Abstract
High-density tiling arrays provide closer view of transcription than regular microarrays and can also be used for annotating functional elements in genomes. The identified transcripts usually have a complex overlapping architecture when compared to the existing genome annotation. Therefore, there is a need for customized tiling array data analysis tools. Since most of the initial tiling arrays were conducted in eukaryotes, data analysis methods are well suited for eukaryotic genomes. For using whole-genome tiling arrays to identify previously unknown transcriptional elements like small RNA and antisense RNA in prokaryotes, existing data analysis tools need to be tailored for prokaryotic genome architecture. Furthermore, automation of such custom data analysis workflow is necessary for biologists to apply this powerful platform for knowledge discovery. Here we describe TAAPP, a web-based package that consists of two modules for prokaryotic tiling array data analysis. The transcript generation module works on normalized data to generate transcriptionally active regions (TARs). The feature extraction and annotation module then maps TARs to existing genome annotation. This module further categorizes the transcription profile into potential novel non-coding RNA, antisense RNA, gene expression and operon structures. The implemented workflow is microarray platform independent and is presented as a web-based service. The web interface is freely available for acedemic use at http://lims.lsbi.mafes.msstate.edu/TAAPP-HTML/.
Key words: transcriptomics, small RNA, operon, prokaryotes, tiling arrays
Introduction
Genomic tiling arrays (overlapping oligonucleotide probes tiled across both strands of genome sequence) provide an unbiased view of genome expression, and have been used to generate transcriptional maps in eukaryotic genomes describing small RNAs (sRNAs), antisense expression, 5′ and 3′ untranslated regions (UTRs) 1, 2, 3. There is increasing appreciation for the significant role that sRNAs play in bacterial adaptation to stress and pathogenesis 4, 5, 6. Computational methods are used for identifying sRNAs, but they still need biological validation 7, 8. Due to the smaller size of prokaryotic genome, tiling arrays are now being used for whole-genome analysis to detect novel transcripts in bacteria 9, 10, 11, 12. Generally, computational tools that automate tiling array data analysis are based on two color arrays 13, 14, and are tailored for eukaryotic genomes. Recently, new tools that focus on prokaryotic genome architecture for probe design and normalization procedures were described 15, 16. However, these tools and other described analysis workflows stop with the identification of transcriptionally active regions (TARs); the end user with little or no computational skills are left with difficult task of mapping these TARs back to the genome and performing feature extraction for knowledge discovery.
Here we describe, for the first time, a computational pipeline named TAAPP (implemented in Perl), which is tailored for prokaryotic tiling array data, and consists of two modules: the first module handles normalized data from single color arrays, identifies expressed probes and then joins them to generate TARs; the second module maps these identified TARs back to the existing genome annotation, facilitating identification of sRNA elements, gene expression, operon structures and antisense RNA. sRNA elements can be identified in the non-coding area of genome where no annotation is available on either strand whereas antisense RNA is usually identified on the opposite strand of any annotated gene/RNA. The design of TAAPP into two separate modules allows data from two color tiling arrays (after analysis into differentially expressed TARs) to be mapped onto the genome directly using the second module. We applied TAAPP to analyze transcriptome of Streptococcus pneumoniae TIGR4 genome using custom high-density tiling arrays. The web interface is freely available for acedemic use at http://lims.lsbi.mafes.msstate.edu/TAAPP-HTML/.
