Abstract
In recent developments in chemistry and genetic engineering, the humble researcher dealing with the origin of life finds her(him)self in a grey area of tackling something that even does not yet have a clear definition agreed upon. A series of chemical steps is described to be considered as the life–nonlife transition, if one adheres to the minimalistic definition: life is self-reproduction with variations. The fully artificial RNA system chosen for the exploration corresponds sequence-wise to the reconstructed initial triplet repeats, presumably corresponding to the earliest protein-coding molecules. The demonstrated occurrence of the mismatches (variations) in otherwise complementary syntheses (“self-reproduction”), in this RNA system, opens an experimental and conceptual perspective to explore the origin of life (and its definition), on the apparent edge of the origin.
Key words: triplet code, origin of life, definition of life, life–nonlife transition
The terms of the problem
A concerted ensemble of reactions should be considered alive if it regenerates and replicates itself and if it is capable of evolving (1). The analysis of the principles determining the transitions from nonliving to living matter (1) pointed out the difficulties encountered in the reconstruction of the path along which these transitions line up.
The accepted definition of life is so worded: “Life is a self-sustained chemical system capable of undergoing Darwinian evolution” (2). The Darwinian notation stresses the intrinsically dynamic nature of the definendum, indicating that we are dealing with a process rather than with a stable system. A process may or may not give rise to emergent properties which, according to the accepted definition of emergence (www.oxforddictionaries.com), result from the collective behavior of the system. Summarizing decades of intense debate: life is the result of the emergence of properties which, by their coming into being, establish the transitions in the process of the evolutionary organization of matter from nonliving to living. Descending to the level of a chemist modelling (or, perhaps, even observing) the transitions, one naturally asks the question: what exactly are those transitions?
Living entities, as we know them, are the inseparable ensemble of genotype and phenotype, the first providing the information for the organization and the embodiment of the second. Being the genotype not only information but an actual form of organization of molecules, it is a form of phenotype on its own merit. Such phenotype, one may say, is not only a collection of enzymatic functions and of organic molecules. The superposition of the two concepts is well illustrated by the relevance of epigenetics, the (partially) transmissible and reversible phenotypic modifications of the genetic information. In the field of interest that goes under the collective name of “studies on the origin of life”, these complexities are not resolved, as indicated by the debate on “genetic-first”, or “metabolism-first”, or “membrane-first”.
This debate is probably parochial, partly influenced by the specific preparation and by the point of view of the proponents. A sound original process possibly had no “first” and the three questions had to be solved jointly before primitive cells could form and start evolving. The origin of primitive cells (3) requires that vesicles could form to be able to perform nutrient intake, that nucleotides could form and concentrate in them, that one or more energy source could be chemically mastered and transformed. It is difficult to envisage that the three processes could take place separately and/or that they were based on incompatible chemical frames.
With more attractive alternatives, in the chemical scenario, in which the three processes could occur within the same time span, at comparable rates and in compatible chemical frames, the occurrence of joint processes would be naturally favored. And the emergence of new properties would also be naturally favored. Thus, leaving the rest of the concomitant processes to their natural self, we examine here in more detail the possibilities connected with the shaping of one of the three processes, the genetic one. If the chemistry and the conditions allowing its development are compatible with those onto which the other processes are based, a reciprocal validation would be obtained. If a single chemical setting is responsible for the evolution of the three processes, the likely possibility that the origin of protocells has been based on this very setting appears to be rather high.
Stability and simplicity
Our operational approach is based on two assumptions: the need of simplicity, and the need of stability. If it is true that (1) generation of transmissible information, (2) mastering of energy transformation, and (3) definition of a reproducible in-and-out (genetics, metabolism and membranes) had to develop all at the same time and in compatible physico-chemical scenarios, the chemistry involved had to be robust and versatile. Hence simplicity.
The reason for that genetic complexity (which is the common denominator for the various forms of living matter we know) emerges from chemical simplicity is admittedly based on a robust principle: increase of stability. This perspective explains why from simple one-carbon atom (i.e., formaldehyde) molecules more relatively complex ones (namely sugars) are formed in the absence of other selection procedures. Increase of stability has the function of a sort of Darwinian selection principle. Paraphrasing from Darwin: the more stable will survive. Or, as elegantly stated by V. Stenger, “something came from nothing because it was more stable than nothing” (4).
