Abstract
Powdery mildew (Pm) caused by the infection of Blumeria graminis f. sp. tritici (Bgt) is a worldwide crop disease resulting in significant loss of wheat yield. To profile the genes and pathways responding to the Bgt infection, here, using Affymetrix wheat microarrays, we compared the leaf transcriptomes before and after Bgt inoculation in two wheat genotypes, a Pm-susceptible cultivar Jingdong 8 (S) and its near-isogenic line (R) carrying a single Pm resistant gene Pm30. Our analysis showed that the original gene expression status in the S and R genotypes of wheat was almost identical before Bgt inoculation, since only 60 genes exhibited differential expression by P = 0.01 cutoff. However, 12 h after Bgt inoculation, 3014 and 2800 genes in the S and R genotype, respectively, responded to infection. A wide range of pathways were involved, including cell wall fortification, flavonoid biosynthesis and metabolic processes. Furthermore, for the first time, we show that sense-antisense pair genes might be participants in wheat-powdery mildew interaction. In addition, the results of qRT-PCR analysis on several candidate genes were consistent with the microarray data in their expression patterns. In summary, this study reveals leaf transcriptome changes before and after powdery mildew infection in wheat near-isogenic lines, suggesting that powdery mildew resistance is a highly complex systematic response involving a large amount of gene regulation.
Keywords: Wheat, Pathogen resistance, Powdery mildew, Microarrays
Introduction
Wheat (Triticum aestivum, AABBDD 2n = 42) is the most widely grown crop in the world, occupying 17% of all the cultivated land and providing approximately 55% carbohydrates for the daily human diet [1]. Powdery mildew (Pm) caused by the infection of Blumeria graminis f. sp. tritici (Bgt) is a common pathogenic disease distributed in many wheat production countries, causing significant loss of yield [2], [3]. By traditional breeding, certain pathogen-resistant genes in wild relatives of wheat have been transmitted to the susceptible common wheat generating a series of isogenic lines showing varying degrees of resistance to a number of Pm types. These Pm-resistant lines have been considered as the most economical and environmentally safe cultivars for disease control, and also have been used as natural laboratory materials for a better understanding of the molecular mechanisms underlying the host-pathogen interaction and defense response in major crop plants [4], [5].
Different approaches have been employed to identify the pathogen-resistant genes in wheat responding to various types of crop diseases [6], [7], [8], [9]. Bernardo et al. showed that 44 genes are differentially expressed at 72 h post inoculation (hpi) in Fusarium head blight resistant and susceptible wheat cultivars [6]. Coram et al. investigated the transcriptional profile of wheat infected by Puccinia striiformis f. sp. tritici, and identified 99 genes specific to high-temperature adult plant resistance [9]. Bhuiyan et al. found that 12 genes in the biosynthesis and supply of methyl units were activated by Bgt infection in the epidermis of diploid wheat (Triticum monococcum), suggesting that genes involved in pathways of producing methyl units are also responsible for the host defense response [7]. He also verified that several genes involved in monolignol biosynthesis were critical for the defense against Pm invasion in wheat [7]. Bruggmann et al. performed tissue-specific cDNA-AFLP analysis of 17,000 transcripts to detect B. graminis f. sp. hordei responsive genes at 6 and 24 hpi and found that 44 and 76 transcripts were specifically up-regulated in epidermis and mesophyll tissues, respectively [8].
Microarray analysis has been used in wheat to determine the transcriptome changes in response to a variety of abiotic stress such as heat, drought and cold [10], [11], [12]. In terms of the biotic stress response in wheat, genome-wide analysis has been done to study the Fusarium graminearum infection, Fusarium pseudograminearum infection, P. striiformis f. sp. tritici infection and P. triticina infection [13], [14], [15], [16]. However, little work has been done to study the transcriptome responses of wheat during the Pm infection on a genomic scale. Thus, identifying novel genes and studying their expression patterns in response to Pm will provide a molecular basis for improving disease resistance in crops, while transcriptome analysis with gene chip could provide us with a plethora of gene expression patterns simultaneously. So using wheat Affymetrix genome arrays, we compared the gene expression patterns of a susceptible cultivar Jingdong 8 (S), and its resistant near-isogenic line (R) carrying a single Pm-resistant gene, Pm30, in leaf tissues, before and after the inoculation with Bgt. Our analysis shows that although the original expression status of the susceptible and resistant wheat was very similar before infection, certain genes with putative pathogen-defense functions demonstrated constitutively higher expression in resistant wheat than in the susceptible wheat. This may play a critical role in building the basal barrier to defend the Pm attack. At 12 hpi, the transcriptomes involved in a wide range of molecular pathways changed dramatically in both the resistant and susceptible cultivars.
Results and discussion
Disease responsive genes were detected after Pm infection
Two wheat lines were used in this study, including Pm-susceptible wheat with highly-susceptible phenotype (S), and its near-isogenic Pm-resistant line with immune phenotype (R). The seedling leaves were harvested at 0 h and 12 hpi, designated as S-0 h/S-12 h and R-0 h/R-12 h, respectively. The leaf samples were collected from three independent biological replicates. We first performed quantile normalization across the twelve slides to remove the systematic bias and used the normalized signal intensity of each sample to evaluate the reproducibility between the replicates. The Pearson correlation coefficients between each two replicates ranged from 0.9784 to 0.9944, indicating the high reproducibility of the microarray experiments (Table S1).
The probe sets prefixed with “AFFX” and “RPTR” were removed and filtered by a fraction call of 100% as described in the Methods, and the ones with a consistent ‘Present’ call in all the three biological replicates were then considered as ‘expressed’ genes. Thus, 39.63% and 44.57% of the 61,127 probe sets were detected as expressed genes in S-0 h and S-12 h samples, indicating that up to 5% more genes were activated after 12 h infection. For the Pm-resistant wheat, 40.13% and 42.43% probe sets were called ‘Present’ in R-0 h and R-12 h samples, respectively. Therefore, about 2% more genes were activated in S genotype than in R genotype of wheat at 12 hpi.
To detect the potential disease responsive (DR) genes, we conducted pairwise comparison between the samples in pairs of S-0 h vs R-0 h, S-0 h vs S-12 h and R-0 h vs R-12 h. With a stringent cutoff of false discovery rate (FDR)-adjusted P = 0.01 and a less stringent cutoff of P = 0.05, we subsequently defined the differentially expressed genes subject to either up- or down-regulation. Between the S-0 h and the R-0 h samples, only 60 genes were detected as differentially expressed using a cutoff of P = 0.01 and 781 genes using a cutoff of P = 0.05. Comparison between S-0 h and S-12 h samples identified 3014 and 7554 differentially expressed genes, while in the R genotype, 2800 and 6906 genes were determined to be differentially expressed at 12 hpi using a P value of 0.01 and 0.05 cutoff, respectively. This analysis is consistent with the previous observation that the pathogen attack can influence a broad range of pathways and a large proportion of the genes in the transcriptional networks are disturbed [9], [14], [15], [16]. Since there were a large number of genes showing response to the Pm infection regardless of the S or R genotype, we decided to use 0.01 as the cutoff to narrow down the range for selecting the possible candidates with the most dramatic changes.
