Abstract
Multivariate analysis of codon and amino acid usage was performed for three Leishmania species, including L. donovani, L. infantum and L. major. It was revealed that all three species are under mutational bias and translational selection. Lower GC12 and higher GC3S in all three parasites suggests that the ancestral highly expressed genes (HEGs), compared to lowly expressed genes (LEGs), might have been rich in AT-content. This also suggests that there must have been a faster rate of evolution under GC-bias in LEGs. It was observed from the estimation of synonymous/non-synonymous substitutions in HEGs that the HEG dataset of L. donovani is much closer to L. major evolutionarily. This is also supported by the higher dN value as compared to dS between L. donovani and L. major, suggesting the conservation of synonymous codon positions between these two species and the role of translational selection in shaping the composition of protein-coding genes.
Key words: Leishmania, relative synonymous codon usage, multivariate analysis, hydropathy, aromaticity
Introduction
Leishmaniasis, an infectious protozoal disease caused by parasites belonging to the genus Leishmania, is still one of the world’s most neglected diseases, affecting mainly developing countries (1). L. major causes the most common form of infection, cutaneous leishmaniasis, while L. donovani and L. infantum are associated with visceral leishmaniasis 2, 3, also known as Kala-azar, in the Indian subcontinent, East Africa, and Mediterranean regions (4). Despite the continuous ongoing efforts in antileishmanial drug discovery and development, there is no effective medicine available so far. The results from current chemotherapeutic drugs available for the treatment of Leishmania infection are not satisfactory (5). The toxic nature of available drugs and the tendency of Leishmania to become resistant reflect the need for discovery of more effective antileishmanial agents (5). Therefore, there is an urgent need to understand the biology of these three Leishmania pathogens. The published genomic details of L. infantum (6) and L. major (7) show that the average GC content was around 59% for both of them. The whole genome of L. donovani has yet not been sequenced, but sequences of some genes and proteins are available online.
Many genes demonstrate a non-random selection of codons in their protein-coding regions. For any given protein we can distinguish at least two sources of bias in codon usage. The first, “amino acid preference”, is the uneven amino acid composition of typical proteins, i.e., some amino acids are used far more frequently than others (8). The second is that once an amino acid has been chosen, there are generally preferences for the use of certain codons. Relative synonymous codon usage (RSCU) and relative amino acid usage (RAAU) are used to measure the non-random usage of specific amino acid. Genes with strong codon bias appear to be expressed at a higher level compared to other genes. Biased codon usage may result from a combination of several factors, namely, biases in the pattern of mutation (9), or translational selection (10) among synonymous codons. Within-species heterogeneity in codon usage has been most clearly elucidated in E. coli (11). The major trend includes a strong bias towards a particular subset of codons in highly expressed genes (HEGs) and more even codon usage in lowly expressed genes (LEGs) 12, 13, 14. Our comparative multivariate analysis of codon and amino acid usage patterns in Leishmania species will provide an insight into the divergence and compositional similarities within and across their genomes and may lead to a better understanding of the biology of the parasites and the development of more effective drug treatments.
Results
Major sources of RSCU variation in the three Leishmania species
Correspondence analysis (COA) was used to explore the variation of RSCU values in the genes from L. donovani, L. infantum and L. major. After plotting genes in 59-dimentional hyperspace, according to the usage of the 59 synonymous sense codons (stop codons and codons with one-to-one mapping to amino acids, i.e., Met, Trp were excluded), COA identifies a series of new orthogonal axes accounting for the greatest variation among genes. The coordinate of each gene on each new axis and the fraction of the total variation accounted for by each axis is generated by COA. Axis 1 and Axis 2 indicate the major trends of variations among genes. Axis 1 accounts for 31.5%, 15.7% and 17.2% of the total variations for RSCU in L. donovani, L. infantum and L. major, respectively (Table 1). In all cases, GC3S (GC content at synonymous codon sites excluding ATG for Met and TGG for Trp) and NC (effective number of codons) exhibited strong correlation with Axis 1. The correlation between GC3S and Axis 1 is negative in L. donovani and L. major but positive in L. infantum. Conversely, the correlation between NC and Axis 1 is positive in L. donovani and L. major but negative in L. infantum. The correlations between GC3S and Axis 1 suggests that highly biased genes, those with G/C-ending codons, are clustered on the negative side in L. donovani and L. major but on the positive side of Axis 1 in L. infantum (Table 1). Also, the high degree of correlation between GC3S and Axis 1 suggests that directional mutational pressure plays a major role in governing the synonymous codon usage. In addition, the low value of NC (Table 1) indicates that HEGs are under translational selection. In L. donovani, GT3S, gravy and aromaticity all significantly contributed to the variation on Axis 2. In L. infantum, both GT3S and gravy significantly correlated with Axis 2 in L. donovani, while in L. major, aromaticity was found to be the only major source of variation on Axis 2.
