Abstract
Recently non-coding RNA (ncRNA) genes have been found to serve many important functions in the cell such as regulation of gene expression at the transcriptional level. Potentially there are more ncRNA molecules yet to be found and their possible functions are to be revealed. The discovery of ncRNAs is a difficult task because they lack sequence indicators such as the start and stop codons displayed by protein-coding RNAs. Current methods utilize either sequence motifs or structural parameters to detect novel ncRNAs within genomes. Here, we present an ab initio ncRNA finder, named ncRNAscout, by utilizing both sequence motifs and structural parameters. Specifically, our method has three components: (i) a measure of the frequency of a sequence, (ii) a measure of the structural stability of a sequence contained in a t-score, and (iii) a measure of the frequency of certain patterns within a sequence that may indicate the presence of ncRNA. Experimental results show that, given a genome and a set of known ncRNAs, our method is able to accurately identify and locate a significant number of ncRNA sequences in the genome. The ncRNAscout tool is available for downloading at http://bioinformatics.njit.edu/ncRNAscout.
Keywords: Genome-wide ncRNA discovery, Sequence motifs, Structural parameters
Introduction
Non-coding RNA (ncRNA) is a term that describes any RNA that is not translated into a protein or any RNA family aside from mRNA. Non-coding RNAs have many important intracellular functions [1], [2]. For example, rRNAs and tRNAs assist in mRNA translation; small nuclear RNAs (snRNAs) splice mRNA; and small nucleolar RNAs (snoRNAs) are involved in the modification of rRNAs [3]. Although ncRNA sequences are abundant within genomes [4], [5] with numbers comparable to those of protein-coding genes [6], many potential ncRNA families are yet to be discovered and their functions are yet to be analyzed. To date, imperfect methods have led to an oversight of ncRNA sequences, even in extensively studied genomes, such as that of Saccharomyces cerevisiae [7], [8]. These ncRNA sequences must be identified using new methodologies. The ability to identify potential ncRNA regions within a genome will allow researchers to further the boundaries of knowledge of yet-to-be discovered ncRNA families and their likely intracellular roles.
The difficulty in discovering ncRNA genes within a given genomic sequence chiefly originates from their primary sequences not being evolutionarily conserved. Hence, methods used in the discovery of protein-coding regions, such as searching for start and stop codons or regions with coding potential, are not effective in search for ncRNA regions [9]. A better method would be to combine sequence and structural features when discovering ncRNA genes [10], [11], [12], [13].
Current tools utilized in ncRNA exploration can be classified into three categories [1], [9]: (i) ncRNA homology search, (ii) ncRNA prediction, and (iii) ab initio ncRNA discovery. This work mainly focuses on the third category. Examples of bioinformatics tools belonging to the first category are BLAST [14], tRNAscan-SE [15], R-Coffee [16], and Infernal [13], [17]. Using a combination of BLAST and tRNAscan-SE allows one to annotate most rRNAs and tRNAs [18]. R-Coffee [16] computes multiple sequence alignments suitable for ncRNA searches. Infernal has been applied to roX1 RNA detection in Drosophila genomes [10]. However, these tools are limited in the numbers of ncRNA families they are able to detect. The tool GotohScan [18] attempts to fix these problems with homology search by utilizing a semi-global alignment approach. GotohScan performs fairly well compared to the aforementioned tools as it is able to identify tRNAs, rRNAs, snRNAs, and many other types of ncRNA, with the exception of microRNAs. Although effective in detecting known ncRNA families, bioinformatics tools that solely rely on homology searches such as GotohScan have not been able to discover novel ncRNAs (i.e., those that may not belong to any known ncRNA families).
Examples of bioinformatics tools in the secondary category designed for ncRNA prediction include QRNA [19], ddbRNA [20], MSARI [21], and EvoFold [22]. The best known tool in this category is perhaps RNAz [5]. RNAz combines multiple alignments of 2–6 sequences with measures of secondary structure conservation and thermodynamic stability of base pairs. RNAz builds on other programs to accomplish its goal. These programs include RNAfold [23] for folding single sequences and RNAalifold [24] for predicting the consensus structure of aligned sequences. Thermodynamic stability is measured by minimum free energy (MFE). RNAz compares the MFE of base pairing within a given sequence to random sequences of the same length and base composition. The tool calculates a z-score, where negative z-scores indicate that a sequence is more stable than expected by chance. The MFE of the consensus structure, as calculated by RNAalifold, is compared to the average MFE of the secondary structures of the individual sequences in a multiple alignment through the usage of a structure conservation index (SCI). The z-score and SCI are combined in an SVM learning algorithm. This SVM algorithm, trained on a set of cross-species ncRNAs, is able to classify an inputted multiple alignment as ncRNA or not.
