Abstract
Despite its efficacy against malaria, the relatively low yield (0.01%-0.8%) of artemisinin in Artemisia annua is a serious limitation to the commercialization of the drug. A better understanding of the biosynthetic pathway of artemisinin and its regulation by both exogenous and endogenous factors is essential to improve artemisinin yield. Increasing evidence has shown that microRNAs (miRNAs) play multiple roles in various biological processes. In this study, we used previously known miRNAs from Arabidopsis and rice against expressed sequence tag (EST) database of A. annua to search for potential miRNAs and their targets in A. annua. A total of six potential miRNAs were predicted, which belong to the miR414 and miR1310 families. Furthermore, eight potential target genes were identified in this species. Among them, seven genes encode proteins that play important roles in artemisinin biosynthesis, including HMG-CoA reductase (HMGR), amorpha-4,11-diene synthase (ADS), farnesyl pyrophosphate synthase (FPS) and cytochrome P450. In addition, a gene coding for putative AINTEGUMENTA, which is involved in signal transduction and development, was also predicted as one of the targets. This is the first in silico study to indicate that miRNAs target genes encoding enzymes involved in artemisinin biosynthesis, which may help to understand the miRNA-mediated regulation of artemisinin biosynthesis in A. annua.
Key words: artemisinin, microRNA, EST, computational prediction, Artemisia annua
Introduction
Malaria is a global health problem with more than 1 billion people living in areas at a high risk of the disease. Artemisinin combination therapies (ACTs) are the recommended treatment regimen for uncomplicated malaria caused by the Plasmodium falciparum parasite (1). It has long been recognized that the problem of artemisinin resistance is best addressed by increasing access to ACTs (2). This approach receives strong support from the global health community. However, there is growing concern that the supply chain will be unable to consistently produce high-quality artemisinin in the quantities that will be required (2). ACT supply remains reliant on the agricultural production of artemisinin, which is a sesquiterpenoid synthesized in the glandular trichomes of the Chinese medicinal plant sweet wormwood (Artemisia annua L.) (3). However, artemisinin concentration in A. annua is low, (in the range of 0.01%-0.8% per dry weight of tissue), which seriously limits the commercialization of the drug (4). Therefore, improved varieties of A. annua for farmers in the developing countries would bring immediate benefits to the existing artemisinin supply chain by reducing production costs, stabilizing supplies, and improving growers’ confidence in the crop (2).
Conventional breeding and genetic engineering approaches would allow construction of A. annua genotypes rich in artemisinin (4). A better understanding of the biochemical pathway leading to the artemisinin synthesis and regulation by both exogenous and endogenous factors is essential for facilitating yield increase (5). With the elucidation of the artemisinin biosynthetic pathway and identification of amorpha-4,11-diene synthase (ADS), which catalyses the first biosynthetic step in artemisinin biosynthesis, it is possible to explore unconventional alternate strategies that are economically viable for the commercial production of artemisinin. Two approaches appear promising. The first approach is to synthesize artemisinin from its simple precursor such as artemisinic acid via semi-synthetic route. Recently, scientific effort is being directed to develop a biological method to supply sufficient and reliable quantities of artemisinic acid, a direct precursor of artemisinin. For example, there was report on engineering of Saccharomyces cerevisiae to produce high titres (up to 100 mg/L) of artemisinic acid using an engineered mevalonate pathway, ADS, and a novel cytochrome P450 monooxygenase (CYP71AV1) from A. annua (6). The second approach is to regulate key enzymes leading to increased artemisinin biosynthesis with metabolic engineering. Certainly, the latter strategy has provided some exciting results and further efforts may accelerate commercialization of this crucial drug. Zhang et al. showed that down-regulation of squalene synthase (SQS), a key enzyme of sterol pathway that is competitive with artemisinin biosynthetic pathway, by hairpin-RNA-mediated gene silencing in A. annua resulted in a 3-fold increase in artemisinin production (7). Furthermore, study on the expression of genes involved in the terpene metabolism also indicated that SQS may significantly compete for farnesyl diphosphate (FDP) in artemisinin-producing tissues of A. annua (8). In addition, higher artemisinin content was reported in induced tetraploid A. annua, which may result from the upregulated expression of some key enzyme genes related to artemisinin biosynthesis including ADS, farnesyl diphosphate synthase (FPS), HMG-CoA reductase (HMGR) and artemisinin metabolite-specific aldehyde hydrogenase 1 (9). Therefore, a better understanding of the molecular mechanisms involved in the artemisinin biosynthesis and regulation will provide better strategies to develop new varieties with a higher content of artemisinin.