Expression of a few DR genes was constitutively higher in resistant wheat than in susceptible wheat before Pm infection
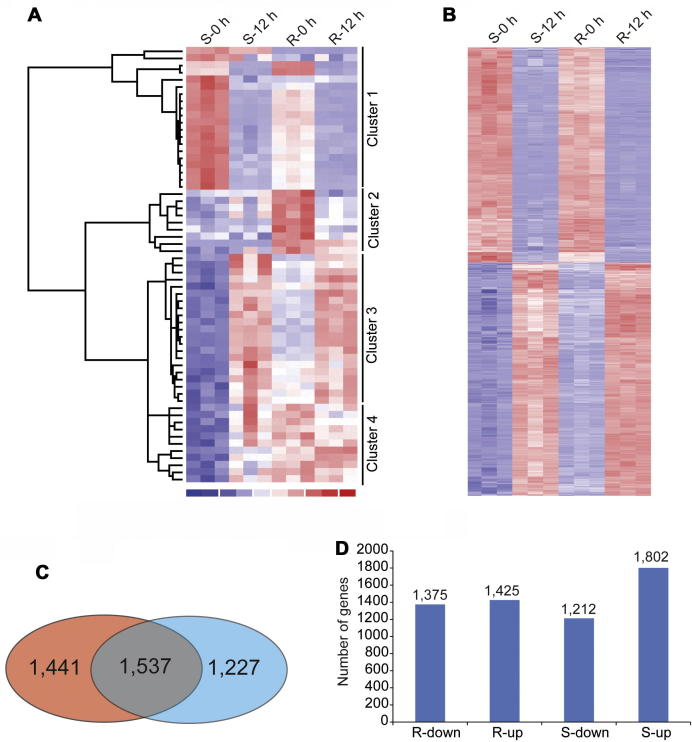
We first performed hierarchical clustering algorithm on the 60 differentially expressed genes between R-0 h and S-0 h samples, and identified four groups of genes (Cluster 1 to 4) exhibiting distinct expression patterns (Figure 1A and Table 1). The genes in Cluster 1 and 3 showed opposite expression trend in S and R genotypes of wheat, containing the genes with various functions. For example, a hydroxyproline-rich glycoprotein family protein which functions in cell wall fortification [17] was up-regulated 12 hpi in Cluster 1. Another two differentially expressed genes were members of lipoxygenases (LOX) family that catalyze the hydroperoxidation of polyunsaturated fatty acids during the first step of producing fatty acid metabolites for the subsequent biological synthesis of antimicrobial compounds in most plants [18], [19]. In Cluster 3, bacterial blight resistance protein was up-regulated in two lines 12 hpi, which might play an important role in basal resistance to powdery mildew. In Cluster 2, there were 9 genes that were expressed relatively high in R-0 h compared to S-0 h, whose expression decreased after the Pm infection.
Figure 1.
Overall analysis of differentially expressed genes in the microarray (A) Hierarchical clustering analysis of the 60 differentially expressed genes detected by P = 0.01 cutoff between the S-0 h and R-0 h samples. (B) Hierarchical clustering analysis of the commonly regulated genes in response to infection identified by P = 0.01 cutoff. Changes of gene expression are displayed from blue (down-regulated) to red (up-regulated). Lighter colors indicate genes with low fold-change and darker colors indicate genes with high fold-change. (C) Venn diagrams showing the common (gray) and unique differentially expressed genes in S (red) and R (blue) genotypes detected by P = 0.01 cutoff. (D) Number of up- or down-regulated genes responsive to Pm in S and R genotypes.
Table 1.
The differentially expressed genes between S-0 h and R-0 h
| Clusters | Probe set ID | Putative function |
|---|---|---|
| Cluster 1 | Ta.3384.1.A1_at | Putative uncharacterized protein |
| Ta.6248.1.S1_at | Putative tetratricopeptide repeat (TPR)-containing protein | |
| Ta.454.1.S1_at | Putative elongation factor Ts family | |
| TaAffx.65068.1.A1_at | Putative MADS box-like protein | |
| Ta.19211.1.A1_x_at | Unknown protein | |
| Ta.408.1.A1_at | Unknown protein | |
| Ta.7830.1.S1_at | Lipoxygenase | |
| Ta.442.1.S1_at | Catalase | |
| Ta.634.1.S1_at | ABC1-like family protein | |
| Ta.15048.1.A1_at | ABC1 family protein | |
| Ta.865.2.A1_at | Hypothetical protein | |
| Ta.23273.1.S1_at | Putative ferredoxin-thioredoxin reductase | |
| Ta.12159.1.A1_at | Hydroxyproline-rich glycoprotein family protein | |
| Ta.28049.1.S1_s_at | MYB20 protein | |
| Ta.10126.3.S1_a_at | Putative zinc finger protein | |
| Ta.3970.1.S1_at | Putative monosaccharide transporter | |
| Ta.5891.2.S1_a_at | Major facilitator superfamily antiporter | |
| Ta.485.1.A1_at | Lipoxygenase 2, chloroplast precursor | |
| Ta.12561.1.S1_at | Unknown protein | |
| TaAffx.97978.1.S1_at | Pentameric polyubiquitin | |
| Cluster 2 | Ta.30913.2.A1_at | Putative phosphate transport protein, mitochondrial |
| Ta.4747.1.S1_x_at | Unknown protein | |
| Ta.5293.1.S1_at | Unknown protein | |
| Ta.29984.1.S1_at | Putative Annexin A8 protein | |
| Ta.23072.1.S1_at | Unknown protein | |
| Ta.632.2.S1_x_at | Putative survival motor neuron-related protein; splicing factor 30 | |
| Ta.8032.2.A1_x_at | Unknown protein | |
| Ta.11519.1.A1_x_at | Unknown protein | |
| TaAffx.110936.1.S1_s_at | Light-inducible protein ATLS | |
| Cluster 3 | TaAffx.28047.3.A1_at | Unknown protein |
| TaAffx.53376.1.S1_at | Bacterial blight resistance protein | |
| TaAffx.50125.2.S1_at | Cytochrome P450 | |
| Ta.4973.2.S1_a_at | Unknown protein | |
| Ta.10740.1.S1_a_at | Putative thioesterase family protein | |
| Ta.25069.1.S1_x_at | Unknown protein | |
| Ta.30628.2.S1_at | Ribosomal protein L36 | |
| Ta.3008.1.S1_a_at | Putative signal peptidase 18 K chain | |
| Ta.2464.1.S1_a_at | Pescadillo-like protein | |
| Ta.11853.1.S1_at | Unknown protein | |
| Ta.380.1.S1_a_at | Unknown protein | |
| Ta.15067.1.S1_at | Unknown protein | |
| Ta.15067.1.S1_x_at | Unknown protein | |
| Ta.9535.1.S1_at | Putative 60S ribosomal protein L28 | |
| Ta.21353.1.S1_a_at | Putative acetone-cyanohydrin lyase | |
| TaAffx.132143.1.S1_s_at | Cyanate hydratase | |
| Ta.6101.1.S1_at | Putative steroid membrane binding protein | |
| Ta.13180.3.S1_at | Unknown protein | |
| Ta.28696.1.S1_x_at | Unknown protein | |
| Ta.30628.1.S1_at | Ribosomal protein L36 | |
| Ta.27331.1.S1_at | Unknown protein | |
| Cluster 4 | TaAffx.70192.1.S1_at | Unknown protein |
| Ta.787.1.S1_x_at | Unknown protein | |
| Ta.9157.1.S1_at | AMP-binding protein | |
| Ta.393.1.S1_at | Unknown protein | |
| Ta.21237.1.S1_x_at | Unknown protein | |
| Ta.9243.1.S1_at | Ankyrin repeat protein | |
| TaAffx.98004.1.S1_at | Multiple stress-associated zinc-finger protein | |
| TaAffx.66205.2.S1_s_at | Encodes protease I (pfpI)-like protein | |
| Ta.8348.1.A1_x_at | DNA-binding protein ABF1 | |
| TaAffx.54209.1.S1_at | Unknown protein | |
These genes were clustered in four groups identified as differentially expressed between S-0 h and R-0 h samples by P = 0.01 cutoff.