A unitary chemical frame for the generation of the precursors of genetic information
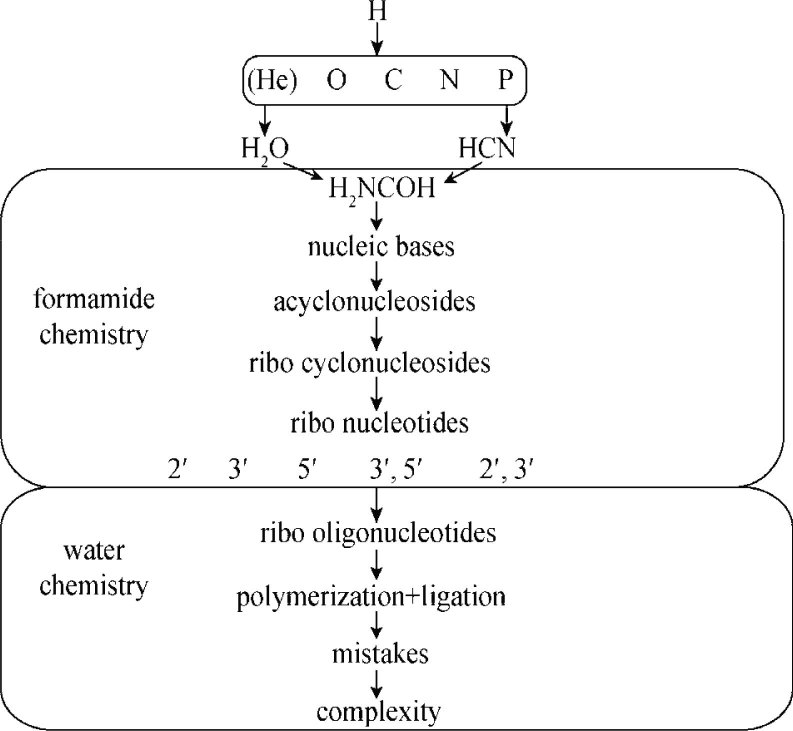
The progression towards increasingly stable states may be traced far back, starting from the transformation of H to more complex atoms. Figure 1 reports, in a concise and rigid flowchart, a possible path going from H to the emergence of pre-genetic complexity. The synthesis of the elements and the formation of stars is a well ploughed and developed field. The fact that one of the prerequisites for life is the presence of elements, such as H, C, O, N, S and P, is well recognized (5), as well as the fact that these are the most abundant elements in space. The reactions of these atoms in space result in a wealth of combinations, whose detection and listing are reported in the website http://astrochemistry.net/. The 151 molecules listed today (08/08/2010) go from molecular hydrogen H2 to the complex cyanodecapentayne HC11N. In a prebiotic perspective based on the principles of stability and simplicity, the relevant facts are that the most abundant interstellar carbon-containing 3-atom molecule is hydrogen cyanide HCN, and that the most abundant inorganic 3-atom is water H2O.
Figure 1.
A flowchart showing from hydrogen to the emergence of sequence complexity. All the first necessary steps may be accomplished in the unitary chemical frame of HCN/formamide chemistry, till the formation of acyclonucleosides (20) and to the closed-ring phosphorylation of nucleosides 15, 21. The abiotic synthesis of nucleosides was reported previously 29, 30. The spontaneous generation of RNA oligomers from purine 3′,5′-cyclic nucleotides (23) and their non-enzymatic ligation in water (24) were described. The figure is modified from previous study (31).