Table 1.
Major trends in synonymous codon usage in L. donovani, L. infantum and L. major as revealed by COA on RSCU of genes on Axis 1 and 2
| Organism | Axis 1 |
Axis 2 |
||||
|---|---|---|---|---|---|---|
| Total variability | Source of variation | Correlation coefficient (r)a | Total variability | Source of variation | Correlation coefficient (r)a | |
| L. donovani | 31.5% | NC | 0.693 | 7.4% | GT3S | –0.371 |
| GC3S | –0.983 | Aromaticity | –0.358 | |||
| Gravy | –0.265 | |||||
| L. infantum | 15.7% | NC | –0.940 | 4.7% | GT3S | 0.621 |
| GC3S | 0.951 | Gravy | –0.131 | |||
| L. major | 17.2% | NC | 0.957 | 4.7% | Aromaticity | –0.117 |
| GC3S | –0.953 | |||||
All correlations are significant at P<0.01.
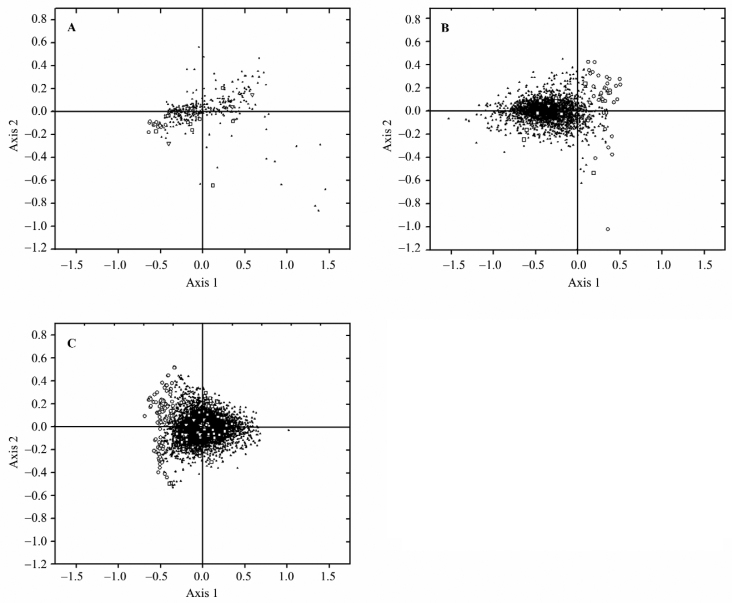
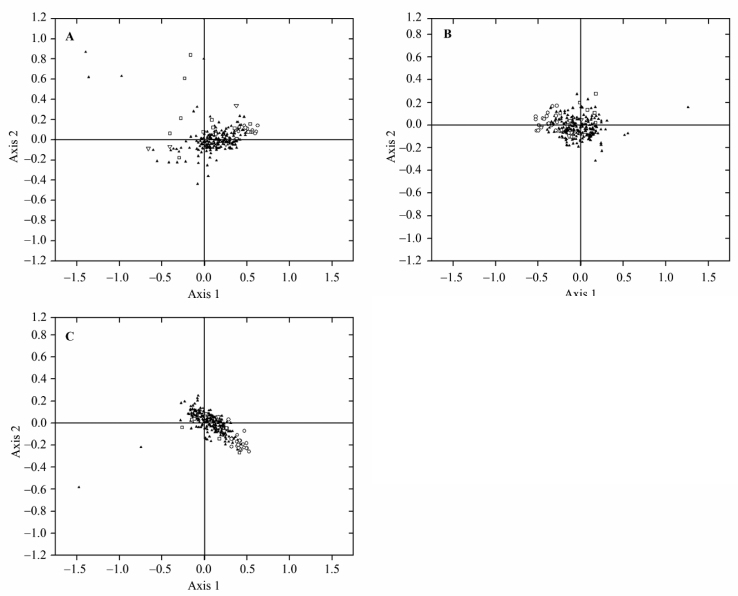
A plot of Axis 1–Axis 2 of each genome under study including L. donovani, L. major, and L. infantum was drawn, showing that HEGs are clustered at one end of Axis 1 (Figure 1, circle), indicating that these genes follow a distinct pattern of synonymous codon usage.
