway (16). Pathway analysis at the KEGG revealed that there was no alternative way for D-glycero-D-manno-heptose 7-phosphate formation in the absence of GmhA. Comparative analysis of human and Leptospira metabolic pathways revealed that LPS biosynthesis pathway was unique to Leptospira. The study coincides with the finding in Mycobacterium tuberculosis (31). Hence, designing novel competitive inhibitor to block GmhA active site would stop LPS biosynthesis, disintegrate the LPS protective barrier, and dissolve the structural integrity of the outer membrane and virulence factor of the pathogen.
Homology modeling
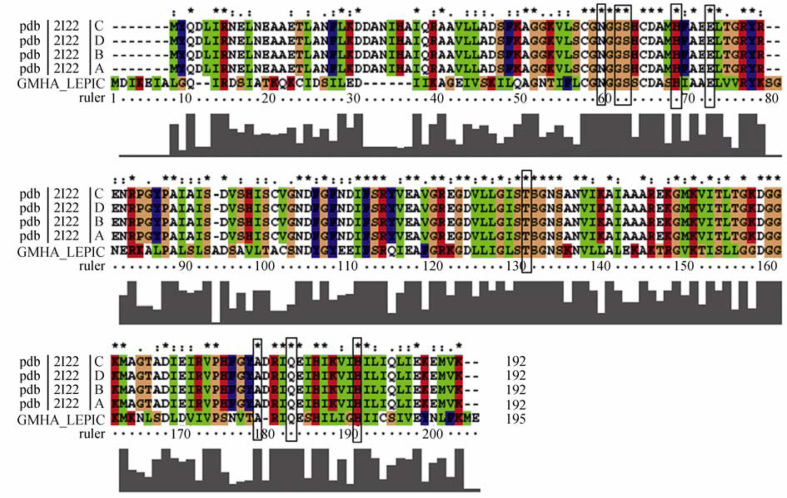
Homology modeling is an efficient method for 3D structure prediction and quick experimental design for docking studies. In general, 30% sequence identity is required for generating useful 3D structure models 32, 33, 34. Crystal structure of Escherichia coli GmhA (PDB ID: 2I22) having 42% identity with the drug target was selected as template. The template protein is a homodimer and the active site residues are present within B and C chains. The active site residues were conserved in both target and template sequences (Figure 1). Twenty quaternary structures of GmhA were generated in Modeller9v7 32, 33. The substrate S7P was incorporated into the model from the template to increase overall model accuracy 32, 33. All models have the same GA341 score of 1.00, which represents that the GmhA protein fold regions were predicted correctly in all 20 models. So, the fourth model having the lowest DOPE score was selected and subjected to model validation 32, 33.
Figure 1.
Multiple sequence alignment of GmhA (four chains) with template 2I22. Dashes represent insertions and deletions. The conserved residues involved in active site are shown in boxes.
Model validation
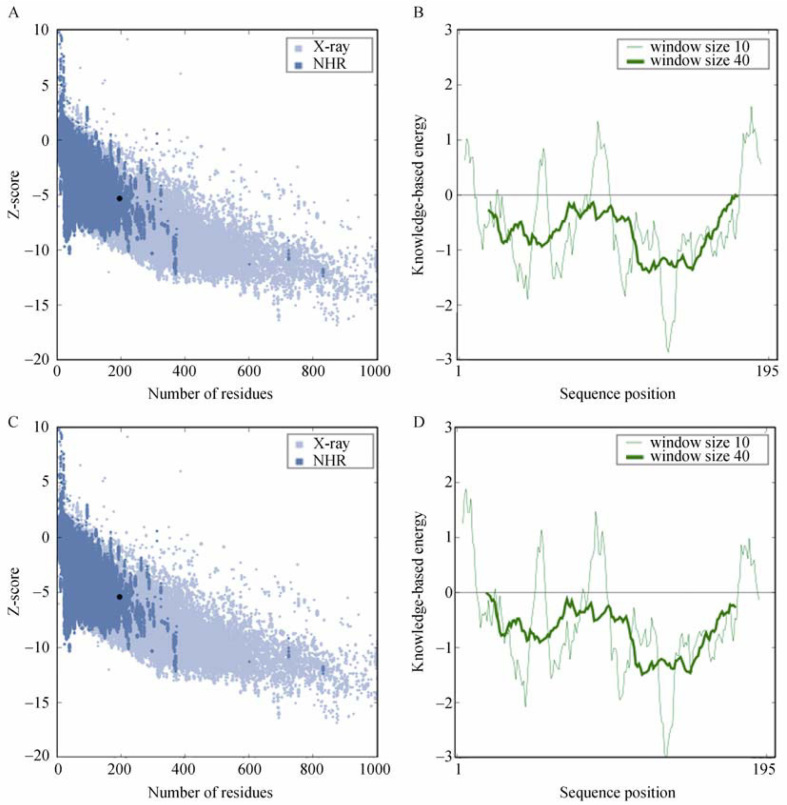
Reliability of the model was checked using diverse techniques. The DOPE profile plots (Figure S1) of leptospires (target) and E. coli (template) GmhA crystal structures were found quite similar 32, 33. The active site residues overlapping in the profile plot showed higher accuracy level of the predicted model (32). The stereochemistry of the model (Procheck analysis) revealed that 91.2% residues were positioned in most favorable region of the Ramachandran plot and was found to compare favorably with data of crystal structure: 2I22 90.1% (Figure S2A and B). Evaluation of GmhA 3D model with ProSA-web revealed a compatible Z score value (A chain = −5.35, D chain = −5.67; B chain = −5.27, C chain = −5.42) within the