Module
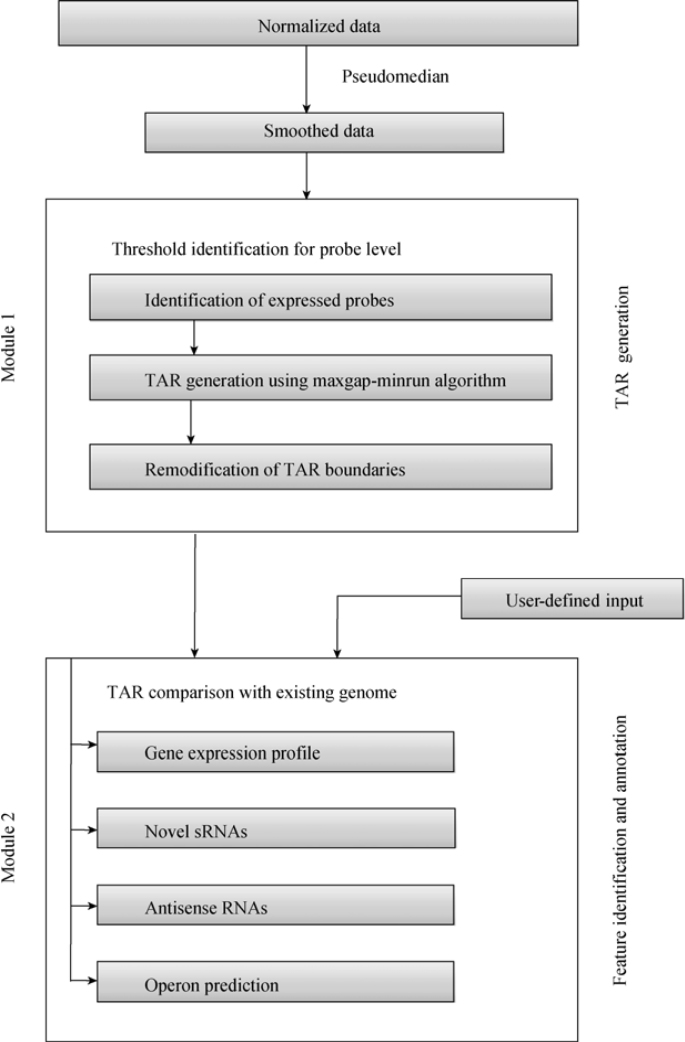
The software consists of two modules. The first module identifies the expressed regions and the second module compares it with existing genome annotation to identify gene expression pattern and novel elements. The flow chart presented in Figure 1 shows the various steps involved in data analysis.
Figure 1.
Flow chart of tiling array analysis and annotation pipeline steps.
Module 1: TAR generation
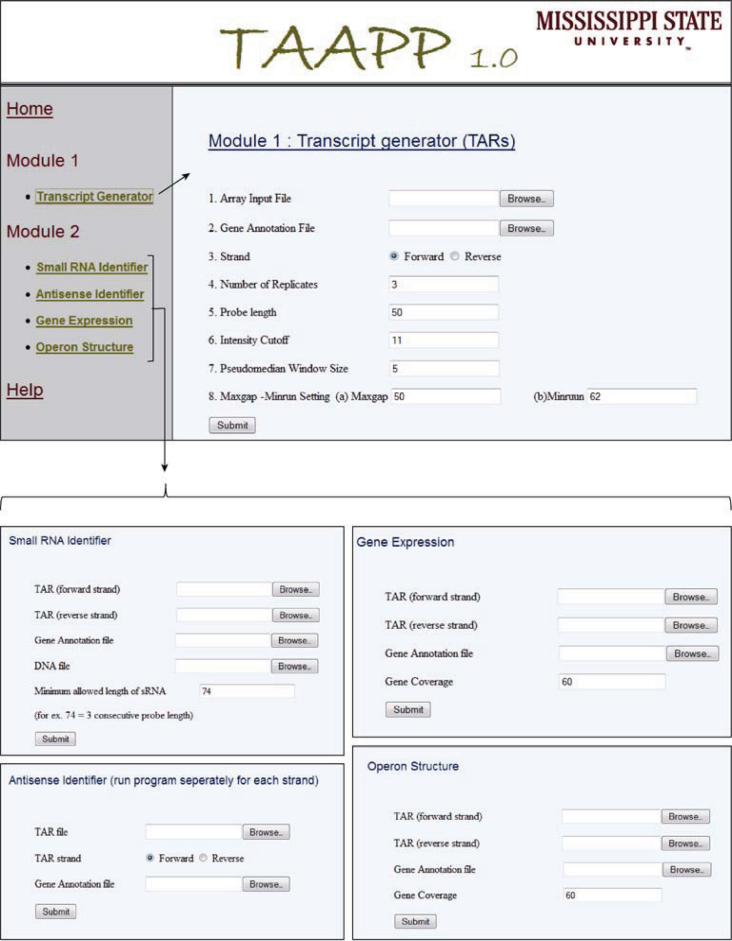
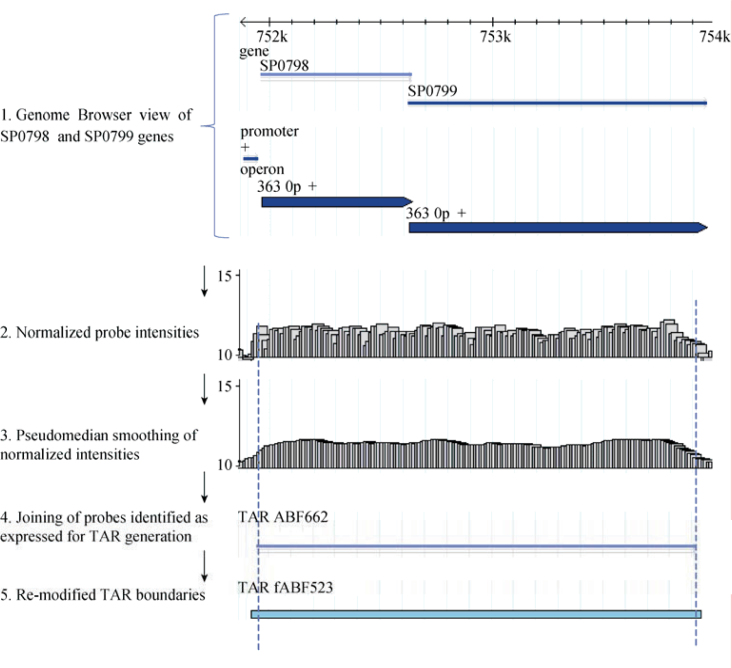
The TAR generation module accepts normalized probe-level data as tab-delimited text file (making the pipeline microarray platform independent) (Figure 2). For classifying the probes as expressed, this module requires the user to input probe intensity cutoff value or supply the files with positive and negative controls for automated calculation. This value is often determined based on the distribution of normalized intensity values for negative and positive control probes on the array and varies with array design (2). A lower cutoff value is associated with higher false positive rates of identification and vice versa. In the absence of experimental controls, user can use the top 90 intensity percentile as a cutoff value (17). To minimize sequence-based effects on probe intensity, a pseudomedian filter is applied, which takes adjacent probe intensities into account and provides smoothing to the data. A pseudomedian filter works by calculating median of all possible pairwise averages in a sliding window and assigning it to the probe at the center (18). The sliding window is then shifted to the next probe and the process