miRNAs are a large family of endogenous small RNAs containing ~22 nucleotides, which are derived from large precursors that are transcribed from non-protein-coding genes (10). Plant miRNAs generally interact with their targets through perfect or near-perfect complementarity and repress translation 11, 12 or cleave targeted mRNAs (13), thus negatively regulate the expression of their target genes (14). Plant miRNAs target a large number of genes with functions in a range of development processes, including meristem cell identity (15), leaf organ morphogenesis 11, 12, polarity and floral differentiation and development (16). miRNAs are also reported to be involved in plant responses to biotic and environmental stresses (17). The metabolite biosynthesis is also regulated by miRNAs (18). A large number of miRNAs have so far been identified in various plant species. However, no miRNA from Asteraceae has been reported yet, reflecting a disparity between the important values of this plant family and insufficient molecular and genetic studies, including small RNA mediated gene regulation, in Asteraceae. To gain insight into miRNAs and their important regulatory functions in artemisinin biosynthetic pathway, we studied miRNA and their targets in A.annua using computational approach.
Computational or bioinformatics approach is one of the many approaches available for miRNA prediction (19), which can discover miRNAs not only from species with full genomic and sufficient EST database available, but also from those with incomplete genomic information while with sufficient EST sequences available (20). In addition, this approach is very useful for predicting miRNAs that usually cannot be detected by the direct cloning, particularly the low-abundance miRNAs. The computational approaches are based on homology search, gene search, neighbor stem-loop search, comparative genomic algorithm or phylogenetic shadowing (21). Homology search, which can be further classified as genome-based search or EST-based search (22), is based on conserved sequences and secondary structures and identifies miRNA genes by searching nucleotide databases using BLAST. Using homology search, orthologues of known miRNAs were revealed in different species, supporting that miRNAs are conserved in different species (23). In addition, hundreds of new miRNAs were also identified using this method from the genomes of model species, such as Arabidopsis (24) and rice (Oryza. sativa) (25).
With the development of computational methods, several computer software programs have been developed to help identify plant potential miRNA target genes in mRNA sequences. Because almost all miRNAs show perfect or near-perfect complementarity with their targets in plants, it is much easier to predict miRNA targets using a BLAST search of mRNA database. More and more studies have shown the success of this powerful approach to select potential miRNA targets in mRNA sequences for experimental validation (21).
Results and Discussion
Identification of potential miRNAs in A. annua
Most mature miRNAs are evolutionarily conserved from species to species within the plant kingdom, which facilitates the prediction of the existence of new miRNA orthologs or homologs in other plant species. In this study, we applied the comprehensive strategy to identify potential miRNAs in A. annua by searching EST against known miRNAs of a dicotyledonous plant Arabidopsis and a monocotyledonous rice.
Following the procedure depicted in Figure 1, 94,724 ESTs from A. annua were searched against 584 mature sequences of miRNAs from Arabidopsis and rice after removal of redundant sequences. In total, six potential miRNAs were predicted from A. annua (Figure 2). The six identified A. annua candidate miRNAs belong to two miRNA families. miR1310 family has one miRNA. miR414 family has five homologs with two from A. thaliana and three from O. sativa, respectively (Table 1).