range of native conformations of crystal structures (35). The ProSA-web analysis showed that the overall residue energies of the GmhA 3D model were largely negative (importantly all active site and its surrounding residues were also largely negative) expect for few peaks. The residue energies including pair energy, combined energy and surface energy were all negative and had similar surface energy tendency with template. The Z score and residue energies of GmhA dimer B, C are illustrated in Figure 2. The prediction efficiency of protein quality predictor (ProQ) tool increases by 15% when the 3D model was evaluated along with its secondary structure. A ProQ LG score >4.0 is necessary for suggesting that a model is of extremely good quality (36). While submitted to ProQ tool, the secondary structure (Figure S3) and the 3D model (Figure 3A) of GmhA showed LG score of 5.285, implying the high accuracy level of the predicted 3D model. Tertiary structure superimposition of template and target using superpose command in Modeller showed Cα RMSD of 0.47 Å and an overall RMSD of 0.76 Å. The low overall RMSD reflects the high structural conservation. Structural alignment between the target and the template showed that the alpha helices and beta sheets matched accurately with each other (Table 2), representing that the GmhA 3D model is highly reliable. Through this assessment and analysis process, we concluded that the GmhA model generated in the present study is reliable to characterize protein-substrate and protein-ligand interactions and to investigate the relation between the structure and function. The validated GmhA model was submitted to protein model database (PMDB), which has accepted the model with less than 3% stereochemical check failures. PMDB ID for the developed GmhA model in complex with S7P was PM0075993.
Figure 2.
ProSA-web Z-scores of modeled protein in PDB determined by X-ray crystallography (light blue) and NMR spectroscopy (dark blue) with respect to their length. Z score is represented in black dot. The energy plots were presented with window size 10 (light green) and window size 40 (dark green). A. The Z-score plot of GmhA chain B. B. Energy plot of GmhA chain B. C. The Z-score plot of GmhA chain C. D. Energy plot of GmhA chain C.
Figure 3.
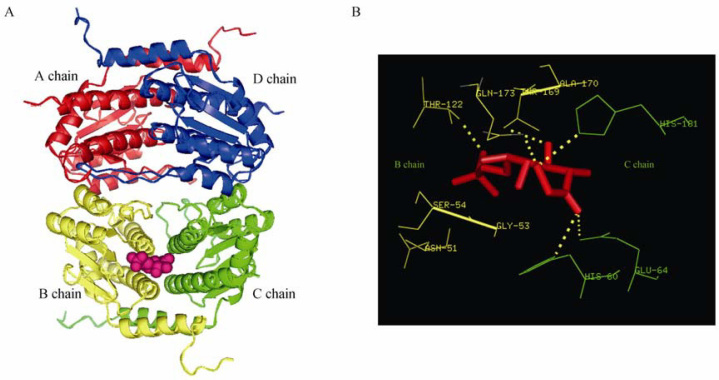
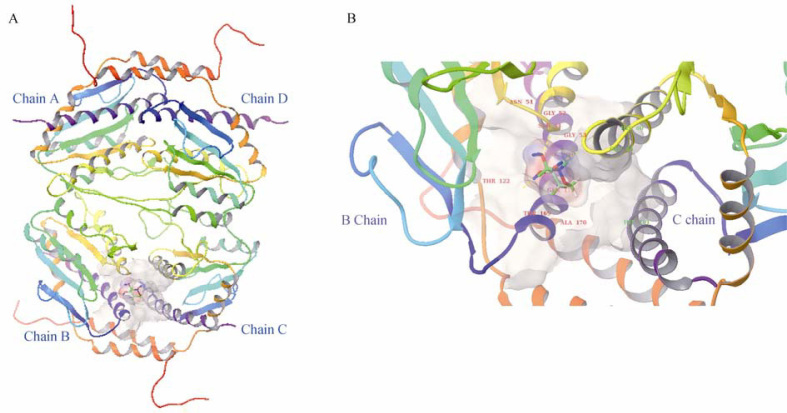
A. GmhA 3D model in complex with substrate S7P (A chain: red; B chain: yellow; C chain: green; D chain: dark blue; Substrate: pink). B. Active site residues of GmhA 3D model (B chain: yellow; C chain: green; Substrate: red).