Figure 1.
Position of genes along the two major axes of variation in COA on RSCU. Position of genes along Axis 1 are plotted against Axis 2 by COA in L. donovani (A), L. infantum (B), and L. major (C). HEG, VSG, and topoisomerase is represented by open circle, square and triangle, respectively. The rest of the genes (LEGs) are indicated by solid triangles. X and Y axis is represented by Axis 1 and Axis 2, respectively. COA: correspondence analysis; HEG: highly expressed gene; VSG: variable surface glycoprotein; LEGs: lowly expressed genes.
A comparison of RSCU values of the HEGs with those of the LEGs shows that in all three parasites examined, a similar subset of synonymous codons, mostly G/C-ending, are preferred by the HEGs (Table S1, codons with bold values). The LEGs exhibit relatively higher usage of A/U-ending codons. But in all three species, even the LEGs prefer to use G/C-endingcodons for most of the amino acids, though the frquencies of such codons are low. This is in agreement with the high GC content in the genes from L. donovani (58.8%), L. infantum (59.3%) and L. major (59.7%). As seen in Table S1, high extent of bias in the synonymous codon usage suggests that the influence of translational selection is strong in all the three Leishmania species.
Codon usage in variant surface glycoproteins, HEGs and the topoisomerase gene
Variable surface glycoproteins (VSGs) have been identified as parasite virulence factors that make possible the survival of Leishmania inside the macro phages (15). DNA topoisomerases are a family of DNA-processing enzymes involved in catalysis of the breakage and rejoining of DNA strands (16). DNA topoisomerase of L. donovani is distinct from other eukaryotic counterparts with respect to its biological properties and preferential sensitivity to many therapeutic agents (17). Due to the therapeutical importance of VSGs and topoisomerases, we have included them separately for analysis of codon and amino acid usage.
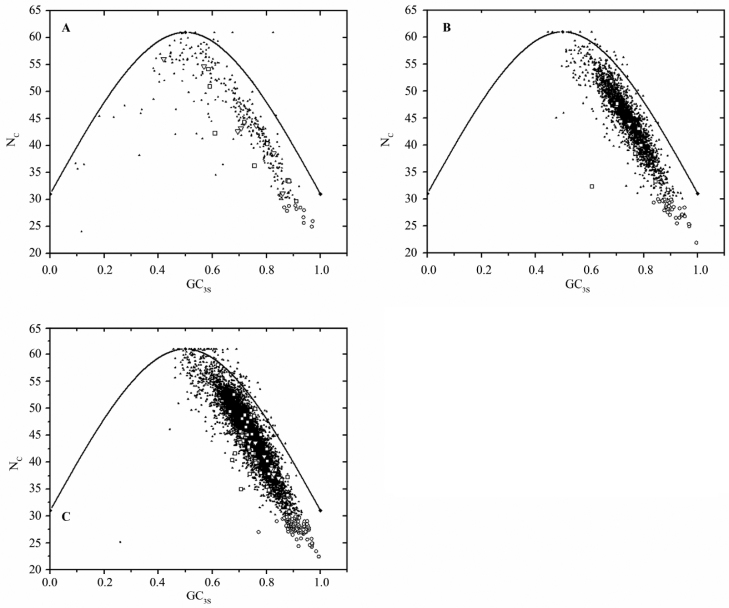
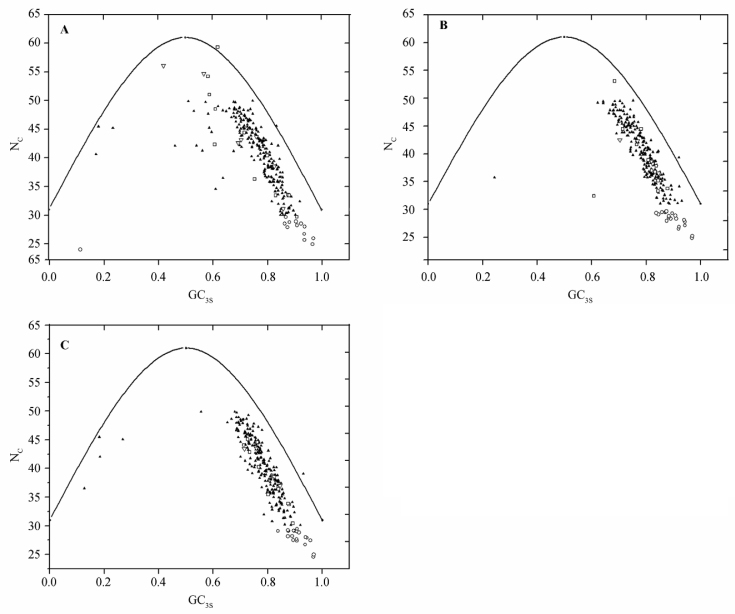
In all the three species of Leishmania examined, genes other than HEGs constitute a single cluster (Figure 1). But this is not the case for some genes, i.e., VSG and topoisomerase genes. Their highly scattered nature on Axis 1–Axis 2 plot suggests that these genes have different codon usage due to mutational pressure or different translational selection. As indicated in Figure 1 and Figure 2, all these genes are also characterized by high GC3S and high NC values.