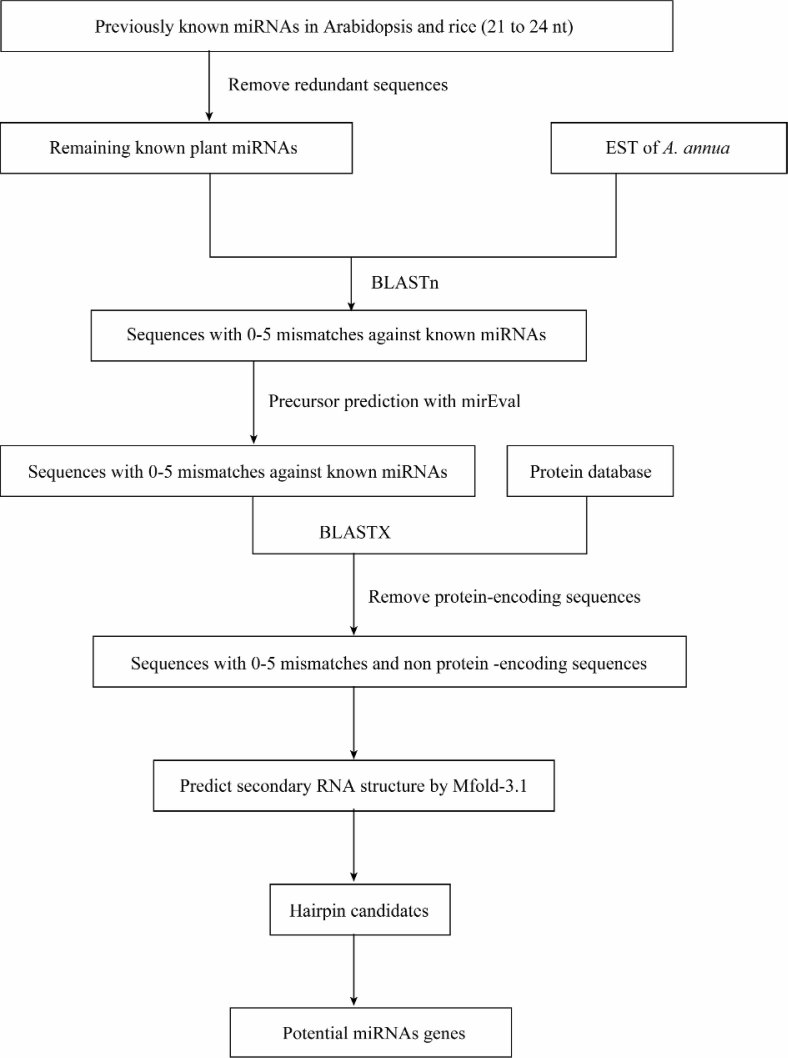
Figure 1.
Schematic diagram for searching potential miRNA genes in A. annua by identifying homologs of previously known plant miRNAs.
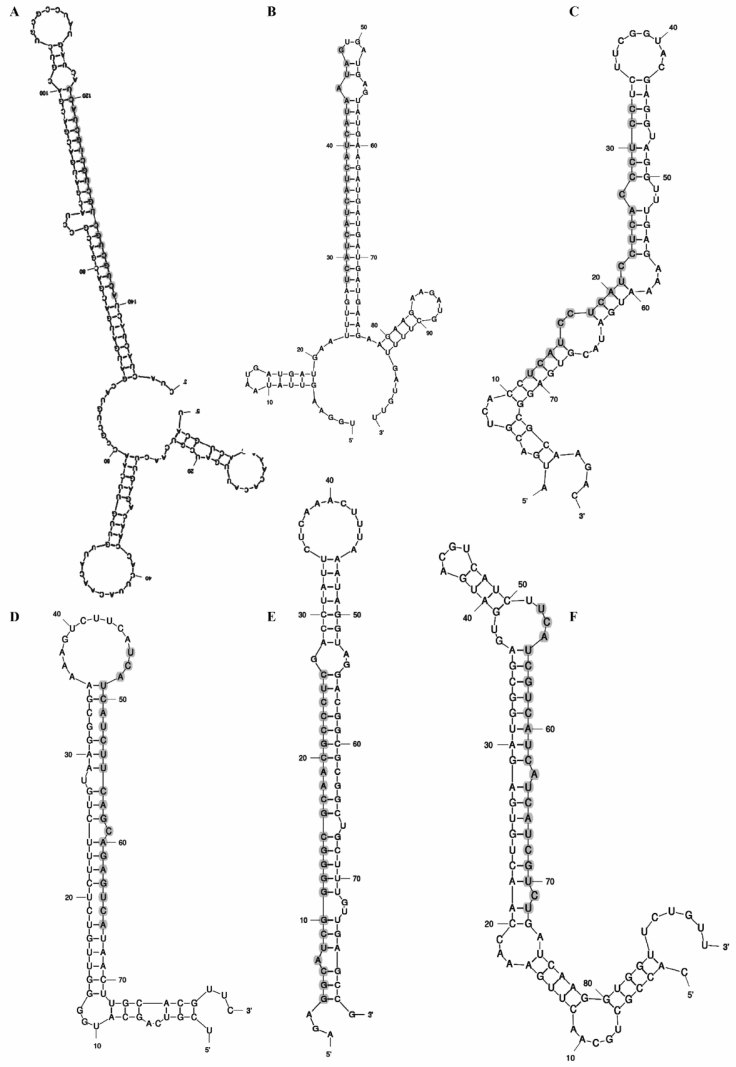
Figure 2.
Mature and precursor sequences and the predicted stem and loop structures of newly identified miRNAs in A. annua. The mature miRNAs are highlighted in gray. A. miR414 (EY064998; homolog of A. thaliana); B. miR414 (EY057163; homolog of O. sativa); C. miR414 (EY082442; homolog of O. sativa); D. miR414 (EY107691; homolog of A. thaliana); E. miR1310 (EY057327; homolog of O. sativa); F. miR414 (EY083764; homolog of O. sativa).