Finally, the most important 10 genes from the comparison of S-0 h and R-0 h were grouped as Cluster 4, showing relatively high expression in R-0 h, which didn’t decrease 12 h after Pm infection, whereas in S genotype, the expression is consistently low both before and after Bgt inoculation. These genes may function to build a constitutive barrier to form the initial resistance defending the pathogen attack. It is worth noting that a transcription factor annotated as a ‘multiple stress-associated zinc-finger protein’ fell into this group. Another important transcription factor was DNA-binding protein ABF1, a member of plant-specific WRKY families, which has been previously shown to be involved in regulating patato responses to the infection of Sonchus yellow net virus (SYNV) and Impatiens necrotic spot virus (INSV) [20]. Another interesting gene in Cluster 4 encodes an AMP-binding protein that has been previously reported in rice for its involvement in the regulation of the defense response through salicylic acid (SA) and/or jasmonic acid/ethylene (JA/ET) signaling pathways [21]. Moreover, we also found several genes of unknown function specifically expressed in Pm-resistant wheat that may be also critical in pathogen defense pathways against powdery mildew, whose functions need to be verified in future.
The transcriptomes of both resistant and susceptible wheat were subject to dramatic changes in response to Pm
Among the 2800 DR genes of R genotype and 3014 genes of S genotype, 1537 of them were found commonly responsive to Pm infection (Figure 1B and C). The heatmap of the clustered DR genes exhibited a consistent trend of up- or down-regulation, indicating that they were responsive to the Pm-infection in a similar manner regardless of the resistant or susceptible characteristics (Figure 1C). Moreover, in the R genotype, 1375 genes and 1425 genes were found subject to respective down- and up-regulation, while in the S genotype, a higher proportion of up-regulated genes (1802) were observed (Figure 1D).
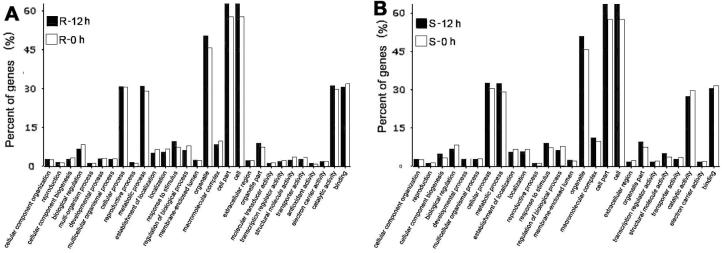
Subsequently, we performed GeneOntology (GO) analysis to test what the functional categories for these genes enriched in R and S genotypic lines after Bgt infection. We used the online tool AriGO that contains the pre-annotated GO term for the Affymetrix wheat microarray [22]. For the 1537 commonly responsive genes in both S and R genotypes, oxidoreductase activity, antioxidant activity, hydrolyase activity and chlorophyll binding in molecular function category, fatty acid catabolic process, organic acid catabolic process, response to stress and ribosome biogenesis in metabolic mechanism category, as well as photosystem I and II, cytoplasm and thylakoid in the cellular component category were differentially regulated. For 1227 R specifically responsive genes, electron carrier activity, molecular transducer activity and developmental processes were included, while for 1441 S specifically responsive genes, leaf senescence, aging, protein and lipid degradation processes respond. The GO analysis showed that even in the resistant and susceptible wheat demonstrating distinct phenotypes in terms of responding to pm infection, most genes and pathways were commonly influenced under pm infection (Figure 2).
Figure 2.
Gene enrichment analysis by GO term classification on the differentially expressed genes in R (A) and S (B) genotypes The GO annotation in wheat was done by blast2go program [73], and the GO enrichment analysis was done by AgriGO online tool [22].
Expression of select candidate genes was validated using experimental qRT-PCR
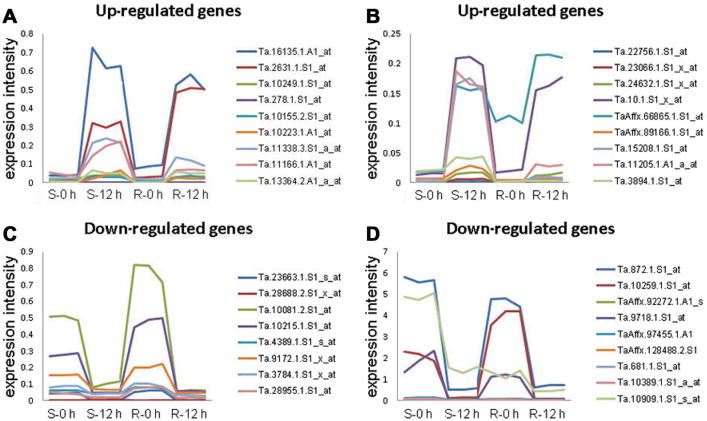
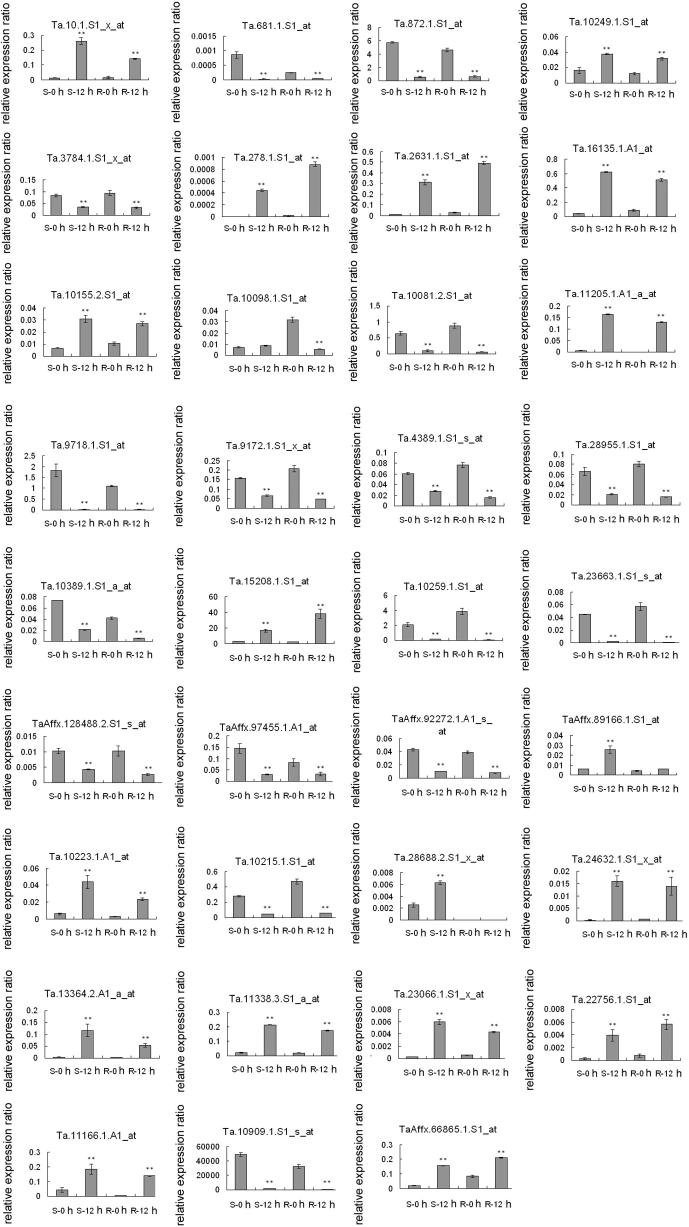
To validate the expression patterns of the DR genes detected by the microarrays, we performed quantitative RT-PCR (qRT-PCR) on 40 candidate genes with significantly changed expression pattern before and after the infection. Expression of 35 (87.5%) out of 40 genes was in good agreement with the microarray results. Among them, 18 genes were up-regulated and the other 17 were down-regulated after 12 h Pm infection in both genotypes, including three well-documented pathogenesis related (PR) proteins (Ta.278.1.S1_at, Ta.4389.1.S1_s_at and Ta.10.1.S1_x_at) during plant disease resistance. Additionally, four genes encoding abiotic stress-related proteins (Ta.10259.1.S1_at, TaAffx.92272.1.A1_s_at, Ta.15208.1.S1_at and Ta.23663.1.S1_s_a) and four redox-reaction related proteins (Ta.872.1.S1_at, Ta.10081.2.S1_at, Ta.10389.1.S1_a_at and Ta.681.1.S1_at) were also validated by qRT-PCR, and showed good agreement with the microarray results (Figure 3 and Figure S1).