is continued for the complete genome sequence. Probes with intensity greater than the cutoff value are classified as expressed probes and consecutive expressed probes are further joined using maxgap-minrun algorithm (2) to generate TARs. The maxgap parameter allows certain number of probes (one or two probes) to be below the cutoff while still being incorporated into the TAR, whereas the minrun parameter defines at least a certain length of the transcript to be considered as TAR (discarding small length transcripts). To accurately identify genes in the densely packed prokaryotic genomes (marked by short intergenic regions), the maxgap parameter is set to zero for the intergenic region. This helps to differentiate transcript of two consecutive genes, which are usually separated by very short intergenic region, if they are not expressed as an operon. Due to the smoothing of dataset generated by pseudomedian filter, slight errors are introduced in the identification of transcript boundaries (start and end). Therefore, we implemented a new step that remodified transcript boundaries using average intensity values (data before pseudomedian calculation). Remodification is conducted by either elongating or shortening transcript ends until the average intensity value of the probe is greater than or equal to the threshold cutoff. Remodified transcripts are again processed using maxgap-minrun method to generate TARs. Expression data for both strands are processed separately to generate TARs. Figure 3 shows the transcriptome snapshot of a short region of S. pneumoniae TIGR4 genome visualized in Genome Browser, during various steps of data analysis.