Figure 2.
The NC plot of genes in the three Leishmania species. NC is plotted against GC3S for L. donovani (A), L. infantum (B) and L. major genes (C). The solid line indicates the expected NC value if bias is due to GC3S alone. HEG, VSG and topoisomerase is represented by open circle, square and triangle, respectively. The rest of the genes (LEGs) are indicated by solid triangles.
Major sources of variation in amino acid usages
To identify the major trends of intra-proteomic variations in amino acid composition in the three Leishhmania species, COA on amino acid usage was performed. The first axis generated by COA accounts for 32%, 24% and 30% of the total variations in L. donovani, L. infantum, and L. major, respectively (Table 2).
Table 2.
Major trends in synonymous codon usage in L. donovani, L. infantum and L. major as revealed by COA on amino acid usage of encoded proteins on Axis 1 and Axis 2
| Organism | Axis 1 |
Axis 2 |
||||
|---|---|---|---|---|---|---|
| Total variability | Source of variation | Correlation coefficient (r)a | Total variability | Source of variation | Correlation coefficient (r)a | |
| L. donovani | 32% | CAI | –0.599 | 20% | Aromaticity | –0.792 |
| NC | 0.540 | Gravy | –0.717 | |||
| GT3S | –0.541 | |||||
| GC12 | 0.767 | |||||
| L. infantum | 24.2% | CAI | –0.645 | 14.3% | Aromaticity | 0.397 |
| NC | 0.430 | Gravy | 0.334 | |||
| GC3S | –0.424 | |||||
| GC12 | 0.946 | |||||
| L. major | 30% | CAI | 0.529 | 14.3% | Aromaticity | 0.669 |
| GC3S | 0.400 | Gravy | 0.531 | |||
| GC12 | –0.862 | |||||
All correlations are significant at P=0.01. GC12: G/C content at first and second codon sites; CAI, codon adaptation index.
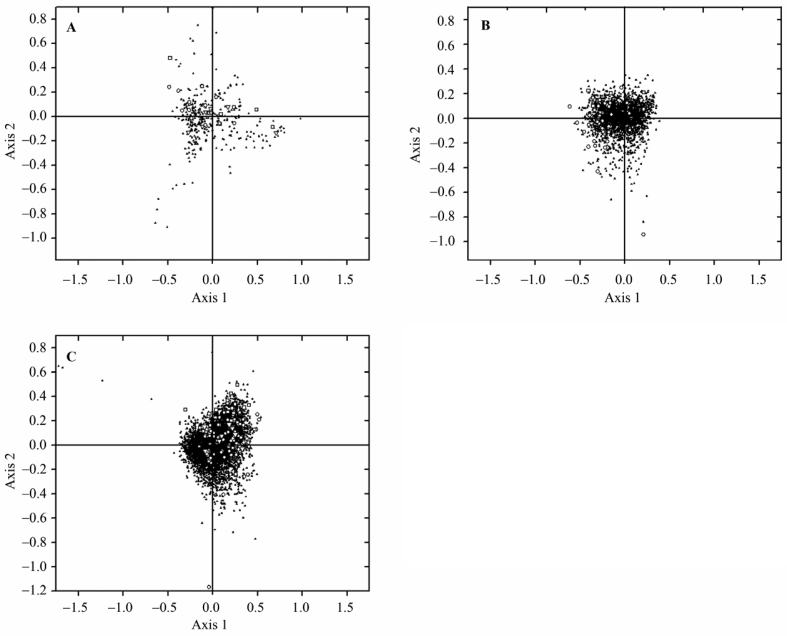
In all three species, codon adaptation index (CAI) and GC12 were common primary sources of intra-proteomic variations in amino acid usage (Table 2). It was also observed that GC3S and NC provide additional trends of variability in all three Leishhmania species. GT3S accounted for the variation on Axis 1 only in L. donovani. Variation on Axis 2 was determined by gravy