Figure 3.
Experimental qRT-PCR validation of selected differentially expressed genes Expression of selected differentially expressed genes subject to up-regulation (A, B) and down-regulation (C, D) after 12 h Pm-infection in the R and S genotypes of wheat was examined using qRT-PCR. In total 35 probe sets were detected and there were three biological replicates for each time point.
Due to the limited genomic information and putative function annotation in wheat, our analysis hereafter was mainly focused on the genes assigned with putative function by HarvEST1.51 (http://www.harvest.ucr.edu/). To extend the manual screening on the DR genes, we filtered candidate genes by the less stringent cutoff value of P = 0.05 and a minimum twofold change between the pair of R-0 h vs R-12 h and the pair of S-0 h vs S-12 h.
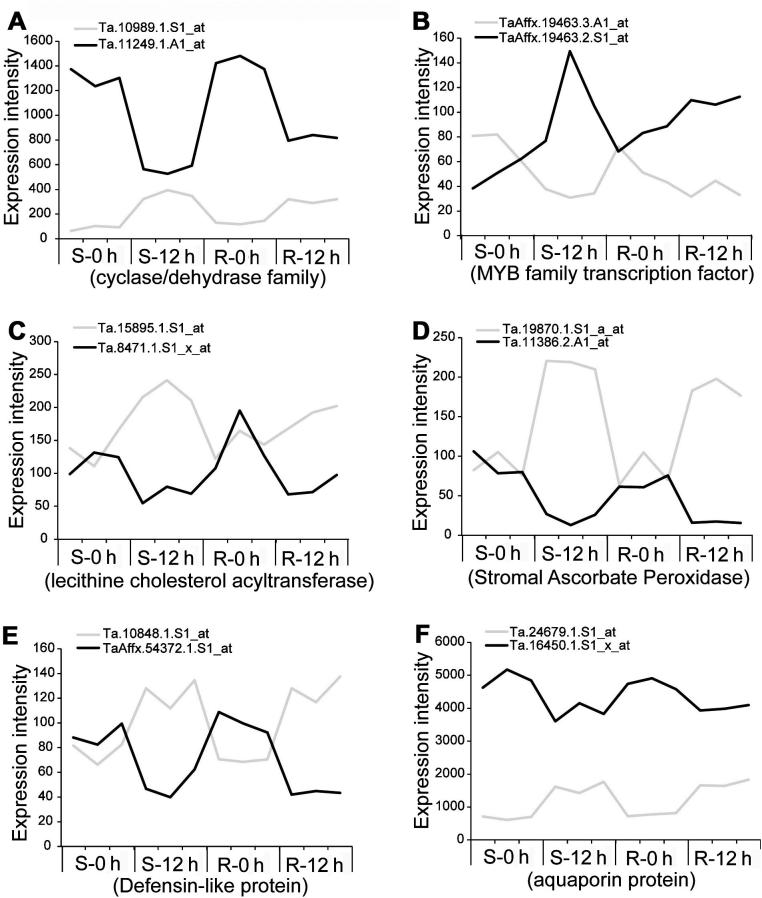
Sense-antisense transcripts showed anti-correlated patterns responding to Pm infection
We have identified ∼100 pairs of sense-antisense transcripts included in the wheat microarray, with a minimum 100 bp overlapping region at the 5′ end showing strong anti-correlation (Pearson correlation coefficient ⩽−0.8) before and after the infection. Six pairs of sense-antisense transcripts, which encode cyclase/dehydrase family, MYB family transcription factors, lecithine cholesterol acyltransferase, stromal ascorbate peroxidase, definsin-like protein and aquaporin protein, have drawn our attention (Figure 4). It has been previously reported that one class of endogenous small RNAs, called natural-antisense-transcript derived siRNAs (nat-siRNAs), are specifically derived from the overlapping region of a pair of sense-antisense transcripts under environmental stress, and can trigger the degradation of the target mRNAs located on the opposite strand by the RNAi silencing machinery [23]. Given the strong anti-correlation of six sense-antisense pair genes, we speculate that some of the Pm responsive genes might be regulated by the antisense RNAs in a similar way, since many miRNAs and siRNAs were previously shown to be activated after the pm infection [24].
Figure 4.
Expression profile of six pairs of selected sense-antisense transcripts The six pairs of genes were selected with significant Pearson anti-correlation of −0.8 at P = 0.05. The potential function of each pair is predicted based on the transcripts that show greater change according to microarray data and a clear functional annotation. There were three replicates for each time point.
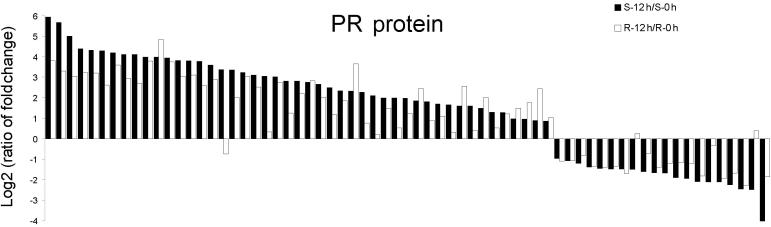
PR genes were highly up-regulated after Pm infection in both resistant and susceptible wheat
The induction of PR proteins has been well documented when plants are exposed to various pathogens, and constitutive expression of PR proteins in transgenic plants can increase the resistance to fungi [25], [26], [27], [28], [29]. It also has been reported that the expression of PR-1, PR-4 and PR-10 were induced to a higher level to trigger the rapid activation of defense-responsive mechanisms in many fungus-challenged plant species, such as wheat, rice, Arabidopsis, European plums and Medicago [30], [31], [32], [33], [34], [35], [36]. Consistent with the earlier reports, 46 genes annotated as PR proteins were up-regulated in both S and R genotypes, including PR-1, PR-2, PR-3, PR-4 and PR-10, with an exceptionally 62-fold change for PR1 (Ta.278.1.S1_at). PR-1 is often used as a marker for the systemic acquired resistance (SAR) [37]. Among the five transcripts encoding PR-1 proteins in the wheat microarray, two were specifically up-regulated only in resistant wheat (Ta.8304.1.S1_a_at and Ta.8304.1.S1_x_at), while the other three were induced in both resistant and susceptible wheat (Ta.278.1.S1_at, Ta.278.1.S1_x_at, and Ta.30739.2.S1_at). We also observed that most PR genes were induced to a higher level in S genotype than in R genotype (Figure 5, Table S2), which is reminiscent of another point of view that PRs are related to the severity of symptom expression rather than to resistance [38].
Figure 5.
Column chart of differentially expressed genes encoding PR proteins All the data used for the figure are the average value of three replicates and are log 2 transformed.
Genes in cell wall fortification pathways responded to Pm
The cell wall, as the first barrier against pathogen attack, reacts to localized stress by directly apposing substances onto the inner surface. It has been suggested that cell wall modification elicited by fungi might represent a disease resistance mechanism by interfering with the invading pathogens [39], [40], [41]. The genes encoding snare protein, syntaxin and WIR1A involved in cell wall fortification can greatly enhance disease resistance in barley, tobacco and Arabidopsis, respectively [42], [43], [44]. Our analysis in wheat showed that many genes responsible for reinforcing cell wall were up-regulated after inoculating the leaves with Bgt. These included nine genes encoding hydroxyproline-rich glycoprotein (HRGP), twelve for proline-rich glycoprotein, six for cellulose synthase, two for syntaxin, one for snare and three for WIR1A (Table S2). The genes annotated as cellulase (Ta.9047.2.S1_a_at) and pectinesterase (Ta.1564.1.S1_at), which play roles in destroying cell walls, were down-regulated in R-12 h to a greater extent than in S-12 h. Indeed, the pectinesterase (Ta.1564.1.S1_at) was up-regulated 13 folds in susceptible wheat. Moreover, the wax layer as the frontline of defence against pathogen invasion is also critically important. We found that the CER1 gene (TaAffx.52653.1.A1_s_at) in wheat, encoding a protein involved in wax biosynthesis, was highly up-regulated in resistant wheat but remained unchanged in susceptible wheat [45]. So we proposed that the increased expression of positive cell wall related protein might reinforce wheat resistance to Pm, while negative protein would play an opposite role. Therefore, the differential expression of these genes between R and S genotypes might lead to different reactions to disease.