Table 2.
Structural alignment data of GmhA 3D model and template 2I22 listing exact matches of alpha helices and beta sheets
| GmhA 3D model |
2I22 |
||
|---|---|---|---|
| Alpha helice residue range | Beta sheet residue range | Exactly matching alpha helices | Exactly matching beta sheets |
| 11-25 | 46-50 | 6-20 | 41-45 |
| 30-41 | 80-82 | 25-36 | 73-75 |
| 54-69 | 116-120 | 49-64 | 95-99 |
| 102-110 | 142-148 | 81-89 | 121-127 |
| 127-139 | 160-164 | 106-118 | 139-143 |
| 169-191 | 148-170 | ||
Active site region
Active site of the model was analyzed to assess presence of catalytic and conserved substrate binding residues. Presence of active site in the interaction site of B and C chains was confirmed through structure visualization using PyMol (Figure 3A and B). Active site residues (Asn51, Gly53, Ser54, Thr122, Thr169, Ala170, Gln173 of B chain, and His60, Glu64, His181 of C chain) were identified by selecting residues within 4 Å of the substrate. The substrate forms six hydrogen bonds with the active site residues: two with Thr169 and one each with His60, Glu64, His181 and Thr122, respectively. The substrate interaction pattern in the GmhA catalytic site contemplates well with the findings in the crystal structure of GmhA of E. coli (16). Glu65 and His180 were the most critical residues for GmhA activity in E. coli 27, 28, 29, 30. The present study revealed that Glu64, His181, Thr169, Thr122 and His60 were identified to be critical for GmhA activity in L. interrogans.
Virtual high-throughput screening
One of the most widely used methods for VHTS is docking of small molecules into active site of protein target and the subsequent scoring. A wide range of different docking programs are available, most of which use semi-rigid docking, where the ligands are treated as flexible and the receptors as rigid. The Glide 5.5 software was used for protein-ligand docking. It offers full spectrum of speed and accuracy from VHTS of millions of compounds to extremely accurate binding mode predictions, providing consistently high enrichment at every level.
Virtual screening from in-house GmhA substrate analog library was performed using Glide 5.5. A total of 12,424 protonation and tautomeric states were generated from 354 substrate analogs using LigPrep, from which 1,919 conformers were retained during post LigPrep. A total of 655 conformers were passed in Lipinski filter and reactive filter from 1,919 conformers. The dataset was further condensed to 65 based on best scoring ligands through VHTS, out of which 14 ligands were identified as lead candidates through careful inspection of the docking poses and possible interactions with the active site for all of the active compounds. All lead molecules satisfies pharmacological properties of 95% drugs.