The C:N triple bond makes HCN highly reactive, hinting to its function as initiator of a cascade of synthetic reactions. When quenched with H2O, HCN yields formamide H2NCOH, more stable but still active, whose most attractive properties in a prebiotic perspective are its liquid state between 4°C and 210°C and the absence of appreciable azeotropic effects. Any dilute solution of formamide in water would rapidly concentrate upon experiencing temperatures higher than 100°C, making it an ideal starting point for synthetic pathways. For mixtures of formamide in other volatile solvents, the result would be similar. Not disregarding the possible direct role of HCN as precursor molecule of nucleic bases, chiefly adenine 6, 7, 8, the thorough investigation of HCN chemistry 9, 10 highlighted the complexity and the sophistication of the chemical pathways involved. As an Occam’s logic-based principle, it is probably safe to assume that if a series of reactions leading to the formation of cytosine differed from the line of the reactions leading to guanine, its prebiotic value is scarce. In order to be effective in terms of genetic origin, the precursors of nucleic acids had to be present at the same time in the same place (or in the same physical-chemical environment) in the appropriate comparable amounts. This would circumvent all the paramount difficulties entailed by differential stabilities and by the necessity of generating precursors that were, all and at the same time, simply and amply activated to be able to spontaneously polymerize in the absence of enzymes 11, 12. Remaining in the greater HCN chemistry area, the synthesis of a large variety of nucleic bases (including adenine, cytosine, uracil and thymine) was reported from its derivative H2NCOH formamide 13, 14, 15, 16. Formamide affords its products simply by heating between 100°C and 160°C in the presence of one or more of a large class of prebiotically available minerals: silica, alumina, zeolites, CaCO3, TiO2, common clays, kaolin, montmorillonites, olivines, phosphate minerals 13, 14, 15, 16, sulphur and iron minerals (17), zirconium-based (18) and boron-based (unpublished) minerals. Guanine can be obtained from formamide by UV irradiation (19) at lower temperatures. The chemical paths leading to the prompt and physically non-fastidious syntheses of all the nucleic bases, and the fact that essentially each of the tested minerals yields a composite panel of nucleic bases show that condensation into nucleic bases is an intrinsic property of formamide itself. In the same formamide chemical frame, acyclonucleosides can be produced by photochemistry in the presence of TiO2 (20), and nucleosides can be phosphorylated in the presence of a phosphate source to yield both open and cyclic nucleotides 15, 21.
In each one of these steps, from formamide to cyclic phosphorylated nucleotides, the same principle applies: the conservation of the most stable molecular forms, relative to the ones that were synthesized and rapidly degraded. In particular, nucleosides can be phosphorylated in every possible position of the sugar moiety (2′, 3′, 5′) but after a sufficiently long time, the more stable 2′,3′- and 3′,5′-cyclic forms remain as the only products of the reaction 15, 21.
From monomers to polymers
We focus on homopolymers, which would have been the least ambitious ground-building beginning in the search for the paths that presumably led to life. Starting from purine 3′,5′-cyclic nucleotides (namely 3′,5′-cAMP and 3′,5′-cGMP), the synthesis of RNA chains was observed (22). The RNA chains, wired by canonical 3′,5′-phosphodiester bonds, formed in water at temperatures between 40°C and 90°C, in the absence of enzymes or cofactors. The polymerization mechanism, presumably based on simple transphosphorylations between purine cyclic nucleotides held in place by purine stacking (22), yielded approximately 25-nt RNA molecules from 3′,5′-cGMPs and up to 120 nt from 3′,5′-cAMPs. The latter were the product of terminal ligation of shorter spontaneously generated poly(A) ribo-oligomers. Apparently, this reaction has been facilitated by formation of complementary A•A base pairs (23). That is, the next step of sophistication that one would hope to somehow introduce—emergence of complementary structure, popped up unsolicited, in guileless natural way. The stacking-based non-enzymatic terminal ligation of parallel poly(A) oligomers (24) allows the formation of tandem-wise oriented RNA polymers from variously sized poly(A) oligomers. Thus, the three basic mechanisms needed in order to at least conceive the spontaneous generation of long RNA sequence (namely generation of oligomers, and their terminal ligation assisted by complementarity) have found their proof-of-principle. Is it possible that these two reactions set the ground for the generation of more complex sequence combinations? The experiments described in the following section show that this is the case.
Formation of RNA sequences and ligation on complementary RNA, with…mistakes
One will never know, perhaps, which were the very first RNAs where the exploration of evolutionary sequence space started from. Nevertheless, keeping this conservative assumption in mind, a solid series of arguments based on statistical, biological, structural and evolutionary considerations have lead to the proposal that the complementary repeating oligomers GGCGGCGGC… and GCCGCCGCC…, and their coding ability for glycine and alanine, were the starters of the triplet code 25, 26. These sequences, however, should have had some prehistory before they came to the (hypothetical) being. Common sense would suggest that the real starters have been simpler, perhaps, just homopolymers. The intrinsic ability of 3′,5′-cGMP to generate up to 30-nt oligomers was reported previously (22). The generation reaction occurs independently of added template but, when a complementary RNA oligo-CCCCC… is present in the cGMP polymerization reaction, the neo-synthesized GGGGG… oligomer terminally ligates to the complementary RNA chain (22). The net result is the creation of a complementary hairpin, one strand of which was provided by the experimenter, the other half was self-generated. The complementarity in this case is of a familiar Watson-Crick type.