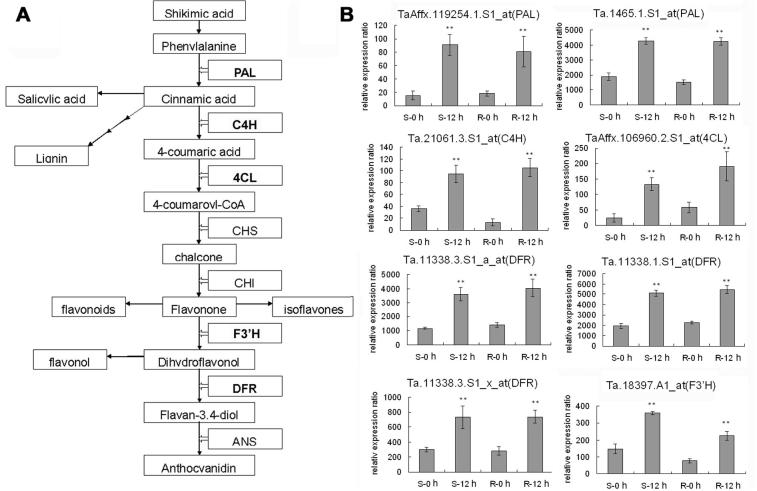
Genes in flavonoids biosynthesis pathways responded to Pm
Flavonoids are ubiquitous secondary metabolites in plants that have been considered to function in a wide range of biological processes, including both abiotic and biotic stress response [46], [47], [48]. Our analysis showed that several key enzymes involved in flavonoid biosynthetic pathways were up-regulated, including phenylalanine ammonia lyase (PAL) (TaAffx.119254.1.S1_at and Ta.1465.1.S1_at) catalyzing the conversion of phenylalanine to cinnamic acid, cinnamate-4 hydroxylase (C4H) (Ta.21061.3.S1_at) required for condensation of 4 coumaric acid, 4-coumarate CoA ligase(4CL) (TaAffx.106960.2.S1_at and TaAffx.106960.1.S1_at) to produce 4-coumaroyl-CoA, flavonoid 3′-hydroxylase(F3′H) (Ta.18397.1.A1_at) converting flavonone to dihydroflavonol and dihydroflavonal-4-reductase (DFR) (Ta.11338.3.S1_a_at, Ta.11338.1.S1_at and Ta.11338.3.S1_x_at) for producing the flavan-3,4-diol (Figure 6A). Experimental validation of these flavonoid biosynthesis related genes by qRT-PCR revealed good agreement with microarray results (Figure 6B). Additionally, proteins participating in flavonoid derivative biosynthesis were also found responsive to Bgt infection, such as caffeic acid O-methyltransferase (COMT), flavonol synthase (FLS), UDP-glucosyltransferase, flavonol 4′-sulfotransferase, flavonol 4′-glucosyltransferase, isoflavone reductase homolog IRL (IFR), UDP-glucose pyrophosphorylase, flavonoid 7-O-glucosyltransferase (FGT). Although their roles in wheat-Bgt interaction remain obscure and no experimental proof has been supplied, we speculated that the flavonoid pathway is a potential participant in wheat resistance to Pm based on our microarray data and qRT-PCR results.
Figure 6.
Analysis of genes involved in anthocyanin and flavonoid biosynthesis (A) Simplified scheme of anthocyanin and flavonoid biosynthesis pathway. (B) Expression patterns of genes related to flavonoid biosynthesis based on microarray analysis. Genes that were up-regulated after Pm infection in R and S genotypes of wheat are highlighted in bold. Student’s t-test was performed to analyze the changes in the gene expression 12 h after Pm infection (∗P < 0.05; ∗∗P < 0.01, compared to 0 h in respective genotype). PAL: phenylalanine ammonia lyase, C4H: cinnamate-4 hydroxylase, 4CL: 4-coumarate CoA ligase, CHS: chalcone synthase, CHI: chalcone isomerase, F3′H: flavonoid 3′-hydroxylase, DFR: dihydroflavonal-4-reductase, ANS: anthocyanin synthase.
Phytohormones in wheat responded to Pm
The roles of phytohormones, such as SA, JA/ET, have been described in plant disease response [49]. We identified numerous genes involved in phytohormone metabolism and signaling pathway that were up-regulated in response to Pm attack, including SA, abscisic acid (ABA), gibberellic acid (GA), ET, Auxin, and cytokinin (CK).
SA induced by ROS, is a well-studied plant signaling molecule responsive to pathogen attack [50]. A number of genes encoding key enzymes in the phenylpropanoid pathway displayed altered expression after Bgt inoculation such as cytochrome P450 monooxygenases which convert benzoic acid to SA.
The ABA is considered as a negative regulator of disease resistance and the expression levels of ABA are anti-correlated with increased disease susceptibility [51], [52]. Zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NECD) and aldehyde oxidase (AO) are the key enzymes in ABA biosynthesis [53]. Our analysis showed that the ZEP (Ta.404.1.S1_x_at), NECD (TaAffx.13292.2.S1_at) and several ABA-responsive proteins (Ta.19973.1.A1_at, Ta.17416.1.S1_at, Ta.27945.1.S1_x_at and TaAffx.132322.1.S1_at) were subject to down-regulation to various extents in resistant and/or susceptible wheat after Pm infection.
In contrast to ABA, GA has a positive effect on plant defense [49]. Ent-kaurene synthase (KS), ent-kaurene oxidase (KO) and GA 20-oxidases (GA20ox) are the key enzymes in GA biosynthesis [54]. All of KS (Ta.8418.1.S1_at), KO (Ta.14904.1.S1_at and Ta.5772.1.A1_at) and GA20ox (Ta.141.1.S1_x_at, Ta.23668.1.S1_at, Ta.3564.1.S1_s_at and Ta.3564.1.S1_at) were up-regulated after Pm infection.
We also demonstrated that the genes involved in ethylene production were up-regulated, such as EIN3 (TaAffx.110715.1.S1_at), a component of ET signaling pathway, and genes encoding ethylene-forming enzymes ACC oxidases and ethylene-responsive element binding proteins (EREBPs) [55]. Although there is a debate regarding the role of ethylene in disease response, our data seems to support the idea that ET signaling is involved in Bgt response in wheat.
Auxin signaling pathway has been found to strengthen the induced immune responses, and the Auxin response factors (ARFs) might act as repressors of plant resistance towards biotrophic pathogens [56]. Three genes encoding ARFs (Ta.25087.1.S1_at, TaAffx.64139.1.S1_at and Ta.6746.1.S1_at) were repressed after Pm attack.
Previous studies have suggested that plant hormones were involved in mediating fungus interaction with plants, and their roles were totally different [49], [50], [51], [52], [53], [54], [55], [56]. Consistent with their reports, our microarray data showed different expression patterns of genes involved in hormone production and signaling in response to Bgt inoculation.