The 14 novel lead molecules were ranked based on the Glide score and tabulated (Table S1). The best ranked lead demonstrated a Glide score of −8.39 Kcal/mol. The active site of the GmhA is present in B and C dimer. Thus, careful observation of the docking poses of 14 novel GmhA competitive inhibitors were made to find the binding modes. Two sets of binding modes were observed in the docking poses. Lead 5, lead 11 and lead 12 were directly interacting with Glu64 of C chain by forming hydrogen bond and at the same time interacting with the active site residues of B chain by forming hydrogen bond network. The remaining inhibitors (except lead 5, lead 11 and lead 12) were mainly confined to B-chain active site residues. Lead 5 showed a similar binding mode as that of substrate S7P. Lead 5 formed three hydrogen bonds with Gly53, Glu64 and Thr169 (Figure 4). The two hydroxyl groups of lead 5 acted as donors for formation of hydrogen bonds with carboxylic group of Glu64 and hydroxyl group of Thr169, respectively. Backbone nitrogen atom of Gly53 acted as hydrogen bond donor to form hydrogen bond with NH2 group of lead 5. The residues Asn51, Gly52, Gly53, Ser54, His60, Glu64, Thr122, Thr169, Ala170, Gln173 and His181 were involved in van der Waal interactions in the scaling 1.0. Lead 5 demonstrated a Glide score of −7.60 Kcal/mol, which is well above the S7P and GmhA Glide score of −3.19 Kcal/mol. Thus, based on the binding mode, lead 5 may be considered as the best lead molecule among the 14 novel inhibitors proposed in the present study for GmhA competitive inhibition.
Figure 4.
Interaction of novel lead (lead 5) in the GmhA active site forming three hydrogen bonds with Glu64, Thr169 and Gly53. A. GmhA whole protein and lead 5 (competitive inhibitor). B. Binding surface and inhibitor surface.
Validation of docking results
Natural substrate S7P was docked flexibly into GmhA active site for validation of identified lead candidates as competitive inhibitor. S7P was docked with a Glide score of −3.41 Kcal/mol. The lower Glide score (−8.39 to −7.41 Kcal/mol) of identified lead candidates compared to S7P docking complexes revealed that the novel leads would bind more competitively into GmhA active site. Glu64, one of the critical residues for isomerase activity of GmhA, was mutated with Asp64. Docking of 14 novel lead molecules and S7P into mutated GmhA model was found to accommodate them in the catalytic cleft with slightly distorted form and lower binding affinity except lead 11 (Table S1). On the other hand, minute changes in the catalytic cleft can be influenced by slight changes in the backbone residues that do not reside in the interface. Also, it is highly plausible that hydrogen bonding of the active site residues holds the key for designing potent selective inhibitors. The result revealed specificity of substrate S7P and identified novel lead molecules towards Glu64 in the active site of GmhA. Thus, the identified 14 lead molecules could be designated as novel inhibitors. GmhA being unique and the active site being 100% conserved to leptospiral serovars, designing competitive inhibitors based on these 14 lead molecules (Table S1) would be highly effective against leptospirosis.
Conclusion
Extensive variation of genomic content in Leptospira strains always remains a challenge to find common inhibitor against leptospirosis. GmhA was identified for its uniqueness in the synthesis of L-glycero-D-manno-heptose as a common drug target to pathogenic Leptospira strains through the subtractive genomic approach. Since GmhA controls the first committed step of LPS biosynthesis, it is of significant interest for novel inhibitor design. The assessment of GmhA modeled structure from L. interrogans revealed that it was of good quality with conserved active site as that of the crystal structure of GmhA in other Gram-negative bacteria. Our approach employing Glide for virtual screening along with QikProp ADME evaluation provided 14 novel inhibitors for GmhA of Leptospira. These 14 inhibitors identified through VHTS using GmhA homology model for Leptospira would be of interest as common inhibitors against leptospiral serovars. Thus, it is hoped that the lead molecules identified in the present study hold promise for anti-leptospiral activities if synthesized and tested in animal models.
Materials and Methods
Hardware and software
The study was carried out on SGI Fuel Workstation with 3.0 GHz processor, 4 GB RAM, 300 GB hard drive and an Nvidia FX 1700 graphics card running in Linux operating system. Bioinformatics softwares, such as Modeller 9v7, Schrodinger software suite 2009 and online resources, were employed to propose the outcomes of the study.
GmhA of Leptospira as drug target
GmhA was selected from the list of 88 common drug targets of L. interrogans serovars Lai and Copenhageni, identified through the substractive genomic approach (13). The GmhA protein sequence was retrieved from UniProt (http://www.uniprot.org/). The involvement of GmhA in leptospiral metabolic pathway was analyzed at the Kyoto Encyclopaedia of Genes and Genome (KEGG) (37). The protein secondary structure was predicted using PSI-PRED (38).