Table 1.
Newly identified miRNAs from ESTs of A. annua
| miRNA | Reference Species | Gene ID | EST length (nt) | NM (nt) | LM (nt) | LP (nt) | Side | A + U (%) | MFE | MEFI |
|---|---|---|---|---|---|---|---|---|---|---|
| miR414 | A. thaliana | EY107691 | 697 | 5 | 21 | 80 | 3′ | 55 | 14.7 | 0.41 |
| A. thaliana | EY064998 | 795 | 4 | 21 | 150 | 3′ | 52 | 46.1 | 0.64 | |
| miR414 | O. sativa | EY057163 | 759 | 5 | 21 | 100 | 5′ | 70 | 26.2 | 0.87 |
| O. sativa | EY083764 | 678 | 2 | 21 | 90 | 3′ | 52.2 | 28.5 | 0.66 | |
| O. sativa | EY082442 | 612 | 3 | 21 | 80 | 5′ | 48.8 | 22.5 | 0.55 | |
| miR1310 | O. sativa | EY057327 | 381 | 3 | 23 | 80 | 5′ | 42.5 | 29.4 | 0.64 |
Note: NM, number of mismatch; LM, length of mature miRNAs; LP, length of precursor; MFE, minimal folding free energy; MFEI, minimal folding free energy index.
It is estimated that in plants, approximate 10,000 ESTs contain 1 miRNA. Therefore, the total of 94,724 ESTs in A. annua examined in this study may contain 9-10 miRNAs. However, in this study, only six miRNAs were predicted even with maximal 5-mismatches were allowed. The average length of ESTs is 654 nt and the longest is 795 nt, while most of miRNA precursors have 80-150 nt as identified by MirEval software (26), suggesting that the EST may contain other element sequences in addition to miRNA precursor sequence (27). The length of miRNA precursors in A. annua varied from 80 to 150 nt, with an average of 97 nt. The different sizes of the identified miRNAs within different families suggest that they may offer unique functions for regulation of miRNA biogenesis or gene expression (28). The diversity of the identified miRNAs could be also found in the location of mature miRNA sequences. It is shown that the sequences of miR1310 and two members of miR414 from O. sativa family were located at the 5’ end of the miRNA precursors, while the other miR414 members were found at the 3’ end. Minimal folding free energy (MFE) is an important characteristic that determines the secondary structure of nucleic acids (DNA and RNA). The lower the MFE is, the higher the thermodynamically stable secondary structure of the corresponding sequence is (29). The MFE index (MFEI) for each sequence was calculated as previously reported (30). In this study, the MEFI values ranged from 0.41 to 0.87. miRNA precursor sequences have significantly higher MFEI value than other non-coding or coding RNAs. To avoid false calling of other RNAs as miRNA candidates, MFEI was also considered when predicting secondary structures (31).
Identifying miRNAs using EST analysis (32) has some advantages over other methods (33). It has been suggested that most of the miRNAs predicted by EST analysis can be recovered by high-throughput deep sequencing (34).
Although we have computationally identified six miRNAs, the number of miRNAs discovered is relatively small. A. annua belongs to Asteraceae, which is the largest plant family of vascular plants on Earth; comprising more than 23,000 genetically diverse and ecologically successful species (http://www.mobot.org/MOBOT/research/APweb/, Angiosperm Phylogeny Website, Version 9, 2011). Unfortunately, not a single miRNA from Asteraceae family has been deposited in the MiRbase (35). We expect that as more miRNAs of this family are publicly available, more miRNAs will be identified in A. annua. Interestingly, a recently published study reported that 151 potentially conserved miRNAs belonging to 26 miRNA families were successfully identified and characterized using qPCR in 11 genus of Asteraceae (36). In addition, another recent article reported 11 highly conserved miRNA precursors from 9 families using gene-oriented clusters of transcript sequences of A. annua with 88,174 UniGenes using a modified computational approach (37). However, no miRNA targets were found among genes encoding enzymes involved in artemisinin biosynthesis, maybe because the conserved miRNAs preform evolutionarily stable functions, or miRNA-mediated regulation of artemisinin synthesis could be exerted primarily by novel or clade-specific miRNAs.