Genes in metabolic pathways responded to Pm
Lipid degradation
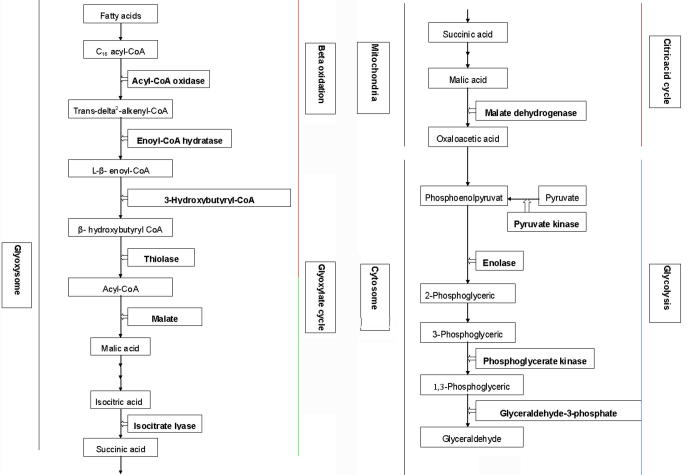
Lipid degradation, particularly the membrane lipids, is one of several biochemical manifestations of cellular senescence [57]. The beta oxidation process is responsible for lipid transformation from fatty acids to acyl-CoA, and acyl-CoA then enters the glyoxylate and tricarboxylic acid circle, which is activated by senescence. Ultimately metabolites turn into carbohydrates through glyconeogenesis [58], [59]. Four enzymes, acyl-CoA oxidase, enoyl-CoA hydratase, thiolase and 3-hydroxybutyryl-CoA dehydrogenase participate in the beta oxidation process. In this study, expression of genes encoding these four enzymes was induced to a higher level in S than in R genotypes. Malate synthase and isocitrate lyase are characteristic enzymes of the glyoxylate circle, which were up-regulated after Pm infection. In S genotype, they were induced by fold changes of 194 (Ta.23970.1.A1_at) and 927 (TaAffx.79139.1.S1_at), while in R genotype, they were only increased by fold changes of 45 and 28, respectively. In addition, genes encoding malate dehydrogenase which catalyze malic acid into oxaloacetic acid were only induced in the S genotype (Figure S2 and Table S3).
Carbohydrate metabolism
Four enzymes, pyruvate kinase, enolase, phosphoglycerate kinase, and glyceraldehyde-3-phosphate dehydrogenase are required for the gluconeogenesis process [60], [61]. We found that most of probe sets which were annotated as genes encoding these enzymes were up-regulated after Pm infection. For example, one gene (Ta.3910.3.S1_a_at) encoding pyruvate kinase was up-regulated only in the S genotype, while another one (TaAffx.81575.1.S1_at) was up-regulated 17 and 8-fold in S and R genotype (Si:Ri = 17:8) (Si: increased fold in S genotype; Ri: increased fold in R genotype), respectively. The Si:Ri for two phosphoglycerate kinase genes was 25:14 (TaAffx.81675.1.S1_at) and 5:4 (TaAffx.6099.1.S1_at), respectively, whereas the Si:Ri for glyceraldehyde-3-phosphate dehydrogenase gene was 134:21 (Ta.15063.1.A1_at) and 153:23 (TaAffx.108685.1.S1_at) (Table S3).
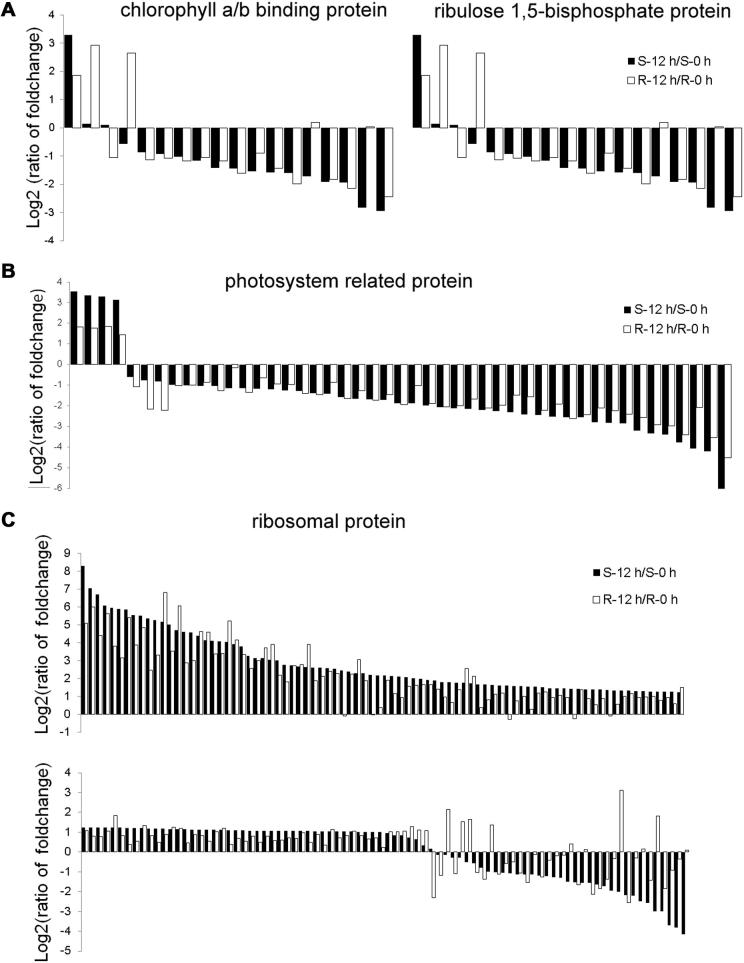
Additionally, we found that 32 genes encoding chlorophyll a/b-binding proteins were responsive to Pm infection. Only two of them were up-regulated, while the others were repressed, and much higher fold changes were observed in S than in R genotype. For instance, TaAffx.114127.1.S1_x_at was reduced up to 300-fold in S genotype, but 150-fold in R genotype at 12 hpi (Figure 7A, Table S3). Most (15/17) genes encoding ribulose-1,5-biophosthate carboxylase and genes (43/47) for photosystem related protein were down-regulated (Figure 7B, Table S3). These results are similar to a previous report by Swarbrick et al. [62].
Figure 7.
Column chart of other differentially expressed genes (A) Chlorophyll a/b binding proteins and ribulose-1,5-bisphosphate carboxylase. (B) Photosystem related proteins. (C) Ribosomal proteins. All the data used for the figure are the average value of three replicates and are log 2 transformed
Proteolytic process
Protein degradation in plants is a complex process involving a multitude of proteolytic pathways, and ubiquitin/proteasome is one of the important pathways [63]. Our study showed that all the genes encoding ubiquitin activating enzyme (E1), one for 26S proteasome and three for ubiquitin conjugating enzyme (E2) were induced only in S genotype. In addition, the Si:Ri for the genes encoding E2 (TaAffx.81408.1.S1_at) and monoubiquitin/carboxy extension protein fusion (Ta.22526.1.S1_at and TaAffx.81696.1.S1_at) was 23:12, 162:37 and 30:20, respectively. Most genes encoding ribosomal protein (132/168) were up-regulated at 12hpi, and the fold change in S genotype was much higher than in R genotype (Figure 7C, Table S3). We speculate that more rapid protein degradation occurs in S genotype, which might be caused by faster senescence in S genotype, as the pathogen develops more quickly and consumes more nutrition. To support this idea, we found that one gene (Ta.15208.1.S1_at) encoding senescence associated protein was up-regulated to a much higher level in S genotype than in R genotype (Si:Ri = 42:7).
Recently, interest in primary metabolism in plants after pathogen infection has been growing. Photosynthesis and nutrient metabolism in several types of plant-pathogen interactions have been investigated [64], [65], [66], [67], [68]. As it has been shown that the induction of defense is cost-intensive, obviously contact with pathogens greatly alters plant primary metabolism [69], [70]. Consistent with these previous studies, a great change was observed in gene expression in both R and S genotypes during the wheat-Pm interaction. Nonetheless, our results showed a considerably greater there was a much more serious change in S genotype than in R genotype of wheat.