Homology modeling of drug target
Homology modeling technique was implemented to predict 3D structure of GhmA. BLASTP (39) analysis of the GmhA protein was performed to obtain homologous entries from the protein data bank (PDB) (40). Co-crystallized structure of GmhA of E. coli with S7P (PDB ID: 2I22) was chosen as template. The BLASTP alignment was further refined using sequence alignment with default parameters in ClustalX (41). Python script was prepared to predict GmhA homodimer (A, D and B, C chain) 3D model incorporating S7P rigidly into the active site using Modeller 9v7 32, 33. A set of 20 models were generated and the structure with the lowest DOPE score was selected for further analysis. To gain better relaxation and more correct arrangement of the atoms, refinement was done on the built GmhA model using Maestro 9.0 by applying OPLS-AA 2001 force field. The energy minimized GmhA model was assessed by Procheck (42), ProSA (35), ProQ (36), DOPE Profile, GA341 score and structure superimposition using SuperPose command 32, 33. DALI pairwise structure alignment tool (43) was used to align secondary structural elements of target 3D model with template crystal structure. The predicted GmhA 3D model was submitted to PMDB (44), which collects 3D models obtained by structure prediction methods.
Ligand-based VHTS
The substrate (S7P) atomic coordinates were downloaded from the PDB 16, 40. The substrate structure was imported to Ligand.Info Meta-database tool and 50 most structurally similar compounds were searched from each of eight sub-databases 45, 46. Consequently, an in-house library of 354 substrate analogs was generated.
Docking and scoring
Docking and scoring calculations were performed using Maestro 9.0. The substrate analogs were prepared using LigPrep with Epik (47) to expand protonation and tautomeric states at 7.0±2.0 pH units. High-energy ionization / tautomer states / ligands not following Lipinski rule of five / ligands with reactive functional group were removed from the generated conformations. A grid box of size 20×20×20 Å was generated on the receptor (modeled GmhA) by picking the active site residues using Glide 5.5 (48). Glide VHTS and standard precision method parameters were checked and set to save 10% of the good scoring ligands (48). The good scoring hits obtained from VHTS were validated based on S7P and GmhA Glide docking energy. Glu64, which was reported as a critical residue for GmhA substrate binding, was mutated with Asp64 and the identified inhibitors and S7P were redocked into the active site to see the change in substrate and inhibitor binding affinity.
Authors’ contributions
AU designed the research project. DP carried out the work to prepare the manuscript. AU conceived of the study and drafted the manuscript. MH gave further insight to the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We are highly thankful to the Department of Biotechnology, Ministry of Science and Technology, India for financial support. We thank Prof. K. Venkateswaralu (SK University, Anantapur) for critically reading the manuscript. We are grateful to Dr. B. Vengamma, Director of Bioinformatics Centre, for providing constant support and encouragement for this research.
Supplementary Material
Figures S1-S3; Table S1
References
- 1.Bharti A.R. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 2.Bourhy P. A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect. Immun. 2007;75:677–683. doi: 10.1128/IAI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trueba G. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int. Microbiol. 2004;7:35–40. [PubMed] [Google Scholar]
- 4.Levett P.N. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuriakose M. Leptospirosis in Kolenchery, Kerala, India: epidemiology, prevalent local serogroups and serovars and a new serovar. Eur. J. Epidemiol. 1997;13:691–697. doi: 10.1023/a:1007300729615. [DOI] [PubMed] [Google Scholar]
- 6.Waitkins S.A. Leptospirosis as an occupational disease. Br. J. Ind. Med. 1986;43:721–725. doi: 10.1136/oem.43.11.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z. Leptospirosis vaccines. Microb. Cell Fact. 2007;6:39. doi: 10.1186/1475-2859-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farr R.W. Leptospirosis. Clin. Infect. Dis. 1995;21:1–6. doi: 10.1093/clinids/21.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Ren S.X. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 10.Nascimento A.L. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 2004;37:459–477. doi: 10.1590/s0100-879x2004000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulach D.M. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA. 2006;103:14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakesh S. In silico approach for future development of subunit vaccines against Leptospira interrogans serovar Lai. Int. J. Bioinformatics Res. 2009;1:85–92. [Google Scholar]