Prediction of potential targets of putative miRNAs in A. annua
Gaining insight into the miRNA targets will help us understand the spectrum of miRNA regulation and elucidate the functional importance of miRNAs. miRNAs may directly target transcription factors which affect plant development and specific genes which control metabolism as well (38). To identify potential regulatory targets, we first searched mRNA database in A. annua and screened for mRNAs complementary to the six miRNAs with less than 4 mismatches. Gaps, G-U and other non-canonical pairs were not allowed and considered as mismatches. By screening against mRNA sequences of A. annua using the six newly identified miRNAs, we found 8 target genes complementary with less than 4 mismatches. Interestingly, one miRNA can be complementary to more than one regulatory target (Table 2). For example, six sequences were detected as targets of miR414 of A. annua.
Table 2.
List of the potential targets of newly identified miRNAs in A. annua
| miRNA | Gene ID | Target Gene ID | Target Protein | Target Function |
|---|---|---|---|---|
| miR414 | EY107691 | GQ468547 | Putative AINTEGUMENTA | Transcription regulation |
| EY064998 | DQ363131 | Putative flavonoid 3′-hydroxylase cytochrome P450 | Oxidation-reduction process | |
| miR1310 | EY057327 | AJ001539 | EPS | Lyase activity |
| miR414 | EY057163 | CAB56499 | Putative SQC | Lyase activity |
| EY083764 | FJ432667, AF327527 | ADS | Biosynthesis of artemisinin | |
| EY082442 | GQ420346 | FPS | Biosynthesis of cholesterol, isoprene, lipid, steroid and sterol | |
| U14625 | HMGR | Oxidation-reduction process | ||
| DQ363131 | Putative flavonoid 3′-hydroxylase cytochrome P450 | Oxidation-reduction process |
Note: EPS, epi-cedrol synthase; SQC, sesquiterpene cyclase; ADS, amorpha-4,11-diene synthase; FPS: farnesyl pyrophosphate synthase; HMGR, HMG-CoA reductase.
One target gene identified for rice miR414 homolog in A. annua (EY082442) is FPS (Table 2). FPS catalyzes two consecutive condensation reactions to produce FDP, which is the starting point of a large variety of essential isoprenoid end products, including artemisinin 39, 40. It is clear that the first dedicated step in the biosynthesis of artemisinin is the cyclization of FDP to form amorpha-4,11-diene catalyzed by ADS, one of the sesquiterpene cyclases (SQCs) (41). The cyclase reaction establishes an important stereochemical framework upon which all other chemical modifications take place (43). Interestingly, another rice miR414 homolog in A. annua, EY083764, is predicted to target ADS (Table 2). Modification of the amorpha-4,11-diene carbon skeleton to produce artemisinin acid was thought to involve a cytochrome P450 enzyme leading to the production of artemisinic alcohol, which could then be oxidized twice by either cytochrome P450 enzymes or dehydrogenases to yield artemisinic acid (44). In our study, we found that both EY082442 and another Arabidopsis miR414 homolog in A. annua (EY064998) were predicted to target putative flavonoid 3′-hydroxylase cytochrome P450 (DQ363131).
In addition, EY082442 was also predicted to target HMGR (U14625), which is a rate-limiting enzyme of the mevalonate pathway. Recently it has been shown that HMGR expression limits artemisinin formation in A. annua (8). Overexpression of ADS and HMGR led to significant increase in the artemisinin yield from the transgenic A. annua (42).
A third rice miR414 homolog of A. annua, EY 57163, targets a putative SQC (CAB56499), while epi-cedrol synthase (EPS, AJ001539), an identified SQC converting FDP to 8-epicedrol, is the only target of miR1310 homolog of A. annua, EY057327 (Table 2). A diagram depicting the involvement of miRNAs and their targets was shown in Figure S1.
Other than the enzymes involved in production of secondary metabolites, our computational result demonstrates that A. annua miR414 (EY107691) targets the mRNA encoding putative AINTEGUMENTA protein (GQ468547) (Table 2), which is a transcription activator that recognizes and binds to the DNA consensus sequence 5′-CAC[AG]N[AT]TNCCNAN G-3′ (45). AINTEGUMENTA is required for the initiation and growth of ovules integumenta, and also involved in organ initiation and development, including floral organs (46). Interestingly, a positive correlation was noticed between the plant age and artemisinin yield (47). Furthermore, artemisinin is present in high concentration in either flowers or leaves but low or zero in stems and roots (48). For A. annua, the highest artemisinin concentration has been reported in leaves and flowers during full bloom stage, in comparison to the pre- and post-flowering stages 49, 50.