There are relatively fewer tools in the third category designed for ab initio ncRNA discovery. Two bioinformatics tools in this category are NCRNASCAN [8] and smyRNA [1]. Given a genomic sequence and a set of ncRNAs, these tools are capable of discovering novel ncRNAs (which may or may not belong to known ncRNA families). While NCRNASCAN relies on parameters within the secondary structure, smyRNA focuses on motifs within the primary sequence. NCRNASCAN had success in detecting microRNAs but failed for other ncRNAs. The developers of NCRNASCAN pointed out that secondary structure alone is generally not statistically significant for the detection of ncRNAs. On the other hand, the more recent tool, smyRNA, which uses sequence motifs, has been shown to discover many novel ncRNAs in genomic sequences. It is therefore reasonable to assume that some sequence motifs may be indicative of the presence of ncRNA.
At the moment, there is no method that combines both sequence motifs and structural parameters for ab initio ncRNA discovery. Since it has been shown that secondary structure alone lacks statistical significance in detecting ncRNAs [8], a hybrid approach is promising. In this study, we present a hybrid method, named ncRNAscout, for ncRNA discovery, and assess its performance relative to smyRNA.
Method
ncRNAscout is an ab initio method, which seeks to improve upon smyRNA by combining both sequence motifs and secondary structure parameters to determine the locations of ncRNA regions within a genome. ncRNAscout adopts three variables: (i) log-likelihood ratio, (ii) t-score of MFE, and (iii) a novel sequential variable probability (SVP). The log-likelihood ratio is used to determine possible regions within the genome that might contain ncRNA sequences. We locate these candidate regions in the genome where the log-likelihood ratio is maximized in a way similar to the scan algorithm employed in smyRNA. Used in a support vector machine (SVM), the t-score and SVP are combined to make a final decision about the presence of ncRNA in each candidate region identified using the log-likelihood ratio.
Log-likelihood ratio
The log-likelihood ratio [1] compares the frequency of a certain k-mer motif within a set of known ncRNAs and the frequency of the k-mer motif within the complete genomic sequence. The number of occurrences of the k-mer motif m in the known ncRNA set and the genome is expressed as and respectively. The frequency of m in the set of known ncRNAs and the frequency of that same motif in the genome are defined respectively as:
The log-likelihood ratio for the specified motif m is then defined as:
For a sequence S within the genome, the sum of all its individual k-mer motif scores will result in the log-likelihood score R for the entire sequence, as shown by:
Here, S[i:i + k − 1] is the k-mer motif in S starting at position i and ending at position i + k − 1. A higher log-likelihood score indicates that S is more common in the set of known ncRNAs and a lower log-likelihood score indicates that S is more common in the genome. A sequence S that is more common in the set of known ncRNAs will be considered more likely to be part of a novel ncRNA.
t-Score of MFE
As mentioned above, ncRNAscout first locates candidate sequences within the genome where the log-likelihood score of the sequences is maximized. The t-score of a candidate sequence’s MFE is used to determine whether or not the candidate sequence is ncRNA. To fold the candidate sequences and to calculate their MFE values, ncRNAscout employs the C libraries of the Vienna RNA package version 1.8.4 [25]. The MFE folding routines of this package were used. If a sequence has a high probability of not being random as determined by the t-score, then it is possible that it is ncRNA. After calculating the MFE of a sequence, the t-score is utilized to normalize the MFE value so that the sequence’s length is not a factor [26].