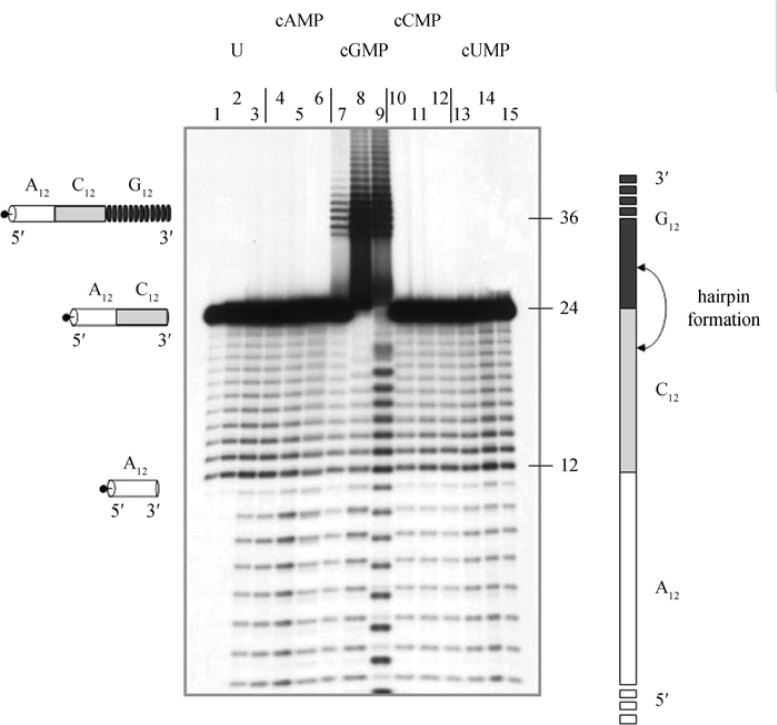
The reaction strictly depends on the polymerizing ability of 3′,5′-cGMP (Figure 2), showing that no oligomers ligating to a poly(C12) stretch are generated from 3′,5′-cAMP, 3′,5′-cCMP or 3′,5′-cUMP. The reverse experiment is not feasible, given that the polymerization conditions for 3′,5′-cCMP are still elusive and the resulting poly(C)-oligomers are only 3-4 nucleotides long (data not published).
Figure 2.
Non-enzymatically generated RNA oligomers terminally ligate to complementary sequences. Formation of homogeneous complementary GC sequences are shown. A 5′-labelled A12C12 oligomer was reacted with no (lanes 1-3) or with increasing amounts (0.1, 1, 10 mM in groups of 3, in increasing order) of respectively 3′,5′-cAMP (lanes 4-6), 3′,5′-cGMP (lanes 7-9), 3′,5′-cCMP (lanes 10-12) and 3′,5′-cUMP ((lanes 13-15). Lane 1: 5′-labelled A12C12 (0.2 μM, 60,000 cpm), untreated; lane 2: same, resuspended and precipitated, mock sample; lane 3: same, 8 h in 10 mM Tris-HCl, pH 5.3, no cyclic triphosphate. The samples were processed for acrylamide electrophoresis in denaturing conditions. For additional experimental details, see Costanzo et al. (22). The fragment length, the labelled position (dot), and the base composition (white: A; light grey: C; darker grey: G) are indicated.
Perfectly matching G:C double strands are the most stable ones, both in their homogeneous form (GGG/CCC) and in alternating combinations of G and C (27). This maximal stability reduces the exposure and the interactive possibilities of the sequence always favoring exact match of G and C. Various mismatches, however, are common events in mutagenesis. “Life is self-reproduction with variations” (28). This aphorism boils down from the analysis of the evolution of the genetic code (25) and accounts for the evolutionary necessity of allowable sequence matching imperfections. It is also in accord with Darwinian primacy of variations in natural selection. Are the sequence matching imperfections compatible with the terminal ligation of abiotically generated sequences and hairpin formation reported above?