Conclusion
A. annua has received increasing attention due to its ability to produce artemisinin, which today is widely used for treatment of malaria. In addition to its anti-malarial properties, artemisinin is cytotoxic for cancer cells. Recent reports demonstrate that artemisinin inhibits the secretion and gene expression of tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6 in a dose-dependent manner (51). The unfortunately low yield of artemisinin and a worldwide shortage of the drug has led to intense research in order to increase the yield of this sesquiterpenoid. Our study is the first in silico study to identify miRNAs and their targets in A. annua, which we hope could help to better understand miRNA-mediated regulation of genes related to artemisinin biosynthesis.
In summary, in the present study, we predicted six miRNAs conserved in A. annua. Furthermore, eight potential target genes were predicted, with functions in a variety of biological processes, including artemisinin biosynthesis, signal transduction and development. Most of the targets are unique to A. annua genome, which encode the enzymes associated with the artemisinin biosynthesis pathway. Interestingly, we also identify one target coding for putative AINTEGUMENTA, a transcription factor involved in developmental process, especially for floral organs, which is coincident with the higher artemisinin content during flowering. The result from the computational prediction will be useful to guide experimental design for biological verification. The next major steps, therefore, are to experimentally analyze the functional categories suggested by our computational approach, determine the analogous molecular functions amongst divergent plant species, and further elucidate any significant correlations between the miRNAs and their target genes. Hopefully all these efforts would help make more artemisinin available at lower costs for more people in the Third World, so people who suffer most from malaria can benefit more from this valuable and effective drug.
Materials and Methods
Databases of miRNAs, ESTs, and mRNA sequences
To search potential miRNAs, a total of previously known 674 miRNAs and their precursor sequences from A. thaliana and O. sativa were obtained from miRNA Registry Database (Release 16.0, October 2010; http://www.mirbase.org/) (52). These miRNAs were defined as the reference set of miRNA sequences. We have referred to the previous work on computational prediction of miRNAs by Zhang et al. (53). To avoid the redundant or overlapping miRNAs, the repeated sequences of miRNAs within the above species were removed and the remaining 584 sequences were used as query sequences for BLAST search. A. annua EST and mRNA databases were obtained from the National Center for Biotechnology Information (NCBI) GenBank nucleotide databases (http://ftp.ncbi.nlm.nih.gov).
Availability of software
Comparative software BLAST-2.2.14 was used from NCBI GenBank. MFOLD 3.1 from website (http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) was used online to analyze secondary structure of RNAs. MirEval (http://tagc.univ-mrs.fr/mireval) was used to predict miRNA precursors (26). BLASTx from NCBI (http://www.ncbi.nlm.nih.gov) was used to analyze potential targets of miRNAs.
Prediction of miRNAs
Procedure for searching potential miRNAs in A. annua is shown in Figure 2. We used the method described by Zhang et al. (53) with some modifications. Briefly, the previously known miRNAs in A. thaliana and O. sativa were screened out, and the redundant sequences were removed. The remaining miRNA sequences were subjected to BLAST search for A. annua miRNA homologs against EST databases.
The mature sequences of all miRNAs from Arabidopsis and rice were subjected to BLASTn search in the A. annua EST databases using BLASTn 2.2.9. The adjusted BLASTn parameter settings were as follows: expect values were set at 1,000; low complexity was chosen as the sequence filter; the number of descriptions and alignments was raised to 1,000. The default word-match size between the query and database sequences was 7. RNA sequences were considered as miRNA candidates only if they fit the following criteria: (1) at least 18 nt length were adopted between the predicted mature miRNAs and (2) allowed to have 0-5 nt mismatches in sequence with all previously known plant mature miRNAs (53). The ESTs that closely match the previously known plant mature miRNAs were included in the set of miRNA candidates and used for additional characterization based on the following criteria: (1) the entire EST sequence was selected to predict the secondary structures and to screen for miRNA precursor sequences; (2) the selected ESTs were further compared with each other to eliminate redundancies; and (3) these sequences were subjected to evaluation for miRNA precursor prediction properties using mirEval software (26). These precursor sequences were used for BLASTx analysis for removing the protein-coding sequences and retaining only the non-protein-coding sequences.