and aromaticity for all three species. Observations from Axis 1–Axis 2 plots of COA on amino acid usage (Figure 3) showed that distribution of the HEGs in L. donovani (Figure 3A) overlapped with that of other genes. In L. infantum most of these genes lie on the left side of Axis 1 (Figure 3B). In the case of L. major, HEGs clustered at the right side of the Axis 1 (Figure 3C). Figure 2 (A and B) and Table 2 together suggest that the HEGs of L. donovani and L. infantum are characterized by relatively high GC12. However, GC12 was low in L. major, which was not expected because of the high GC content in L. major. This may be due to the effect of mutational pressure on L. major.
Figure 3.
Position of genes along the two major axes of variation in COA on RAAU. Position of genes along Axis 1 are plotted against Axis 2 by COA on RAAU in L. donovani (A), L. infantum (B) and L. major (C). HEG, VSG, and topoisomerase is represented by open circle, square and triangle, respectively. The rest of the genes (LEGs) are indicated by solid triangles. X and Y axis is represented by Axis 1 and Axis 2, respectively.
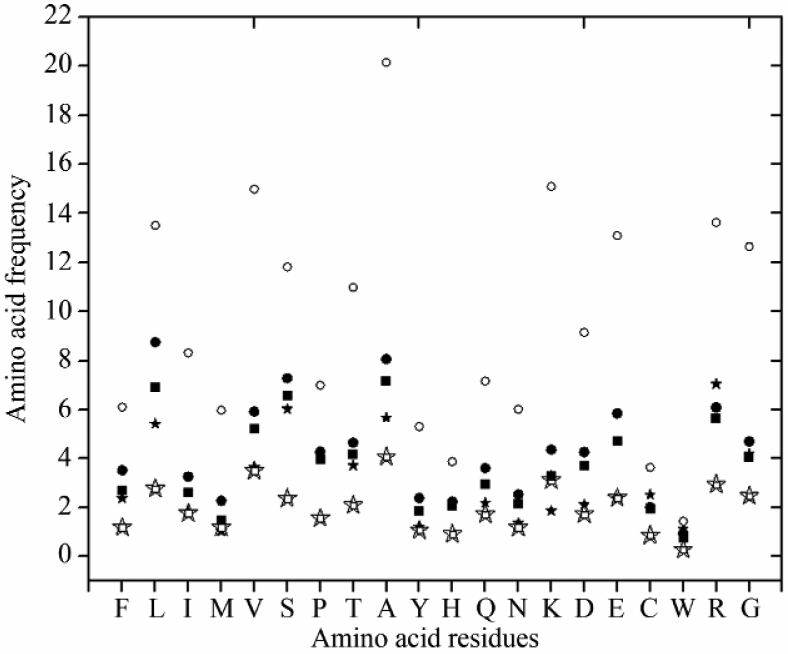
GC1 (G/C content at first codon sites) and GC2 (G/C content at first codon sites) of HEGs are similar in all three species (Table S2). GC1 and GC2 of HEGs in L. donovani are lower than those in LEGs in all species, which could be due to mutational bias in L. donovani, suggesting the higher AT content in LEGs in L. donovani. Figure 4 shows the average amino acid frequencies in proteins encoded by the HEGs and LEGs in the three parasites under study. The frequency of many amino acids differs in these two sets of genes in L. major and is distributed widely, whereas GC-rich codons are dominant in HEGs as compared to LEGs (Figure 4, open and solid circles). But this distribution is restricted to one extreme end in the case of L. donovani (Figure 4, open and solid stars) and L. infantum (Figure 4, open and solid squares). These data suggest that there is a major variation in selecting the codons for amino acids usage.
Figure 4.