Controllable way to monitor the mismatches and their effects is to replace the oligo(C) starter in the previous scheme by an imperfect version of it, while monitoring incorporation of G, the only free monomer in the reaction mixture. Figure 3 shows the results of terminal ligation of neo-generated oligo(G) on imperfectly matching sequences: GCCGCCGCC (Figure 3A) and GCCGCCGCA (Figure 3B and C). If the ligation and further elongation of oligo(G) on these starters occur, that would mean that the mismatches G•G and G•A are allowed by this system, since G is the only monomer present. The experiment, thus, consists of non-enzymatic polymerization of 3′,5′-cGMP in the presence of the indicated 5′-labelled oligomer, followed by electrophoretic analysis in strongly denaturing conditions. The neo-generated oligomers that are terminally ligated to the partially complementary sequences are seen in the upper part of the gel field. Although with an efficiency lower than that observed with homogeneous matching (Figure 2), ligation and chain growth occurs also on this heterogeneous combinations with the mismatches.
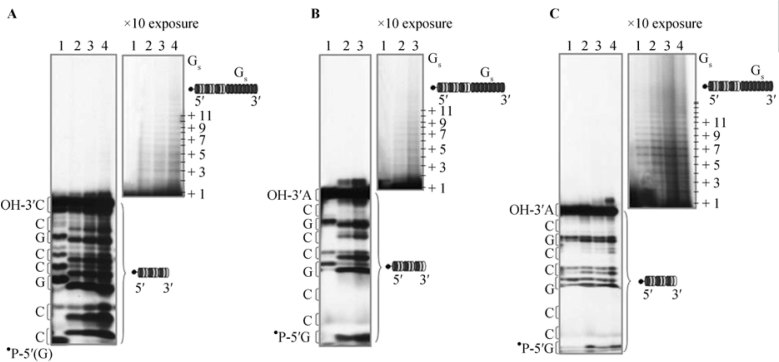
Figure 3.
Non-enzymatically generated RNA oligomers terminally ligate to degenerate complementary sequence. A. Synthesis of oligo(G) from 3′,5′-cGMP as a function of increasing amount of 5′-P-labelled GCCGCCGCC. Lane 1: 0.16 µM of 5′- ‘PGCCGCCGCC, no 3′,5′-cGMP. Lanes 2-4: 6 mM 3′,5′-cGMP in the presence of 0.16 μM, 0.3 μM and 0.6 μM of GCCGCCGCC, respectively. After reaction for 8 h at 60°C in 10 mM Tris-HCl, pH 5.3, the samples were processed for denaturing electrophoresis on acrylamide. For additional experimental details, see Brahms et al. (23). Left panel: autoradiographic exposure for 5 h; right panel: autoradiographic exposure for 50 h, only the upper part is shown. The double bands corresponding to each identified fragment of the template oligo are due to the hydrolytic process, resulting first in a 2′,3′-phosphate cyclic bridge (upper band), which is successively open (lower band). The presence of 3′,5′-cGMP increases the rate of opening. The neo-synthesized and terminally ligated oligo(G) is visible at higher exposure (left panel), as indicated. B. The same as Figure 3A for 5′-P-labelled GCCGCCGCA. Lane 1: no 3′,5′-cGMP; lane 2: 6 mM 3′,5′-cGMP with 0.16 μM of template oligo; lane 3: same with 0.3 µM. C. The same as Figure 3A for 5′-•PGCCGCCGCA (0.6 µM) reacted with 3′,5′-cGMP (6 mM) for 0, 0.5, 2 and 18 h, respectively.
Figure 3A shows the synthesis of oligo(G) as a function with increasing concentration of a 5′-labelled GCCGCCGCC template (as detailed in the legend to Figure 2). In the assay conditions (60°C, 8 h) the 9-mer undergoes partial degradation (lane 1). In the presence of increasing amounts of template (lanes 2-4) the neo-synthesized G oligomer ligates at the 3′ extremity with canonical 3′-5′ phosphoester bonds [(as shown by RNase analyses (22)] reaching a length >10 nt. With perfectly matching sequences neo-synthesized oligo(G) on preformed oligo(A12C12) (matching 12G to 12C, Figure 2), the efficiency of reaction is high, and at the monomer/template ratio of 5×103 (1 mM monomer/0.2 μM template) (lane 8), all the template oligomer is transformed into the corresponding hairpin structure. With the partially matching sequences [neo-synthesized oligo(G) on GCCGCCGCC, matching 6/9] at the ratio of 1×104 (lane 4), the efficiency is at least two orders of magnitude lower. Figure 3B shows that the same principle applies on molecular combinations with lower matching [neo-synthesized oligo(G) on GCCGCCGCA, matching 5/9].