Prediction of secondary structure
Precursor sequences of these potential miRNA homologs were subjected to hairpin structure predictions using the Zuker folding algorithm with Mfold-3.1. The following parameters were used in predicting the secondary structures: (1) linear RNA sequence; (2) folding temperatures fixed at 37°C; ionic conditions of 1 M NaCl without divalent ions; (3) percent suboptimality number of 5; (4) maximum interior/bulge loop size of 30; (5) the grid lines in energy dot plot turned on. All other parameters were set with default values. In brief, the following criteria were applied in designating the RNA sequence as an miRNA homolog as described by Wang et al. (54): (1) the sequence could fold into an appropriate stem-loop hairpin secondary structure; (2) the small RNA sits in one arm of the hairpin structure; (3) no more than 6 mismatches are between the predicted mature miRNA sequence and its opposite miRNA (miRNA*) sequence in the secondary structure; (4) no loop or break is in the miRNA or miRNA* sequences, and (5) predicted secondary structure has higher MFEI and negative MFE.
The MFEI was calculated using the following equation (54):
MEFI= [(MEF/length of the RNA sequence) × 100] / (G+C)%
MFE denotes the negative folding free energies (ΔG).
Prediction of mRNA targets of miRNAs
Previous study has shown that most known plant miRNAs bind to the protein-coding region of their mRNA targets with perfect or nearly-perfect sequence complementarity, and degrade the target mRNA in a way similar to RNA interference 10, 12. This suggests a powerful approach to predict miRNA targets in plants by simply using homology search (Figure 3). In this study, we used homology search to predict miRNA targets in A. annua. The number of allowed mismatches at complementary sites between miRNA sequences and potential mRNA targets was no more than 4 and no gaps were allowed at the complementary sites.
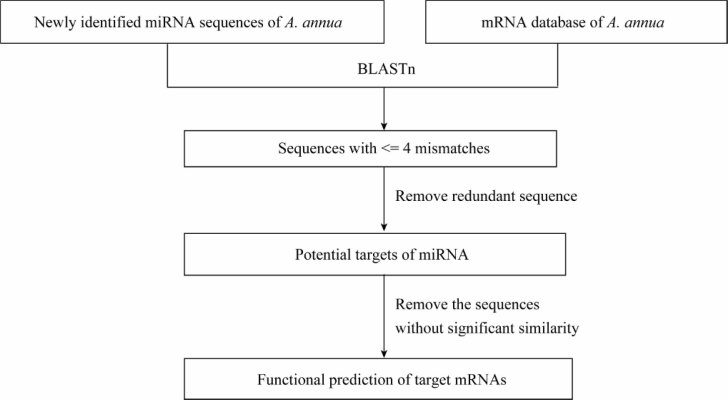
Figure 3.
Schematic diagram for searching potential target mRNA of miRNAs by blasting mRNA database of A. annua with newly identified miRNA sequences.
Authors’ contributions
RKM conceived the project. AP and RKM collected the data and conducted the computational analysis. NB and PKN supervised the work. AP and RKM interpreted the data and RKM prepared the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
AP and RKM thank Dr. Mrutyunjay Suar, Director of School of Biotechnology in KIIT University, for his encouragement and support during the course of study.
Supplementary Material
Figure S1
References
- 1.W.H.O. WHO Press, World Health Organization; Geneva, Switzerland: 2010. World Malaria Report. [Google Scholar]
- 2.Committee on the Economics of Antimalarial Drugs . Antimalaria drug and drug resistance. In: Arrow K.J., editor. Saving Lives, Buying Time: Economics of Malaria Drugs in an Age of Resistance. National Academies Press; Washington D.C., USA: 2004. pp. 252–300. [PubMed] [Google Scholar]
- 3.Klayman D.L. Qinghaosu (artemisinin): an anti-malarial drug from China. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- 4.Abdin M.Z. Artemisinin, a novel antimalarial drug: biochemical and molecular approaches for enhanced production. Planta Med. 2003;69:289–299. doi: 10.1055/s-2003-38871. [DOI] [PubMed] [Google Scholar]
- 5.Weathers P.J. Artemisinin: the biosynthetic pathway and its regulation in Artemisia annua, a terpenoid-rich species. In Vitro Cell. Dev. Bio. Plant. 2006;42:309–317. [Google Scholar]
- 6.Ro D.K. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L. Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Biochem. 2009;52:199–207. doi: 10.1042/BA20080068. [DOI] [PubMed] [Google Scholar]
- 8.Olofsson L. Relative expression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol. 2011;11:45. doi: 10.1186/1471-2229-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X. Enhancement of artemisinin content in tetraploid Artemisia annua plants by modulating the expression of genes in artemisinin biosynthetic pathway. Biotechnol. Appl. Biochem. 2011;58:50–57. doi: 10.1002/bab.13. [DOI] [PubMed] [Google Scholar]
- 10.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Aukerman M.J., Sakai H. Regulation of flowering time and floral organ identity by a microRNAs and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwab R. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Allen E. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Mallory A.C. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juarez M.T. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 17.Kasschau K.D. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 18.Robert-Seilaniantz A. A biotic or abiotic stress? In: Pareek A., editor. Abiotic Stress Adaptation in Plants, Physiological, Molecular and Genomic Foundation. Springer; Dordrecht, Netherlands: 2009. pp. 113–116. [Google Scholar]