To calculate the t-score, the values of mean and standard deviation of random sequences of a certain nucleotide length were calculated. We generated x samples, each having y random nucleotide sequences, to calculate these values. We set x to 15 and y to 250, to create a sufficiently large sample size to represent the set of random sequences. The lengths of these sequences ranged from 50 to 750 nucleotides (nt), in steps of 50. Each sequence has the same GC content as the genome used as input. This then allows ncRNAscout to create and use a linear model to calculate the mean and standard deviation of the MFE values of nucleotide sequences at various lengths. While searching for ncRNA regions in the genome, the MFE e of potential ncRNA regions is calculated, the sample mean and sample standard deviation are obtained from the model and the t-score is calculated by:
Sequential variable probability (SVP)
SVP is another variable that utilizes sequence patterns. It contains four components: (i) GC content, (ii) continuous CG content, (iii) continuous CC content, and (iv) continuous GG content. The continuous X1 X2 content is the percentage of the sequence that has the X2 nucleotide directly following the X1 nucleotide on a DNA molecule. GC content has been previously used to identify ncRNA genes within AT-rich genomes such as those of the Archaea Pyrococcus furiosus and P. abyssi [27], because A-T pairings are detrimental to the thermal stability of an RNA secondary structure [28]. Consequently, G and C should be significantly more common within ncRNA sequences. Therefore it can be hypothesized that the continuous CG content, continuous CC content and continuous GG content should be higher in ncRNA sequences than in non-ncRNA sequences.
To get the SVP value, each component was assigned a weight, i.e., WGC, WCC, WGG, and WCG so that better indicators will have a larger presence in the value. Next, for each known ncRNA, we generate a corresponding nucleotide sequence of the same length and with the same GC content as the genome. The mean and standard deviation of the four components are then calculated and are used as the SVP model. During runtime, the four components of the SVP value, namely SGC, SCC, SGG, and SCG, are calculated for each possible ncRNA region; the SVP is calculated such that:
The individual components are calculated by getting a t-score of the SXX value in the random sequence distribution. Then, the resulting probability calculated using Boost C++ t-distribution libraries (http://www.boost.org/) is the SXX value.
Evaluation
Performance measures
To evaluate and compare ncRNAscout with smyRNA, four measures are used: (i) sensitivity, (ii) positive prediction value (PPV), (iii) Matthews correlation coefficient (MCC), and (iv) percentage of known ncRNAs detected. These performance measures are used to determine how accurately a method is able to locate the regions of known ncRNAs within a genome. Sensitivity focuses on the number of true positives (TP) and the number of false negatives (FN), PPV focuses on TP and the number of false positives (FP), and MCC is a combination of both. A TP is defined as a location within the genome that is determined by a method to be part of an ncRNA and that is actually part of an ncRNA. A FP is defined as a location within the genome that is determined by the method to be part of an ncRNA when it is not. A FN is defined as a location within the genome that is determined to not be ncRNA when it is ncRNA.
To calculate the performance measure values, the known ncRNA genes used in the study and the ncRNA sequences detected by the method are rewritten as a series of ‘ones’ and ‘zeroes’. A ‘one’ represents a known or detected ncRNA location and a ‘zero’ represents a portion of the genome that is not a known ncRNA or not determined by the method to be ncRNA. An AND operation is conducted upon the two sequences to determine the number of TPs and an XOR operation is used to determine the number of FPs and FNs. Sensitivity, PPV and MCC are then defined as:
Since MCC provides a balance between the sensitivity and PPV values as it takes both FP and FN into account, it is therefore used as the evaluation measure. In this work, an MCC of 0.5 is set as a cutoff for an acceptable prediction.
Evaluation of false discovery rates
To compute the false discovery rate (FDR) of a method, either wet-lab experiments must be conducted to identify every known ncRNA in the genome or an in silico experiment is conducted on shuffled genomes [11]. These shuffled genomes are created so that no unknown ncRNA exists in their nucleotide sequences. To shuffle the genome, we use the same algorithm as in [1], which consists of the following steps:
-
1.
Extract and remove all known ncRNA genes and store them in an array.
-
2.
Generate a number i that is larger than the nucleotide length of the genome G without the extracted genes.
-
3.
Repeat steps 4 and 5 i number of times.
-
4.
Generate two random integers x, y.
-
5.
Swap G[x] and G[y].
-
6.
Insert the previously extracted ncRNA genes back into the newly formed sequence.