Frequency of amino acid residues in proteins encoded by HEGs and LEGs in three Leishmania species. Open stars, squares and circles represent HEGs in L. donovani, L. infantum and L. major, respectively and solid stars, squares and circles indicate LEGs in L. donovani, L. infantum and L. major, respectively. The amino acid (single letter code) residues are showed on X axis and the frequency of amino acid occurring in proteins encoded by LEGs and HEGs is indicated on Y axis.
Conservation of HEGs
Estimation of dS (number of synonymous substitutions per synonymous sites) and dN (non-synonymous substitutions per non-synonymous site) on the orthologs of HEGs in L. donovani with L. infantum and L. major was performed to investigate the evolution of amino acid substitution. Pairwise alignment was done between the orthologs of HEGs of L. donovani–L. infantum and L. donovani–L. major, and the total numbers of synonymous substitutions and non-synonymous substitutions are calculated. Table 3 shows that dN is higher than dS in both the groups. It is noteworthy that the dS and dN values of L. donovani–L. major are lower, while the dN/dS ratio is higher than those of L. donovani–L. infantum. This means that L. infantum has deviated at the synonymous and non-synonymous codon positions at a much faster rate than L. major.
Table 3.
dS and dN in orthologs of HEGs
| Ortholog pairs | dS | dN | dN/dS |
|---|---|---|---|
| L. donovani–L. infantum | 0.094 | 0.12 | 1.27 |
| L. donovani–L. major | 0.056 | 0.074 | 1.32 |
Codon and amino acid usage analysis for homologous genes
According to COA on RSCU, Axis 1 accounts for 30.54%, 26.47% and 32.53% of the total variations due to GC3S and NC in three species (Table 4). On Axis 2, GT3S and aromaticity account for the major trends of variation. NC is correlated with Axis 1 positively in L. infantum but negatively in L. donovani and L. major, while an opposite trend was observed for the correlation between GC3S and Axis 1 in these three species, suggesting that the genes with G/C-ending codons are clustered on the right side but on the negative side in L. infantum due to negative correlation (Figure 5, Axis 1–Axis 2 plot of homologous genes). It has also been noted (Table 4) that NC is negatively correlated with Axis 1 in L. donovani and L. major, which may be due to the decrease in codon bias among the genes lying towards the right side of Axis 1. This high correlation suggests that directional mutational pressure is dominating for governing synonymous codon usage.
Table 4.
Major trends in synonymous codon usage of homologous genes in three Leishmania species as revealed by COA on codon usage of encoded proteins on Axis 1 and 2
| Organism | Axis 1 |
Axis 2 |
||||
|---|---|---|---|---|---|---|
| Total variability | Source of variation | Correlation coefficient (r)a | Total variability | Source of variation | Correlation coefficient (r)a | |
| L. donovani | 30.54% | NC | –0.797 | 7.74% | GT3S | 0.338 |
| GC3S | 0.982 | Aromaticity | 0.125 | |||
| L. infantum | 26.47% | NC | 0.891 | 8.13% | GT3S | 0.237 |
| GC3S | –0.958 | Aromaticity | –0.309 | |||
| Gravy | –0.408 | |||||
| L. major | 32.53% | NC | –0.761 | 6.95% | GT3S | –0.223 |
| GC3S | 0.976 | Aromaticity | –0.288 | |||
| Gravy | –0.221 | |||||
All correlations are significant at P<0.01.
Figure 5.
Position of homologous genes along the two major axes of variation in COA on RSCU. Position of homologous genes along Axis 1 were plotted against Axis 2 by COA on RSCU in L. donovani (A), L. infantum (B), and L. major (C). HEGs, VSG, and topoisomerase are represented by circle, square, and triangle, respectively. The rest of the genes (LEGs) are indicated by solid triangles.
NC–GC3S plot (Figure 6) indicates that HEGs constitute a single cluster, but VSG and topoisomerase genes demonstrate different codon usage pattern and are characterized by high NC and GC3S for L. infantum and L. major, while topoisomerase genes in L. donovani are distributed randomly (range 0.42- 0.86). COA on amino acid usage has been performed for teomic variability. Axis 1 accounts for 33.51%, 39.3% and 31.59% of total variation in the three species of Leishmania (Table 5). CAI is the common source of intra-proteomic variation in all species. GC12 accounts for the additional variation in L. donovani and L. infantum, while for L. major, gravy and aromaticity contribute to variation besides CAI. Variation on Axis 2 is determined by gravy and aromaticity in L. donovani and L. infantum, while for L. major, GC12 and GT3S were the main contributors for the intra-proteomic variation on Axis 2. In all three species, HEGs, when plotted on Axis 1–Axis 2 (Figure S1), were scattered, which was not expected because the average GC content of these species is high. This discrepancy may be due to the influence of mutational pressure.