- 19.Xie F.L. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Letters. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B.H. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B.H. Evidence that miRNAs are different from other RNAs. Cell Mol. Life Sci. 2006;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 23.Adams M.D. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 24.Wang X.J. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y. Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim. Biophys. Sin. 2005;37:75–87. (Shanghai) [PubMed] [Google Scholar]
- 26.Ritchie W. Mireval: a web tool for simple micro-RNA prediction in genome sequences. Bioinformatics. 2008;24:1394–1396. doi: 10.1093/bioinformatics/btn137. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B. Conservation and divergence of plant microRNAs genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 29.Prabu G.R., Mandal A.K.A. Computational identification of miRNAs and their target genes from expressed sequence tags of tea (Camellia sinensis) Genomics Proteomics Bioinformatics. 2010;8:113–121. doi: 10.1016/S1672-0229(10)60012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Z. Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum) Gene. 2008;414:60–66. doi: 10.1016/j.gene.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B.H. Identification of cotton miRNA and their targets. Gene. 2007;397:26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Frazier T.P. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011 doi: 10.1007/s12033-011-9387-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang B.H. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- 34.Kwak P.B. Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics. 2009;10:457. doi: 10.1186/1471-2164-10-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monavar Feshani A. Identification and validation of Asteraceae miRNAs by the expressed sequence tag analysis. Gene. 2011 doi: 10.1016/j.gene.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Quintero A.L. Mining of miRNAs and potential targets from gene oriented clusters of transcripts sequences of the anti-malarial plant, Artemisia annua. Biotechnol. Lett. 2011 doi: 10.1007/s10529-011-0808-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B.H. Plant microRNAs: a small regulatory molecule with big impact. Dev. Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Banyai W. Overexpression of farnesyl pyrophosphate synthase (FPS) gene affected artemisinin content and growth of Artemisia annua L. Plant Cell Tissue Organ Cult. 2010;103:255–265. [Google Scholar]
- 40.Newman J.D., Chappell J. Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Crit. Rev. Biochem. Mol. Biol. 1997;34:95–106. doi: 10.1080/10409239991209228. [DOI] [PubMed] [Google Scholar]
- 41.Mercke P. Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 2000;381:173–180. doi: 10.1006/abbi.2000.1962. [DOI] [PubMed] [Google Scholar]
- 42.Alam P., Abdin M.Z. Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep. 2011;30:1919–1928. doi: 10.1007/s00299-011-1099-6. [DOI] [PubMed] [Google Scholar]
- 43.Mercke P. Cloning, expression and characterization of epi-cedrol synthase, a sesquiterpene cyclase from Artemisia annua L. Arch. Biochem. Biophys. 1999;369:213–222. doi: 10.1006/abbi.1999.1358. [DOI] [PubMed] [Google Scholar]
- 44.Teoh K.H. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 45.Klucher K.M. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliott R.C. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh A. Evaluation of Artemisia annua strains for higher artemisinin production. Planta Med. 1988;54:475–476. doi: 10.1055/s-2006-962515. [DOI] [PubMed] [Google Scholar]
- 48.Charles D.J. Germplasm variation in artemisinin content of Artemisia annua using an alternative method of artemisinin analysis from crude plant extracts. J. Nat. Prod. 1990;53:157–160. [Google Scholar]
- 49.Baraldi R. Distribution of artemisinin and bioactive flavonoids from Artemisia annua L. during plant growth. Biochem. Syst. Ecol. 2008;36:340–348. [Google Scholar]
- 50.Mannan A. Effects of vegetative and flowering stages on the biosynthesis of artemisinin in Artemisia species. Arch. Pharm. Res. 2011;34:1657–1661. doi: 10.1007/s12272-011-1010-6. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y. The anti-malarial artemisinin inhibits pro-inflammatory cytokines via the NF-κB canonical signaling pathway in PMA-induced THP-1 monocytes. Int. J. Mol. Med. 2011;27:233–241. doi: 10.3892/ijmm.2010.580. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B. Identification of cotton microRNAs and their targets. Gene. 2007;397:26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 54.Wang L. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genomics. 2011;12:154. doi: 10.1186/1471-2164-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1