Conclusion
In this study, we used Affymetrix wheat genome microarrays to profile the transcriptomes of susceptible and resistant wheat cultivars in response to powdery mildew. Our analysis identified a number of candidate genes showing higher expression levels in resistant wheat than in susceptible wheat before the Pm infection, indicating their potential roles in building the basal barrier for the pathogen defense and flavonoid pathway could play a crucial role in disease resistance. For the first time, we report that sense-antisense pair genes are potentially involved in disease resistance in wheat, and based on our analysis, susceptible wheat may display greater degradation of lipid, protein and carbohydrate than resistant wheat after Bgt inoculation. Several candidate genes with potential pathogen defense functions in response to Bgt infection were also experimentally validated by qRT-PCR. Future functional analysis of these Bgt responsive genes is expected to help towards a better understanding of the molecular mechanisms of pathogen-defense in wheat.
Materials and methods
Plant growth and tissue collection
Seeds of S genotype) and R genotype were planted in 8–10 cm diameter pots. Seedlings were manually inoculated when the first leaf was fully expanded, with a locally-prevalent Bgt isolate E09. Inoculation was performed by dusting conidia from neighboring sporulating susceptible seedlings onto the test seedlings. Leaf samples were collected from both lines at 0 h and 12 hpi, respectively, followed by freezing in liquid nitrogen for subsequent RNA extraction.
RNA extraction and microarray hybridization
Total RNAs were extracted using Trizol reagent (Invitrogen). First, mRNAs were enriched from 80–90 μg total RNAs using the RNeasy Plant Mini Kit (QIAGEN), and then were reversely transcribed to double-stranded cDNAs using the GeneChip® Two-Cycle cDNA Synthesis Kit. The biotin-labeled cRNAs were made using the GeneChip® IVT Labeling Kit (Affymetrix, CA, USA). Twenty micrograms of cRNA samples were fragmented and hybridized for 16 h at 45oC to the Affymetrix Wheat Genome Array containing 61,127 probe sets representing 55,052 transcripts based on the ESTs and full-length cDNAs collection in wheat. After washing the microarrays using the Genechip® Fluidics Station 450, the microarrays were scanned using the Genechip® 3000 Scanner located in the bioinformatics facility at the China Agriculture University.
Microarray data analysis
The chip images were scanned and the hybridization intensities of the probe sets were extracted to generate the CEL files by the Affymetrix GeneChip Operating Software (GCOS 1.2). The resulting CEL files were imported into the software dChip for data processing and analysis, including background adjustment, normalization of the raw data, summarizing gene expression signals and detecting the differentially expressed genes [46]. In order to assess the reproducibility of microarray data, the normalized signal intensities from the three replicates of each sample were used to calculate correlation coefficients. After removing the probe sets prefixed with “AFFX” and “RPTR” by fraction call 100%, statistical identification of DR genes was performed by the t-test functional module in dChip, followed by the false discovery rate (FDR) adjustment [46], [71], [72]. The wheat microarray annotation was obtained from HarvEST (http://www.harvest.ucr.edu/). The microarray data has been submitted to NCBI GEO database under the accession number GSE27339.
Quantitative real-time PCR (qRT-PCR) analysis
A portion of seedling leaves was used for qRT-PCR analysis to validate the expression patterns detected by microarrays. Total RNAs were extracted using Trizol reagent (Invitrogen, USA). Two microgram total RNAs of each sample were used to synthesize the first-strand cDNA by using oligo (dT)18 primer with M-MLV reverse transcriptase (Promega, USA) according to the manufacture’s instructions. The product of reverse transcription was tested by amplifying a wheat actin gene (Ta-actin) fragment, which was used as the endogenous control. PCR primers were designed using DNAMAN software, and the primer pairs used to amplify probe set-specific products were listed in Table S4. qRT-PCR was performed using the cDNA samples in a 10 μL mixture containing 1× LightCycler-FastStart DNA master SYBR Green I. PCR was performed as follows: initial denaturation for 10 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 45 s at 55 to 60 °C, 30 s at 72 °C, and 72 °C for 5 min as the last step. The threshold cycles (Ct) of each test target were averaged for triplicate reactions and the values were normalized according to the Ct of Ta-actin. Each PCR product was evaluated in at least three independent experiments, and the value of 2−△△ct from three replicates were then subjected to student’s t test. Only the genes, which showed more than 2-fold change with significant differences (P < 0.01), were defined as differentially expressed.
Authors’ contributions
MX and XW performed major data analyses and drafted the manuscript. HP, YY and YH collected the materials. ZN designed the study. CX and QS supervised the project and co-wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgement
This work was supported by the State High-Tech Program (Grant No. 2006AA10A104) of the Ministry of Science & Technology of China, the National Natural Science Foundation of China (Grant No. 30871528) and China Transgenic Research Program (Grant Nos. 2008ZX08002-001 and 2008ZX08009-002).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.05.002.
Contributor Information
Chaojie Xie, Email: xiecj127@126.com.
Qixin Sun, Email: qxsun@cau.edu.cn.
Supplementary data
Supplementary Figure 1.
Supplementary Figure 2.
References
- 1.Gill B.S. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics. 2004;168:1087–1096. doi: 10.1534/genetics.104.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffey C.A. Effectiveness of adult-plant resistance in reducing grain yield loss to powderymildew in winter wheat. Plant Dis. 1993;77:618–622. [Google Scholar]
- 3.Leath S., Bowen K.L. Effects of powdery mildew, triadimenolseed treatment, and triadimefon foliar sprays on yieldof winter wheat in North Carolina. Phytopathology. 1989;79:152–155. [Google Scholar]
- 4.Bhullar Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA. 2009;106:9519–9524. doi: 10.1073/pnas.0904152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J. 2011;10:398–409. doi: 10.1111/j.1467-7652.2011.00670.x. [DOI] [PubMed] [Google Scholar]
- 6.Bernardo A. Fusarium graminearum-induced changes in gene expression between Fusarium head blight-resistant and susceptible wheat cultivars. Funct Integr Genomics. 2007;7:69–77. doi: 10.1007/s10142-006-0028-1. [DOI] [PubMed] [Google Scholar]
- 7.Bhuiyan N.H. Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol Biol. 2007;64:305–318. doi: 10.1007/s11103-007-9155-x. [DOI] [PubMed] [Google Scholar]
- 8.Bruggmann R. Analysis of epidermis- and mesophyll-specific transcript accumulation in powdery mildew-inoculated wheat leaves. Plant Mol Biol. 2005;58:247–267. doi: 10.1007/s11103-005-3099-9. [DOI] [PubMed] [Google Scholar]
- 9.Coram T.E. Transcriptome analysis of high-temperature adult-plant resistance conditioned by Yr39 during the wheat-Puccinia striiformis f. sp. tritici interaction. Mol Plant Pathol. 2008;9:479–493. doi: 10.1111/j.1364-3703.2008.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laudencia-Chingcuanco D. Genome-wide gene expression analysis supports a developmental model of low temperature tolerance gene regulation in wheat (Triticum aestivum L.) BMC Genomics. 2011;12:299. doi: 10.1186/1471-2164-12-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aprile A. Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics. 2009;10:279. doi: 10.1186/1471-2164-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin D.D. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics. 2008;9:432. doi: 10.1186/1471-2164-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golkari S. QTL-specific microarray gene expression analysis of wheat resistance to Fusarium head blight in Sumai-3 and two susceptible NILs. Genome. 2009;52:409–418. doi: 10.1139/g09-018. [DOI] [PubMed] [Google Scholar]
- 14.Bolton M.D. Lr34-mediated leaf rust resistance in wheat: transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Mol Plant Microbe Interact. 2008;21:1515–1527. doi: 10.1094/MPMI-21-12-1515. [DOI] [PubMed] [Google Scholar]