This methodology ensures that the unknown ncRNA sequences located within the genome are broken up and will cause the method to search only for the known ncRNAs within the shuffled genome. An unshuffled genome is defined as a genome that was not put through these six steps. The FDR of a method is then defined as:
where TP and FP are calculated using a shuffled genome.
Classification of sequences
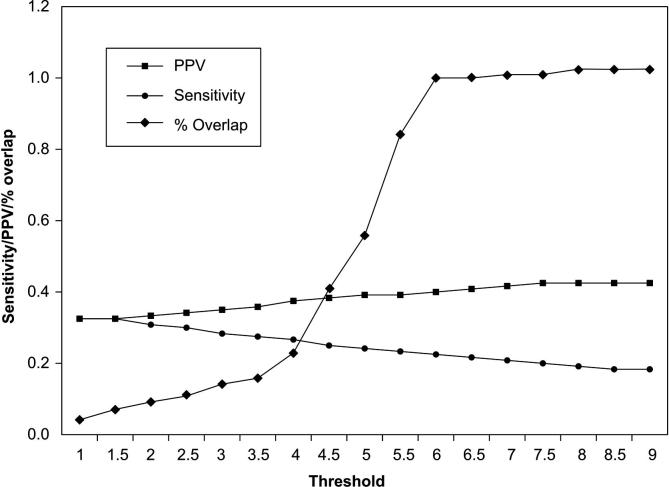
ncRNAscout performs a double checking on each sequence to make sure it is a potential ncRNA. First, the log-likelihood ratio of the sequence must be over a certain threshold. In smyRNA, a threshold value of 11.0 was used; however, this threshold value can be lowered to 6.0, a more lenient value that allows more regions to pass to the next stage in which the sequences are run through a SVM [29]. For ncRNAscout, the threshold value of 6.0 provided the best trade-off between sensitivity, PPV, and percent of findings/detections that overlap with known ncRNA sequences on a shuffled Escherichia coli genome (Figure 1).
Figure 1.
Sensitivity, PPV and percentage of detections that overlap with known ncRNA genes for ncRNAscout on a shuffled E. coli genome Half of the shuffled genome was used as training data and the other half was used as test data. ncRNAscout demonstrated the best performance at a threshold of 6.0 with a PPV of 0.393, sensitivity of 0.213, and percentage of overlap with known ncRNA genes of 1.0.
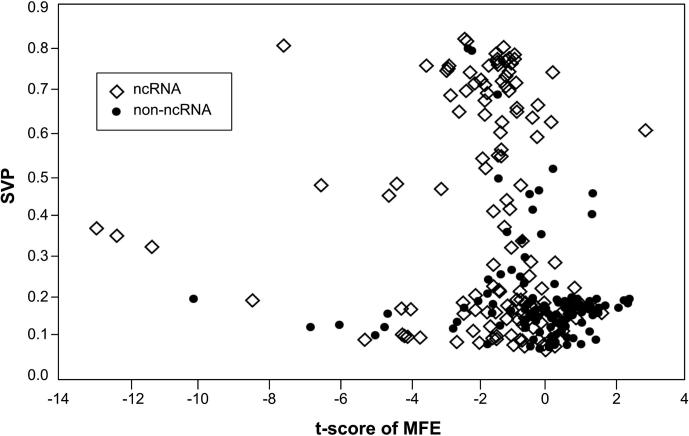
The SVM used in this study was the LIBSVM downloaded from http://www.csie.ntu.edu.tw/~cjlin/libsvm/ [30]. ncRNAscout uses a premade model trained on the genome of E. coli str. K-12 substr. DH10B (GenBank accession No. CP000948). The training set, which contains positive and negative examples, can be downloaded from http://bioinformatics.njit.edu/ncRNAscout. Positive sequences are ncRNAs on the E. coli genome that are obtained from the Rfam database v10.1 [31]. Negative sequences are non-ncRNAs that are randomly generated as done in producing shuffled genomes. Each training record contains two features, t-score of MFE and SVP, extracted from a sequence. A cross-plot is presented in Figure 2, which shows the distribution of the positive and negative examples in the training set.
Figure 2.
Cross plot showing the distribution of positive (ncRNA) and negative (non-ncRNA) examples in the training set.