Figure 6.
The NC plot of homologous genes in the three Leishmania species. NC is plotted against GC3S for L. donovani (A), L. infantum (B) and L. major genes (C). The solid line indicates the expected NC value if bias is due to GC3S alone. HEG, VSG, and topoisomerase is represented by open circle, square and triangle, respectively. The rest of the genes (LEGs) are indicated by solid triangles.
Table 5.
Major trends in synonymous codon usage of homologous genes in 3 Leishmania species as revealed by COA on amino acid usage of encoded proteins on Axis 1 and 2
| Organism | Axis 1 |
Axis 2 |
||||
|---|---|---|---|---|---|---|
| Total variability | Source of variation | Correlation coefficient (r)a | Total variability | Source of variation | Correlation coefficient (r)a | |
| L. donovani | 33.51% | CAI | 0.569 | 19.51% | Gravy | 0.629 |
| GC12 | –0.761 | Aromaticity | 0.751 | |||
| GT3S | 0.519 | |||||
| L. infantum | 39.3% | CAI | 0.478 | 8.13% | Aromaticity | 0.667 |
| GC12 | –0.557 | Gravy | 0.550 | |||
| L. major | 31.59% | CAI | 0.443 | 19.5% | GT3S | –0.460 |
| Gravy | 0.567 | Aromaticity | –0.242 | |||
| Aromaticity | 0.700 | GC12 | 0.715 | |||
All correlations are significant at P ≤ 0.01.
Discussion
The present study reveals the major trends involved in the selection of gene/protein composition of the three Leishmania species examined. The analysis of synonymous codon usage and amino acid variations shows that genomes of all the three Leishmania species are under mutational bias and translational selection.
In all three species, the lower GC12 and higher GC3S in HEGs as compared to LEGs suggest that the ancestor of the HEGs might have been relatively rich in AT-content. Previous studies have suggested a universal AT mutational bias, because many types of spontaneous mutations (e.g., the deamination of cytosine) cause GC to AT changes (18). This also suggests that the LEGs have evolved at a faster rate and become GC-rich. The lower dS and dN values in L. donovani–L. major than those in L. donovani–L. infantum suggests that the HEG dataset of L. donovani is evolutionarily much closer to L. major. The higher value of dN as compared to dS shows that synonymous positions are more conserved between L. donovani and L. major, and mutational bias plays a major role in shaping the composition of protein-coding genes. Additionally, optimal codons in all three Leishmania species are G/C-ending in HEGs but A/T-ending in LEGs. This supports the fact that translational selection works more strongly on synonymous sites of HEGs 19, 20, 21. As a result, the HEGs of these three species are characterized by low GC12 and high GC3S in comparison to the LEGs. The HEGs may further be explored to identify the essential genes, for example, by applying in silico subtracting genomic approach, and could be helpful in searching potential therapeutic drug targets for curing leishmaniasis.
Materials and Methods
Sequence dataset
Complete protein-coding gene sequences of L. infantum and L. major were extracted from the Sanger database (http://www.sanger.ac.uk/) while protein-coding sequences of L. donovani were obtained from NCBI (http://www.ncbi.nlm.nih.gov/), which contain 2,655, 9,159 and 368 (till April 30, 2011) protein-coding genes, respectively. To minimize sampling error, genes with less than 100 codons, internal stop codons, not-translatable codons, incomplete start and stop codons, and pseudogenes were excluded from the analysis. Therefore, finally 2,559 and 8,132 genes were included for analysis for L. infantum and L. major, respectively. No such filter was applied for L. donovani due to fewer gene sequences.
Homologs for L. donovani were searched using BLAST. For this purpose the E-value cut-off was set to e-100 and genes with E-value less than e-100 were considered as homologs. According to this criterion, a total of 341 genes from L. infantum and 340 genes from L. major were found as homologs for 347 genes from L. donovani.