Figure 2.
Web interface of TAAPP modules and sub-modules.
Figure 3.
Snapshot of a short region of S. pneumoniae TIGR4 genome visualized in Genome Browser. Track 1 shows the operon region containing two genes SP0798 and SP0799 along with the predicted promoter. Tracks 2 to 5 show the probe intensity corresponding to the region depicted in Track 1 at various steps of tiling array data analysis.
Module 2: feature extraction and annotation
This module maps the identified TARs generated from module 1 (or any other tiling analysis workflow) with the existing genome annotation. Mapping TARs to annotated open reading frames (ORFs) helps identify the basal transcription of the genome under experimental conditions. On the other hand, TARs identified outside the ORF boundaries are potential novel expressed regions missed by the initial annotation. Module 2 is further divided into four separate sub-modules.
Sub-module 1: sRNA identifier
To identify sRNAs, TARs were mapped onto the intergenic regions of the S. pneumoniae. Intergenic regions within operons, small 5′ and 3′ UTR of mRNAs, and non-unique regions (mobile genetic elements and repetitive regions) of the genome were excluded for this analysis. Transcripts expressed (greater than the specified minimum length) from the intergenic regions were classified as novel sRNA. The results for sRNAs include the start and end coordinates along with the DNA sequence.
Sub-module 2: antisense identifier
This sub-module generates a list of TARs (called antisense RNAs) that are found on the non-coding strand of a gene. The antisense RNAs for genes show different kinds of expression patterns. For example, a gene might have many antisense RNAs or an antisense RNA may overlap the whole gene. Apart from listing all the genes that had detectable antisense RNA, the module classifies them into four different categories—5DASH overlap (antisense transcript overlapping 5′-end of gene), 3DASH overlap (antisense transcript overlappling 3′-end of gene), PART (antisense transcript as a small part located between gene ends), OVERLAP (antisense transcript fully overlappling the gene). Earlier studies have shown that 5′/3′ antisense overlaps are likely to be involved in regulatory activities (19).
Sub-module 3: gene expression
Due to experimental variations, probes for a given genomic region may not be always expressed. Therefore, a gene region may be represented as a mixed set of expressed and non-expressed probes. A gene is considered as expressed if it has relatively higher proportion of expressed probes. The default cutoff value is taken as 70%, which represents the proportion of probes classified as expressed (P<0.001 in a binomial test) (20). The program generates a list of expressed genes based on the default selection criteria.
Sub-module 4: operon struture
Since tiling arrays measure expression in the intergenic regions of the genomes, they can be used to identify operon structures in bacteria. Two or more consecutive genes are considered to be part of an operon, if they fulfill the following criteria: (1) they are expressed; (2) they are transcribed in the same direction; and (3) the intergenic region between the genes is identified as a single expressed transcript that overlaps the genes in both directions. Overlapping pairs of genes are joined together to identify large operon structures.
TAAPP is implemented in Perl. The software is available as a web server, so it does not need any special software installation. The two TAAPP modules are independent of each other and their simple input/output format makes them suitable for any microarray platform. An extensive help file with sample input dataset is provided online.
Application
Whole-genome tiling arrays are used to study transcriptional pattern in eukaryotes as well as prokaryotic species. Many conventional tiling array analysis programs exist for the design and analysis of tiling array datasets, but most of them were developed for eukaryotic genomes (13). The majority of these programs do not work for single color tiling arrays or customized tiling arrays. Very few software tools were described in literature for prokaryotic tiling array data 15, 16. However, these tools mainly focus on tiling array probe design and data normalization. To our knowledge, there is no software tool for prokaryotes, which performs transcript comparison with genome annotation and helps in the identification of novel features. In prokaryotes, tiling arrays can also be used to identify operon structures in bacteria, which is not possible in eukaryotic genomes.