We used the radial basis function (RBF) kernel provided in the LIBSVM package. The RBF kernel achieved the best results among all kernel functions included in the package. The optimal parameter values for the RBF kernel were determined by grid search using 10-fold cross validation with the grid.py utility supplied by LIBSVM running on the training set (best C = 13.4543426441, γ = 8). The testing set contains sequences taken from four other genomes explained below, which are different from E. coli. The SVM took the t-score of MFE and the SVP value of a test sequence as input, and classified the sequence as either ncRNA or not.
Performance comparison
To effectively compare ncRNAscout and smyRNA using the aforementioned performance measures, both methods were run on four different microbial genomes: Acholeplasma laidlawii PG-8A (GenBank accession No. CP000896), Candidatus Methanoregula boonei 6A8 (GenBank accession No. CP000780), Brucella suis 1330 chromosome 1 (GenBank accession No. AE014291), and Acidovorax sp. JS42 (GenBank accession No. CP000539). These four genomes constitute the testing set. The genomes were chosen so that there would be a variety in both their nucleotide length and their GC content. This way, the usage of these four genomes allows the methods to be tested on a variety of situations including AT-rich, GC-rich, long, and short genomes.
To make sure the genome files are current and accurate, the complete records of the genomes were downloaded from NCBI’s GenBank in FASTA format. The downloaded FASTA files were then processed to remove all non-ATCG characters, such as the new line character and other ASCII code ‘00’ characters, thus ensuring that both the GC content of the genomes, as well as the other SVP components, can be calculated accurately. Next, for each of these four genomes, the set of known ncRNAs was downloaded from the Rfam database v10.1 [31] as a generic feature format (GFF) file. This file was then parsed to retrieve the start position, end position, and aliases of all entries of type “ncRNA” which were subsequently used to create an “rnapos” file. A trimmed version of this file without the aliases was used in testing ncRNAscout and smyRNA. Another trimmed rnapos file for E. coli without the aliases was used to train smyRNA according to the methodology of Salari et al. [1].
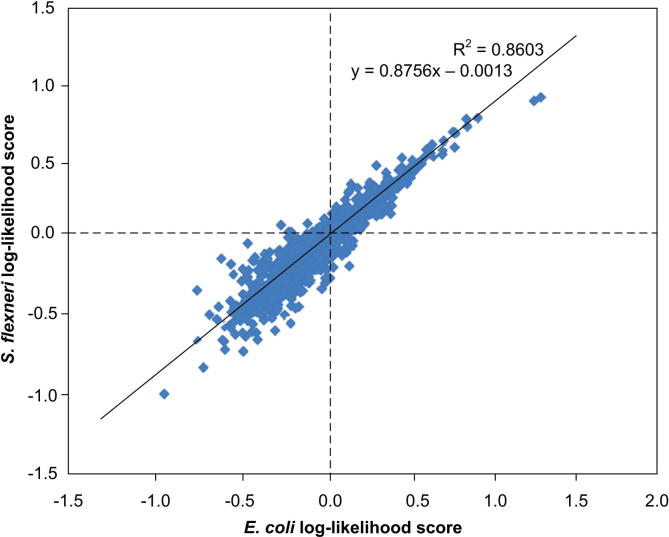
Like smyRNA, ncRNAscout’s log-likelihood ratio algorithm was set to search for pentamers (k-mer motif with k = 5) because Salari et al. [1] found the best results using this k value. To verify that the two methods of calculating log-likelihood ratios produce identical outcomes, the log-likelihood scores for E. coli and Shigella flexneri (GenBank accession No. AE005674) were compared. With smyRNA’s algorithm, a linear relationship was found between the log-likelihood scores of these two genomes. If ncRNAscout was accurately calculating the log-likelihood scores, the same relationship should be observed. Figure 3 supports this hypothesis.
Figure 3.
Correlation of log-likelihood scores between E. coli and Shigella flexneri With a correlation coefficient of approximately 0.928 and an R2 value of 0.8603 when using a linear fit, a linear relationship does exist for log-likelihood scores between E. coli and S. flexneri. This relationship demonstrates that log-likelihood score algorithms of ncRNAscout and smyRNA produce similar outcomes.