Although with lower efficiency, in these molecular combinations with mismatches, G chains do grow and ligate. Similar results were observed with GCCGCCGCU and GCCGCCGCG (data not shown). Figure 3C shows the kinetics of RNA oligo(G) chain growth and terminal ligation on 5′-PGCCGCCGCA. The peak of the synthetic reaction is reached after two hours, when hydrolytic degradation starts. The combinatorial limits of this assay are currently matter of detailed analysis.
The overall result is, however, clear: not only the mismatches are allowed to some degree in the syntheses, but, more generally, the generation of complex sequences is possible in principle. The efficiency of the synthesis of complementary hairpin-forming RNA oligonucleotides depends on the details of the physico-chemical environment, but RNA does replicate, with…mistakes.
Green light to direct self-reproduction with variations
The demonstration above is not yet literally letter and spirit of the minimalistic definition of life, as it is not self-reproduction yet. However, since the self-reproduction in a similar easy-to-design system as simple as the setting above would be, obviously, based on complementary syntheses, both components of the definition (in this case complementary synthesis and sequence variations) are in place. The study, therefore, opens the green light to direct creation of artificial life in its simplest RNA world version. An exciting experimental perspective is to watch evolution of the replicating system towards best efficiency leading, perhaps, to the predicted emergence of the GCC tandem repeats, known to be the best expandable sequences in the triplet expansion diseases.
There is plenty of room for a sceptic, who would deny the proposed self-reproducing and evolving system for the status of living entity. The power of evolution inherent in the system is, however, as immense as combinatorial potential of RNA sequences, and subsequent emergences one would expect from such evolving system may well turn to qualify as creation of life of an undeniable status.
Authors’ contributions
SP collected the datasets and conducted data analyses. ET and EM supervised the project and co-wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank Silvia Lopizzo for helpful contributions. This work was supported by Grant ASI-INAF n. I/015/07/0 “Esplorazione del Sistema Solare”.
References
- 1.Rasmussen S. Evolution. Transitions from nonliving to living matter. Science. 2004;303:963–965. doi: 10.1126/science.1093669. [DOI] [PubMed] [Google Scholar]
- 2.Joyce J. Foreword. In: Deamer D.W., Fleischaker G.R., editors. Origins of Life: the Central Concepts. Jones and Bartlett; Boston, USA: 1994. pp. xi–xii. 233. [Google Scholar]
- 3.Meierhenrich U.J. On the origin of primitive cells: from nutrient intake to elongation of encapsulated nucleotides. Angewandte Chemie International Edition. 2010;49:3738–3750. doi: 10.1002/anie.200905465. [DOI] [PubMed] [Google Scholar]
- 4.Stenger V.J. The Comprehensible Cosmos: Where Do the Laws of Physics Come from? Prometheus Books; Amherst, USA: 2006. [Google Scholar]
- 5.Spaans M. The synthesis of the elements and the formation of stars. In: Ehrenfreud P., editor. Astrobiology: Future Perspectives. Kluwer Academic Publishers; Dordrecht, Netherlands: 2004. pp. 1–16. [Google Scholar]
- 6.Orò J., Kimball A.P. Synthesis of adenine from ammonium cyanide. Biochem. Biophys. Res. Commun. 1960;2:407–412. [Google Scholar]
- 7.Orò J. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive earth conditions. Nature. 1961;191:1193–1194. doi: 10.1038/1911193a0. [DOI] [PubMed] [Google Scholar]
- 8.Orò J., Kimball A. Synthesis of purines under possible primitive earth conditions. I. Adenine from hydrogen cyanide. Arch. Biochem. Biophys. 1961;94:217–227. doi: 10.1016/0003-9861(61)90033-9. [DOI] [PubMed] [Google Scholar]