Here we described a set of programs tailored for prokaryotic genome architecture that identifies expressed transcripts from normalized data and performs feature extraction. We implemented TAAPP on a custom S. pneumoniae TIGR4 single color Nimblegen tiling array dataset (Roche NimbleGen, Madison, USA), obtained from Gene Expression Omnibus database at NCBI (GSE12636). Initial data processing was done using NMPP module, which is used for preprocessing of Nimblegen specific microarray chips (21). Normalized data were used as the input for TAAPP. The TAR generation module identified 1,324 TARs in the forward (+) strand and 1,190 TARs in the reverse (−) strand with default settings. The feature identification module identified a set of 50 novel non-coding sRNAs in the intergenic regions. In total, 994 genes were expressed out of 2,015 annotated genes. The operon identifier sub-module identified 202 operon structures, consisting of 520 genes. These results for sRNA identification and operon prediction along with more analyses and RT-PCR validation were published in a separate manuscript (22). A descriptive help file is also provided with sample input and output files, along with instructions for executing and interpreting the results of the two modules.
TAAPP automates the analysis of prokaryotic tiling array datasets and is provided as an easy-to-use web interface. The future work includes addition of confidence scores to identified novel regions and inclusion of features (like promoter and terminator) to identified transciptional elements. Another possible improvement could be the modification of module 1 to facilitate the input of deep sequencing data.
Authors’ contributions
All authors contributed to the development and design of TAAPP. RK wrote all the scripts for the implementation and drafted the manuscript. BN, MLL and SCB edited the draft manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This project was partially supported by a grant from the National Science foundation of USA (Mississippi EPSCoR-0903787). We acknowledge the Institute for Genomics, Biocomputing and Biotechnology (IGBB), Mississippi State University for assistance with the article-processing charges for the manuscript. We thank Tony Arick and IGBB, Mississippi State University, for hosting the TAAPP web server.
References
- 1.He H. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17:1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kampa D. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada K. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 4.Narberhaus F., Vogel J. Regulatory RNAs in prokaryotes: here, there and everywhere. Mol. Microbiol. 2009;74:261–269. doi: 10.1111/j.1365-2958.2009.06869.x. [DOI] [PubMed] [Google Scholar]
- 5.Romby P. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Toledo-Arana A. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Livny J., Waldor M.K. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni R.V., Kulkarni P.R. Computational approaches for the discovery of bacterial small RNAs. Methods. 2007;43:131–139. doi: 10.1016/j.ymeth.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Akama T. Whole-genome tiling array analysis of Mycobacterium leprae RNA reveals high expression of pseudogenes and noncoding regions. J. Bacteriol. 2009;191:3321–3327. doi: 10.1128/JB.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyakoshi M. High-resolution mapping of plasmid transcriptomes in different host bacteria. BMC Genomics. 2009;10:12. doi: 10.1186/1471-2164-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo-Arana A. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 12.Tsui H.C. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 2010;192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X.S. Getting started in tiling microarray analysis. PLoS Comput. Biol. 2007;3:1842–1844. doi: 10.1371/journal.pcbi.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z.D. Tilescope: online analysis pipeline for high-density tiling microarray data. Genome Biol. 2007;8:R81. doi: 10.1186/gb-2007-8-5-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillippy A.M. Efficient oligonucleotide probe selection for pan-genomic tiling arrays. BMC Bioinformatics. 2009;10:293. doi: 10.1186/1471-2105-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomassen G.O. Custom design and analysis of high-density oligonucleotide bacterial tiling microarrays. PLoS One. 2009;4:e5943. doi: 10.1371/journal.pone.0005943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertone P. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 18.Royce T.E. An efficient pseudomedian filter for tiling microrrays. BMC Bioinformatics. 2007;8:186. doi: 10.1186/1471-2105-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 20.David L. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X. NMPP: a user-customized NimbleGen microarray data processing pipeline. Bioinformatics. 2006;22:2955–2957. doi: 10.1093/bioinformatics/btl525. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics. 2010;11:350. doi: 10.1186/1471-2164-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]