Parameters used for identifying trends of variations
For each protein-coding gene under study, the following parameters were calculated, which include RSCU, RAAU, CAI, GC12, GC3S at synonymous codon sites excluding ATG for Met, TGG for Trp and stop codons, average hydropathy (22) and aromaticity (23) of the gene products.
Datasets of HEGs and LEGs
Datasets of putative HEGs and LEGs were obtained by taking genes from the two extreme ends of Axis 1 of COA on RSCU in all three parasites.
Statistical analyses
The program CodonW 1.1.4 (Peden, J., 1999. available at http://sourceforge.net/projects/codonw/) was used to analyze codon usage, COA (24), GC3S, RSCU (22), and CAI 14, 18. A 2×2 contingency table χ2 was used to detect the significant differences in codon and amino acid usage.
Estimation of non-synonymous and synonymous substitutions in HEGs
Orthologs for HEGs (genes lying at the one extreme end of Axis 1 of COA) of L. donovani were extracted using BLAST. The cut-off E-value for searching orthologs was set to e-50 so the homologs with E-value less than e-50 were considered as orthologs. Pairwise alignments between the orthologs and estimation of dS and dN were carried out using MEGA4 program (25).
Authors’ contributions
NC and RP were involved in this study on all aspects, contributed to the design of the project and wrote the manuscript. ASV performed synonymous/non-synonymous substitutions analysis. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
The authors are thankful to the Sub-Distributed Information Center (BTISnet SubDIC) and Department of Biotechnology, BIT, Mesra, Ranchi for their kind support.
Supplementary Material
Tables S1 and S2; Figure S1
References
- 1.World Health Organization . WHO Press; Geneva, Switzerland: 2010. Control of the leishmaniases: report of a meeting of the WHO Expert Committee on the Control of Leishmaniases. WHO technical report series (no. 949) [Google Scholar]
- 2.Minodier P., Parola P. Cutaneous leishmaniasis treatment. Travel Med. Infect. Dis. 2007;5:150–158. doi: 10.1016/j.tmaid.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Gibson M.E. The identification of kala-azar and the discovery of Leishmania donovani. Med. Hist. 1983;27:203–213. doi: 10.1017/s0025727300042691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desjeux P. Leishmaniasis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Singh S. New developments in diagnosis of leishmaniasis. Indian J. Med. Res. 2006;123:311–330. [PubMed] [Google Scholar]
- 6.Peacock C.S. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nature Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivens A.C. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayhoff M.O. Atlas of Protein Sequence and Structure. In: Hunt L.T., editor. Vol 5 Supplement 3. National Biomedical Research Foundation; Washington D.C, USA: 1978. [Google Scholar]
- 9.Levin D.B., Whittome B. Codon usage in nucleopolyhedroviruses. J. Gen. Virol. 2000;81:2313–2325. doi: 10.1099/0022-1317-81-9-2313. [DOI] [PubMed] [Google Scholar]
- 10.Grantham R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elton B. Doublet frequencies and codon weighting in the DNA of Escherichia coli. J. Mol. Evol. 1976;8:117–135. doi: 10.1007/BF01739098. [DOI] [PubMed] [Google Scholar]
- 12.Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 14.Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhuri G. Surface acid proteinase (gp63) of Leishmania mexicana. A metalloenzyme capable of protecting of liposome-encapsulated proteins from phagolysosomal degradation by macrophages. J. Biol. Chem. 1989;264:7483–7489. [PubMed] [Google Scholar]
- 16.Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;6:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 17.Cheesman S.J. The topoisomerases of protozoan parasites. Parasitol. Today. 2000;7:277–281. doi: 10.1016/s0169-4758(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 18.Sharp P.M., Li W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birdsell J.A. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 20.Iida K., Akashi H. A test of translational selection at ‘silent’ sites in the human genome: base composition comparisons in alternatively spliced genes. Gene. 2000;261:93–105. doi: 10.1016/s0378-1119(00)00482-0. [DOI] [PubMed] [Google Scholar]
- 21.Lafay B. Absence of translationally selected synonymous codon usage bias in Helicobacter pylori. Microbiology. 2000;146:851–860. doi: 10.1099/00221287-146-4-851. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Lobry J.R., Gautier C. Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res. 1994;22:3174–3180. doi: 10.1093/nar/22.15.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenacre M.J. Academic Press; New York, USA: 1984. Theory and Applications of Correspondence Analysis. [Google Scholar]
- 25.Tamura K. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2; Figure S1