- 9.Orgel L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 10.Delaye L., Lazcano A. Prebiological evolution and the physics of the origin of life. Physics Life Rev. 2005;2:47–64. doi: 10.1016/j.plrev.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Van Holde K.E. The origin of life: a thermodyanic critique. In: Halvorson H.O., Van Holde K.E., editors. The Origins of Life and Evolution. Alan R. Liss; New York, USA: 1980. pp. 31–46. [Google Scholar]
- 12.Alberty R.A. Thermodynamic properties of enzyme-catalyzed reactions involving guanine, xanthine, and their nucleosides and nucleotides. Biophys. Chem. 2006;121:157–162. doi: 10.1016/j.bpc.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Saladino R. On the prebiotic synthesis of nucleobases, nucleotides, oligonucleotides, pre-RNA and pre-DNA molecules. Topics in Current Chemistry. 2005;259:29–68. [Google Scholar]
- 14.Saladino R. Formamide chemistry and the origin of informational polymers. Chem. Biodivers. 2007;4:694–720. doi: 10.1002/cbdv.200790059. [DOI] [PubMed] [Google Scholar]
- 15.Saladino R. From formamide to RNA: the roles of formamide and water in the evolution of chemical information. Res. Microbiol. 2009;160:441–448. doi: 10.1016/j.resmic.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Ciciriello F. Spontaneous generation revisited at the molecular level. In: Pontarotti P., editor. Evolutionary Biology: Concept, Modeling and Application. Springer; Berlin, Germany: 2009. pp. 3–22. [Google Scholar]
- 17.Saladino R. Synthesis and degradation of nucleic acid components by formamide and iron sulfur minerals. J. Am. Chem. Soc. 2008;130:15512–15518. doi: 10.1021/ja804782e. [DOI] [PubMed] [Google Scholar]
- 18.Saladino R. The role of the formamide/zirconia system on the synthesis of nucleobases and biogenic carboxylic acid derivatives. J. Mol. Evol. 2010;71:100–110. doi: 10.1007/s00239-010-9366-7. [DOI] [PubMed] [Google Scholar]
- 19.Barks H.L. Guanine, adenine, and hypoxanthine production in UV-irradiated formamide solutions: relaxation of the requirements for prebiotic purine nucleobase formation. Chembiochem. 2010;11:1240–1243. doi: 10.1002/cbic.201000074. [DOI] [PubMed] [Google Scholar]
- 20.Saladino R. One-pot TiO2-catalyzed synthesis of nucleic bases and acyclonucleosides from formamide: implications for the origin of life. Chembiochem. 2003;4:514–521. doi: 10.1002/cbic.200300567. [DOI] [PubMed] [Google Scholar]
- 21.Costanzo G. Nucleoside phosphorylation by phosphate minerals. J. Biol. Chem. 2007;282:16729–16735. doi: 10.1074/jbc.M611346200. [DOI] [PubMed] [Google Scholar]
- 22.Costanzo G. Generation of long RNA chains in water. J. Biol. Chem. 2009;284:33206–33216. doi: 10.1074/jbc.M109.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brahms J. Adenylate oligomers in single and double-strand conformation. J. Mol. Biol. 1966;15:467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- 24.Pino S. Nonenzymatic RNA ligation water. J. Biol. Chem. 2008;283:36494–36503. doi: 10.1074/jbc.M805333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trifonov E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene. 2000;261:139–151. doi: 10.1016/s0378-1119(00)00476-5. [DOI] [PubMed] [Google Scholar]
- 26.Trifonov E.N. Distinct stages of protein evolution as suggested by protein sequence analysis. J. Mol. Evol. 2001;53:394–401. doi: 10.1007/s002390010229. [DOI] [PubMed] [Google Scholar]
- 27.Serra M.J., Turner D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 28.Trifonov E.N. The origin of the genetic code and of the earliest oligopeptides. Res. Microbiol. 2009;160:481–486. doi: 10.1016/j.resmic.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Bean H.D. Formation of a beta-pyrimidine nucleoside by a free pyrimidine base and ribose in a plausible prebiotic reaction. J. Am. Chem. Soc. 2007;129:9556–9557. doi: 10.1021/ja072781a. [DOI] [PubMed] [Google Scholar]
- 30.Powner M.W. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 31.Di Mauro E. On the emergence of pre-genetic information. J. Cosmol. 2010;10:3381–3387. [Google Scholar]