- 13.Umamaheswari A. In silico identification of common putative drug targets in Leptospira interrogans. J. Chem. Biol. 2010;3:175–187. doi: 10.1007/s12154-010-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umamaheswari A. In silico putative drug targets in Leptospira interrogans and homology modeling of UDP-N-acetylglucosamine 1-carboxyvinyltransferase MurA. Genomic Med. 2008;2:295. [Google Scholar]
- 15.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor P.L. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J. Biol. Chem. 2008;283:2835–2845. doi: 10.1074/jbc.M706163200. [DOI] [PubMed] [Google Scholar]
- 17.Adler B., Faine S. The genus Leptospira. In: Dworkin M., editor. The Prokaryotes. Springer; New York, USA: 2006. p. 297. [Google Scholar]
- 18.Farrelly H.E. Opsonic monoclonal antibodies against lipopolysaccharide antigens of Leptospira interrogans serovar hardjo. J. Med. Microbiol. 1987;23:1–7. doi: 10.1099/00222615-23-1-1. [DOI] [PubMed] [Google Scholar]
- 19.Jost B.H. A monoclonal antibody reacting with a determinant on leptospiral lipopolysaccharide protects guinea pigs against leptospirosis. J. Med. Microbiol. 1986;22:269–275. doi: 10.1099/00222615-22-3-269. [DOI] [PubMed] [Google Scholar]
- 20.Midwinter A. Vaccination of mice with lipopolysaccharide (LPS) and LPS-derived immuno-conjugates from Leptospira interrogans. J. Med. Microbiol. 1990;33:199–204. doi: 10.1099/00222615-33-3-199. [DOI] [PubMed] [Google Scholar]
- 21.Vinh T.U. Characterization and taxonomic significance of lipopolysaccharides of Leptospira interrogans serovar hardjo. J. Gen. Microbiol. 1989;135:2663–2673. doi: 10.1099/00221287-135-10-2663. [DOI] [PubMed] [Google Scholar]
- 22.Yanagihara Y. Identification of 4-O-methylmannose in cell wall polysaccharide of Leptospira. Microbiol. Immunol. 1983;27:711–715. doi: 10.1111/j.1348-0421.1983.tb00634.x. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yethon J.A., Whitfield C. Lipopolysaccharide as a target for the development of novel therapeutics in gram-negative bacteria. Curr. Drug Targets Infect. Disord. 2001;1:91–106. doi: 10.2174/1568005014606143. [DOI] [PubMed] [Google Scholar]
- 26.Onishi H.R. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 27.Valvano M.A. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology. 2002;148:1979–1989. doi: 10.1099/00221287-148-7-1979. [DOI] [PubMed] [Google Scholar]
- 28.Kneidinger B. Biosynthesis pathway of ADP-L-glycero-beta-D-manno-heptose in Escherichia coli. J. Bacteriol. 2002;184:363–369. doi: 10.1128/JB.184.2.363-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eidels L., Osborn M.J. Lipopolysaccharide and aldoheptose biosynthesis in transketolase mutants of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 1971;68:1673–1677. doi: 10.1073/pnas.68.8.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kneidinger B. Biosynthesis of nucleotide-activated D-glycero-D-manno-heptose. J. Biol. Chem. 2001;276:20935–20944. doi: 10.1074/jbc.M100378200. [DOI] [PubMed] [Google Scholar]
- 31.Anishetty S. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput. Biol. Chem. 2005;29:368–378. doi: 10.1016/j.compbiolchem.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Eswar N. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Paramasivan R. Prediction of 3-dimensional structure of salivary odorant-binding protein-2 of the mosquito Culex quinquefasciatus, the vector of human lymphatic filariasis. In Silico Biol. 2007;7:1–6. [PubMed] [Google Scholar]
- 35.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallner B., Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryson K. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;1:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman H.M. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J.D. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskowski R.A. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 43.Holm L. Unification of protein families. Curr. Opin. Struct. Biol. 1998;8:372–379. doi: 10.1016/s0959-440x(98)80072-9. [DOI] [PubMed] [Google Scholar]
- 44.Castrignano T. The PMDB Protein Model Database. Nucleic Acids Res. 2006;34:D306–D309. doi: 10.1093/nar/gkj105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Grotthuss M. Ligand-Info, searching for similar small compounds using index profiles. Bioinformatics. 2003;19:1041–1042. doi: 10.1093/bioinformatics/btg117. [DOI] [PubMed] [Google Scholar]
- 46.Umamaheswari A. Virtual screening for potential inhibitors of homology modeled Leptospira interrogans MurD ligase. J. Chem. Biol. 2010;3:165–173. doi: 10.1007/s12154-010-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks W.H. Computational validation of the importance of absolute stereochemistry in virtual screening. J. Chem. Inf. Model. 2008;48:639–645. doi: 10.1021/ci700358r. [DOI] [PubMed] [Google Scholar]
- 48.Friesner R.A. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S3; Table S1