Once all the necessary data was prepared, ncRNAscout and smyRNA were used to scan the genomes. The results were then analyzed to determine the accuracy of both methods in identifying known ncRNA sequences. Each genome had its nucleotide length, GC content, and number of known ncRNAs recorded in addition to its potentially detectable ncRNA regions. We say that a method is able to detect a known ncRNA if the method can correctly identify at least one nucleotide in the known ncRNA sequence. That is, the putative ncRNA predicted by the method overlaps with the known ncRNA sequence. The results are summarized in Table 1, which shows that ncRNAscout is able to detect more of the known ncRNAs than smyRNA and with more accuracy as well.
Table 1.
ncRNAs detected in four genomes by ncRNAscout and smyRNA, respectively
| Genome source | Nucleotide length (nt) | GC content (%) | No. of known ncRNAs (No. of results detected by ncRNAscout, smyRNA) | Percentage of known ncRNAs detected (%) | No. of detections with MCC > 0.5 |
|---|---|---|---|---|---|
| Acholeplasmalaidlawii PG-8A | 1,496,992 | 31.92 | 42 (291, 389) | 92.857, 95.238 | 9, 12 |
| Acidovorax sp. JS42 | 4,448,856 | 66.17 | 134 (753, 3088) | 96.269, 70.149 | 20, 13 |
| Brucella suis 1330 chromosome 1 | 2,107,794 | 57.21 | 49 (239, 838) | 89.796, 71.429 | 34, 5 |
| Candidatus Methanoregula boonei 6A8 | 2,542,943 | 54.51 | 23 (76, 467) | 73.913, 56.522 | 10, 3 |
Note: In columns 4, 5, and 6, results from ncRNAscout are in bold and results from smyRNA are in italics.
For example, refer to the Acidovorax genome in Table 1. There are 134 known ncRNAs in the Acidovorax genome. ncRNAscout detects 96.269% (i.e., 129) known ncRNAs in the genome. In total, ncRNAscout detects 753 putative ncRNAs in the Acidovorax genome, among which 753–129 (i.e., 624) are unknown ncRNAs. On the other hand, smyRNA detects 70.149% (i.e., 94) known ncRNAs. Furthermore, ncRNAscout detects 20 ncRNAs with MCC > 0.5, while 13 ncRNAs with MCC > 0.5 were detected by smyRNA, which indicates that ncRNAscout is more accurate than smyRNA.
Details concerning the genomic locations of the putative ncRNAs detected by ncRNAscout can be found in Supplementary Material, available for download at http://bioinformatics.njit.edu/ncRNAscout. We also conducted experiments to compare ncRNAscout with smyRNA using different criteria where a method is said to detect a known ncRNA if the method correctly identifies at least three (five, respectively) nucleotides in the known ncRNA sequence. The results obtained based on these different criteria are the same as those given in Table 1.
We next examined which ncRNA types can be found using ncRNAscout and smyRNA respectively. To run this experiment, the Acidovorax genome was used because it contained the highest amount of known ncRNAs (134) and the largest number of ncRNA types (9) among the four genomes studied here. This provides a broader and more diverse dataset that can be analyzed. The results are summarized in Table 2, which shows that smyRNA is able to detect 8 types of ncRNA and ncRNAscout is able to detect 7 types. Neither method was able to detect every type of ncRNA.
Table 2.
Different types of known ncRNAs detected by ncRNAscout and smyRNA in the Acidovorax genome
| ncRNA type | Total No. in each type | No. detected by ncRNAscout | No. detected by smyRNA |
|---|---|---|---|
| SSU_rRNA_bacteria | 3 | 3 | 3 |
| PtaRNA1 | 1 | 0 | 1 |
| Bacteria_small_SRP | 1 | 0 | 1 |
| CRISPR_DR4 | 75 | 75 | 59 |
| tRNA | 46 | 43 | 46 |
| PK-G12rRNA | 3 | 3 | 3 |
| RNaseP_bact_a | 1 | 1 | 0 |
| tmRNA | 1 | 1 | 1 |
| 5S_rRNA | 3 | 3 | 3 |
Finally, an experiment was performed to estimate the FDR of ncRNAscout and smyRNA. Notice that results from an unshuffled genome could not be analyzed for FDR because there could be unknown ncRNAs in the genome that have yet to be discovered. This prompted the use of a shuffled version of the Acidovorax genome. Table 3 compares the FDRs of smyRNA and ncRNAscout, and shows the amount of their detections that overlapped with known ncRNA regions on the shuffled genome.