- 15.Wan Probing plant–pathogen interactions and downstream defense signaling using DNA microarrays. Funct Integr Genomics. 2002;2:259–273. doi: 10.1007/s10142-002-0080-4. [DOI] [PubMed] [Google Scholar]
- 16.Bozkurt T.O. Cellular and transcriptional responses of wheat during compatible and incompatible race-specific interactions with Puccinia striiformis f. sp. Tritici. Mol Plant Pathol. 2010;11:625–640. doi: 10.1111/j.1364-3703.2010.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Showalter A.M. Accumulation of hydroxyproline-rich glycoprotein mRNAs in response to fungal elicitor and infection. Proc Natl Acad Sci USA. 1985;82:6551–6555. doi: 10.1073/pnas.82.19.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang I.S., Hwang B.K. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010;152:948–967. doi: 10.1104/pp.109.147827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feussner I., Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 20.Senthil G. Specific and common changes in Nicotiana benthamiana gene expression in response to infection by enveloped viruses. J Gen Virol. 2005;86:2615–2625. doi: 10.1099/vir.0.81043-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X.C. Molecular characterization of a defense-related AMP-binding protein gene, OsBIABP1, from rice. J Zhejiang Univ Sci B. 2009;10:731–739. doi: 10.1631/jzus.B0920042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Z. AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katiyar-Agarwal S. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin M.M. Diverse set of miRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.) BMC Plant Biol. 2010;10:123. doi: 10.1186/1471-2229-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutt J.R. Disease response to tobacco mosaic virus in transgenic tobacco plants that constitutively express the pathogenesis-related PR1b gene. Virology. 1989;173:89–97. doi: 10.1016/0042-6822(89)90224-9. [DOI] [PubMed] [Google Scholar]
- 26.Datta K. Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci. 2001;160:405–414. doi: 10.1016/s0168-9452(00)00413-1. [DOI] [PubMed] [Google Scholar]
- 27.Ding C.K. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta. 2002;214:895–901. doi: 10.1007/s00425-001-0698-9. [DOI] [PubMed] [Google Scholar]
- 28.Silva H. Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol Plant Microbe Interact. 1999;12:1053–1063. doi: 10.1094/MPMI.1999.12.12.1053. [DOI] [PubMed] [Google Scholar]
- 29.White R.F. The chemical induction of PR (b) proteins and resistance to TMV infection in tobacco. Antiviral Res. 1986;6:177–185. doi: 10.1016/0166-3542(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 30.Friedrich L. Pathogenesis-related protein 4 is structurally homologous to the carboxy-terminal domains of hevein, Win-1 and Win-2. Mol Gen Genet. 1991;230:113–119. doi: 10.1007/BF00290658. [DOI] [PubMed] [Google Scholar]
- 31.Breda C. Defense reaction in Medicago sativa: a gene encoding a class 10 PR protein is expressed in vascular bundles. Mol Plant Microbe Interact. 1996;9:713–719. doi: 10.1094/mpmi-9-0713. [DOI] [PubMed] [Google Scholar]
- 32.Thomma B.P. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 33.Bertini L. Molecular and functional analysis of new members of the wheat PR4 gene family. Biol Chem. 2006;387:1101–1111. doi: 10.1515/BC.2006.136. [DOI] [PubMed] [Google Scholar]
- 34.Makandar R. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact. 2006;19:123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- 35.Quilis J. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact. 2008;21:1215–1231. doi: 10.1094/MPMI-21-9-1215. [DOI] [PubMed] [Google Scholar]
- 36.El-kereamy A. Expression analysis of a plum pathogenesis related 10 (PR10) protein during brown rot infection. Plant Cell Rep. 2009;28:95–102. doi: 10.1007/s00299-008-0612-z. [DOI] [PubMed] [Google Scholar]
- 37.Friedrich L. NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol Plant Microbe Interact. 2010;14:1114–1124. doi: 10.1094/MPMI.2001.14.9.1114. [DOI] [PubMed] [Google Scholar]
- 38.Edreva A.M. Induction of ‘pathogenesis-related’ proteins in tobacco leaves by physiological (non-pathogenic) disorders. J Exp Bot. 1990;41:701–703. [Google Scholar]
- 39.Israel H.W. Cell wall appositions and plant disease resistance. Acoustic microscopy of papillae that block fungal ingress. Proc Natl Acad Sci USA. 1980;77:2046–2049. doi: 10.1073/pnas.77.4.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brisson L.F. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez-Blanco C. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins N.C. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 43.Assaad F.F. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell. 2004;15:5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalde M. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA. 2007;104:11850–11855. doi: 10.1073/pnas.0701083104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aarts M.G. Molecular characterization of the CER1 gene of arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 1995;7:2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 47.Li J. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walia H. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bari R. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 50.Leon J. Induction of benzoic acid 2-hydroxylase in virus-inoculated tobacco. Plant Physiol. 1993;103:323–328. doi: 10.1104/pp.103.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauch-Mani B., Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Fan J. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009;150:1750–1761. doi: 10.1104/pp.109.137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkelstein R. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olszewski N. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feys B.J., Parker J.E. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- 56.Navarro L. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 57.Thompson J. Lipid metabolism during leaf senescence. Prog Lipid Res. 1998;37:119–141. doi: 10.1016/s0163-7827(98)00006-x. [DOI] [PubMed] [Google Scholar]
- 58.Paliyath G. Regulation of membrane lipid in senesencing carnation flowers. Plant Physiol. 1987;71:503–511. [Google Scholar]
- 59.Gerhardt B. Fatty acid degradation in plants. Prog Lipid Res. 1992;31:417–446. doi: 10.1016/0163-7827(92)90004-3. [DOI] [PubMed] [Google Scholar]
- 60.O’Brien R.W. Enzymatic analysis of the pathways of glucose catabolism and gluconeogenesis in Pseudomonas citronellolis. Arch Microbiol. 1975;103:71–76. doi: 10.1007/BF00436332. [DOI] [PubMed] [Google Scholar]
- 61.Yu J.P. Pathway of glycogen metabolism in Methanococcus maripaludis. J Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swarbrick P.J. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006;29:1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- 63.Mykles D.L. Intracellular proteinases of invertebrates: calcium-dependent and proteasome/ubiquitin-dependent systems. Int Rev Cytol. 1998;184:157–289. doi: 10.1016/s0074-7696(08)62181-6. [DOI] [PubMed] [Google Scholar]
- 64.Wright D. Source–sink relationships in wheat leaves infected with powdery mildew. Physiol Mol Plant Pathol. 1995;47:237–253. [Google Scholar]
- 65.Chou H. Infection of Arabidopsis thaliana leaves with Albugo candida causes a reprogramming of host metabolism. Mol Plant Pathol. 2000;1:99–113. doi: 10.1046/j.1364-3703.2000.00013.x. [DOI] [PubMed] [Google Scholar]
- 66.Herbers K. Regulation of carbohydrate partitioning during the interaction of potato virus Y with tobacco. Mol Plant Pathol. 2000;1:51–59. doi: 10.1046/j.1364-3703.2000.00007.x. [DOI] [PubMed] [Google Scholar]
- 67.Scharte J. Photosynthesis and carbohydrate metabolism in tobacco leaves during an incompatible interaction with Phytophthora nicotianae. Plant Cell Environ. 2005;28:1421–1435. [Google Scholar]
- 68.Berger S. Plant physiology meets phytopathology: plant primary metabolism and plant–pathogen interactions. J Exp Bot. 2007;58:4019–4026. doi: 10.1093/jxb/erm298. [DOI] [PubMed] [Google Scholar]
- 69.Swarbrick P.J. Metabolic consequences of susceptibility and resistance in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006;29:1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- 70.Heil M., Bostock R.M. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot. 2002;89:503–512. doi: 10.1093/aob/mcf076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McClintick J.M., Edenberg H.J. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benjamini Y. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 73.Conesa A. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.