Table 3.
Results from ncRNAscout and smyRNA on the shuffled Acidovorax genome
| Method | No. of detections | Detections overlapping with ncRNAs (%) | FDR (%) | PPV | Sensitivity | MCC |
|---|---|---|---|---|---|---|
| ncRNAscout | 56 | 14.3 | 88.794 | 0.112 | 0.9696 | 0.3296 |
| smyRNA | 2894 | 0.584 | 99.883 | 0.00117 | 0.239 | 0.0168 |
Discussion
For the bacterial genomes analyzed in this work, ncRNAscout was able to discover significantly more known ncRNAs that surpass the MCC threshold of 0.5, compared to smyRNA (Table 1). This suggests that ncRNAscout is more accurate in its detections. On average, ncRNAscout detects approximately 88.21% of all the known ncRNAs in the four genomes in Table 1 with a standard deviation of 9.89%. In comparison, smyRNA detects approximately 73.33% of all the known ncRNAs in Table 1 with a standard deviation of 16.1%.
From Table 2, smyRNA appears to be able to detect more ncRNA types than ncRNAscout. Out of the 9 different types of known ncRNA, smyRNA detected 8 types and ncRNAscout was able to detect 7 types. Interestingly, ncRNAscout performed much better than smyRNA at detecting the CRISPR_DR4 ncRNA type and only slightly worse at detecting tRNA. However, ncRNAscout could be used in conjunction with tRNAscan-SE [15] to detect all tRNAs.
In addition, because GC content makes up 50% of the SVP variable, it would be reasonable to assume that if GC content were only significant in AT-rich genomes then there would be a correlation between the percentage of known ncRNAs detected and GC content. However, from ncRNAscout’s results, there appears to be no correlation between the two numbers (Table 1). These results indicate that SVP can be effectively used to detect ncRNA genes, and ncRNAscout is not only limited to AT-rich genomes. It is interesting to point out, though, that smyRNA performs its best in the AT-rich genome of Acholeplasma laidlawii PG-8A by detecting around 95% of all known ncRNAs (Table 1). In the other three GC-rich genomes in Table 1, known ncRNA detection rates ranged from 56% to 71% for smyRNA. At the same time, results generated by ncRNAscout remained relatively consistent.
Currently, a problem with ab initio ncRNA sequence discovery tools is the number of false positives they produce. It is impractical to do wet-lab experiments to verify each result detected by these tools. In our proposed approach, a reduction of FDR is necessary, and hence a second layer of verification is added to the process. From the shuffled genome results (Table 3), ncRNAscout has a lower FDR value of 88.794% compared to 99.883% with smyRNA. This number thus proves that the addition of the second layer of verification by the SVM using structural parameters and sequence patterns does reduce the FDR of the program. Future research will hopefully be able to utilize more accurate structural and sequential parameters to lower the FDR value even further. In addition to the FDR, ncRNAscout demonstrates improvement in its detection algorithm as it has higher PPV, sensitivity, and MCC values as well, as shown in Table 3.
Even though Rivas and Eddy [8] have shown that secondary structure itself is not significant in detecting ncRNA regions, and Klein et al. [27] have only been able to use GC content in AT-rich genomes, ncRNAscout is able to use both sequence motifs and structural parameters to detect ncRNAs with moderate success. Both ncRNAscout and smyRNA utilize similar pattern-searching methods to look for the initial regions where ncRNA may exist. While smyRNA only uses a threshold (with a value of 11.0) for the log-likelihood ratio, ncRNAscout lowers this threshold (with a value of 6.0) and uses it in combination with a SVM that takes t-scores of MFE and SVP values as input. Our experimental results demonstrated the effectiveness of this hybrid approach.
It should be pointed out that both ncRNAscout and smyRNA are designed for ab initio ncRNA discovery, which, given a genomic sequence and a set of ncRNAs, are capable of discovering novel ncRNAs (which may not belong to any known ncRNA families). These tools differ from the ncRNA homology search methods surveyed in the Introduction section since the latter aim to find members of known ncRNA families and are not applicable to novel ncRNA discovery. There have been efforts to use some of the ncRNA prediction tools surveyed in the Introduction section, such as RNAz, for novel ncRNA searches. However, the discovery ability of RNAz depends on the quality of multiple alignments inputted to the program [1].
Conclusion
In this paper we present a new ab initio method (ncRNAscout) for ncRNA discovery that seeks to merge sequence motifs with structural parameters. Our experimental results show that ncRNAscout is able to accurately identify more known ncRNAs than the closely related method, smyRNA. Both methods detect a large number of unknown ncRNAs (Table 1), suggesting that the two tools could be used together for novel ncRNA discovery. Since ncRNAscout utilizes the same basic concept as smyRNA, we conclude that the additional parameters employed by ncRNAscout, including t-scores of MFE and SVP values, are what make ncRNAscout more accurate than smyRNA. Together, sequence motifs and structural parameters have the potential to contribute to the building of leading methods for genome-wide ncRNA discovery.
Authors’ contributions
MB designed and implemented the algorithm, conducted experiments and drafted the manuscript. MCC and JW conceived the idea of using this approach and assisted with manuscript preparation. LZ verified the computational procedures and results. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank the anonymous reviewers for many suggestions. This research was supported in part by US National Science Foundation (Grant No. IIS-0707571).
References
- 1.Salari R. SmyRNA: a novel ab initio ncRNA gene finder. PLoS One. 2009;4:e5433. doi: 10.1371/journal.pone.0005433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki M., Hayashizaki Y. Mouse-centric comparative transcriptomics of protein coding and non-coding RNAs. Bioessays. 2004;26:833–843. doi: 10.1002/bies.20084. [DOI] [PubMed] [Google Scholar]
- 3.Mattick J.S., Makunin I.V. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 4.Khaladkar M. RADAR: a web server for RNA data analysis and research. Nucleic Acids Res. 2007;35:W300–W304. doi: 10.1093/nar/gkm253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washietl S. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cawley S. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 7.Olivas W.M. Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res. 1997;25:4619–4625. doi: 10.1093/nar/25.22.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas E., Eddy S.R. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000;16:583–605. doi: 10.1093/bioinformatics/16.7.583. [DOI] [PubMed] [Google Scholar]
- 9.Meyer I.M. A practical guide to the art of RNA gene prediction. Brief Bioinform. 2007;8:396–414. doi: 10.1093/bib/bbm011. [DOI] [PubMed] [Google Scholar]
- 10.Byron K. Mining roX1 RNA in Drosophila genomes using covariance models. Int J Comp Biosci. 2010;1:22–32. [Google Scholar]
- 11.Khaladkar M. Mining small RNA structure elements in untranslated regions of human and mouse mRNAs using structure-based alignment. BMC Genomics. 2008;9:189. doi: 10.1186/1471-2164-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J. A method for aligning RNA secondary structures and its application to RNA motif detection. BMC Bioinformatics. 2005;6:89. doi: 10.1186/1471-2105-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nawrocki E.P. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe T.M., Eddy S.R. TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilm A. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nawrocki E.P., Eddy S.R. Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput Biol. 2007;3:e56. doi: 10.1371/journal.pcbi.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertel J. Non-coding RNA annotation of the genome of Trichoplax adhaerens. Nucleic Acids Res. 2009;37:1602–1615. doi: 10.1093/nar/gkn1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivas E., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.di Bernardo D. DdbRNA: detection of conserved secondary structures in multiple alignments. Bioinformatics. 2003;19:1606–1611. doi: 10.1093/bioinformatics/btg229. [DOI] [PubMed] [Google Scholar]
- 21.Coventry A. MSARI: multiple sequence alignments for statistical detection of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:12102–12107. doi: 10.1073/pnas.0404193101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen J.S. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofacker I.L. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 25.Gruber A.R. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spirollari J. Predicting consensus structures for RNA alignments via pseudo-energy minimization. Bioinform Biol Insights. 2009;3:51–69. doi: 10.4137/bbi.s2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein R.J. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc Natl Acad Sci USA. 2002;99:7542–7547. doi: 10.1073/pnas.112063799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakovchuk P. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 30.Fan R.-E. Working set selection using the second order information for training SVM. J Mach Learn Res. 2005;6:1889–1918. [Google Scholar]
- 31.Gardner P.P. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]