Some 15 years ago, Gane (now Dr. Gane Ka-Shu Wong, Professor and iCORE Chair in Biosystems Informatics, University of Alberta, Canada) and I were staring at a set of plots and scratching our heads, wondering why there was a negative GC-content gradient when we aligned human transcripts from 5′ to 3′ to the genome. Until we had published several papers on other more interesting issues based on analyses of human genome sequences and variations [1], [2], [3] and found the same phenomenon from the rice genes a few years later [4], [5], had we realized the importance of this nearly universal feature albeit variable from bacteria to human [6]. Thinking along the line, we also did another exploratory experiment at University of Washington, taking the advantage of the Environmental Genome Project supported by NIH’s National Institute of Environmental Health Sciences, to re-sequence a couple hundred genes, of course including some interesting introns, especially those that are small in size (a median of 78 bp; also called minimal intron). The effort led to a realization of natural selection on functional sequence elements [7] in addition to just protein-coding sequences that can be evaluated with different methods [8], [9], [10].
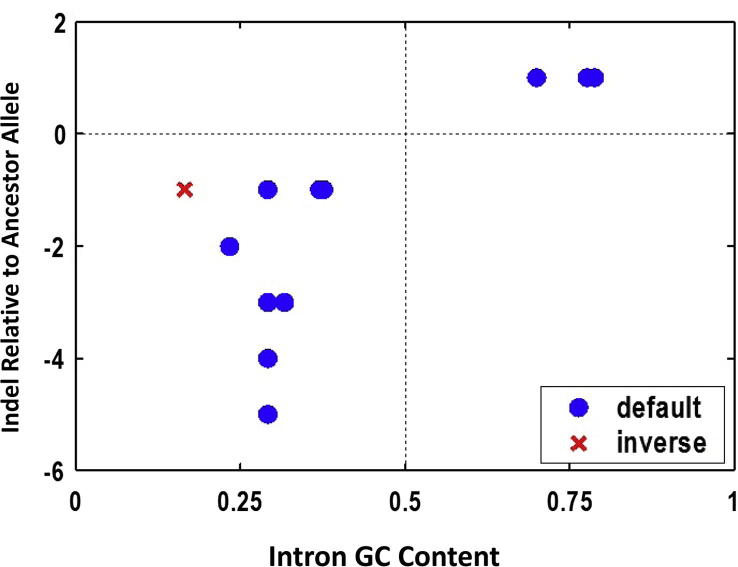
However, there were two pieces of the puzzles for which we did not have explanations at the time. One was the relatedness of GC content to indels found in the minimal introns (Figure 1; see the figure legend for more details) and the other was the GC gradient at the 3′-end, albeit weaker as compared to that of the 5′ end. Thanks to several of my hard working graduate students, as Gane and I joked some decade ago—let us leave these enchanting projects to our future graduate students—when we were limited by manpower for new initiatives. We are now getting very close to understanding both [6], [11], [12], [13], [14], [15], [16], [17]. The two examples are just “the tip of the iceberg” of other dimensions of gene regulation that leaves sequence signatures in the genome sequence in the context of populations and lineages.
Figure 1.
Insertion-deletion (indel) relative to the major allele as a function of GC content. For a total of 12 (10 rare alleles and 2 common alleles; for more details please see reference 7) indels, the partition of insertions (above the horizontal dashed line) and deletions (below the horizontal dashed line) into GC-rich and GC-poor minimal introns is almost absolute other than a single exception (red; the major allele is a deletion rather than an insertion). The rule that GC-rich and GC-poor minimal introns tend to have more insertions and more deletions, respectively, is by and large correct for human minimal introns [9].
The challenges are multifold and we can only discuss a few examples here. First, the far biggest challenge is how to evaluate transcript-centric mutations that usually behave differently among species and lineages, such as GC-rich (vertebrates and grasses) and GC-poor (most unicellular organisms) genomes [4], [15]. Transcripts can be defined as the sole component of the gene-space and contain both protein-coding exons and non-coding introns; they comprise either the greater majority (over 90%) in animal genomes or variable fractions in plant genomes (from 50% in the rice genome and less than 10% in the wheat or barley genomes). The signature at the nucleotide composition level for transcript-centric mutations often exhibits as a GC-content gradient that shows uneven mutation rates along the length of transcripts as opposed to replication-centric mutations that are relatively evenly distributed over the entire genome [6], [15]. Second, at the gene structural level, an optimal size for the minimal intron is another example, and only one type of the variations, short indels, are sensitive to natural selection [7], [11]. Third, at the gene organization level, we know that most of vertebrate genes are in fact organized as clusters rather than distributed stochastically [13], [18], [19]. Some may form tighter clusters and other may break out easily over time in different lineages. And a significant fraction of them may be regulated in some unique ways, such as in circadian rhythms. Fourth, regardless of what is the fraction of the protein-coding sequences in a given genome, the rest is left alone without legitimate and systematic ways to be evaluated within a neo-Darwinian framework. And this significant rest is often over 98% of the mammalian genomes and 90–99% of the plant genomes. Thousands of transcripts, not encoding proteins, arise from it [20]; large number of uncharacterized chromosomal sequence elements are “hidden treasures” for further exploitations [21], [22]; and most of all, the historic relics of all genomes are buried in it.
On the one hand, if long-term natural selection is constantly fixing everything that includes beneficial and weakly deleterious mutations to a genome, the genome sequence and its variations carried by a population of the species must have been leaving some kinds of recognizable sequence signatures to be scrutinized as we are armed with powerful and efficient sequencing tools. On the other hand, the more the sequences are functional, the more obvious the sequence signatures should be. Therefore, we have at least two goals here: one is to identify the potentially functional sequence elements and the other is to recognize the sequence signatures as well as the beneficial and weakly deleterious variations for the assessment of their functional implications or molecular mechanisms within or among organisms and lineages.
There is one complication for us to think deeply—we need to be able to predict the nature of the variations in a signature. It may be protein-coding or non-coding; it must function as genetic or epigenetic alleles at a minimum; and the nature of such ascertainments is certainly data-intensive and statistical. Therefore, we can think of two “tracks”, where exon and intron sequences, transcribed and non-transcribed genome segments, informational and operational RNAs, genetic and epigenetic mechanisms, or Darwinian and Lamarckian interpretations are all distinguished. Although there are certainly overlaps in terms of mechanisms between the two tracks, we can at least use the narrower definitions first. Of course, what in the informational track has been well studied and regarded as the common dogma. However, what in the operational track is apparently not yet formulated let alone fully exploited.
To define the operational track, we need to employ a framework; it does not have to be Lamarckian in the classic sense but has to be non-genetic or epigenetic. We are not entombing older dogma but seeking new thoughts and new lines of evidence to solve our yet more puzzling biology and its many unsolved mysteries. We need new paradigms and new concepts more than ever. In a way, we are actually going to make links between the Darwinian and the Lamarckian frameworks at molecular level while we are making distinctions between the two. For instance, we need to think about how genetic defects or benefits of a molecular mechanism, such as splicing (especially that for the spliceosomal introns, which seems to be abandoned and altered many times to different extents in the history of genome evolution), which are not measurable by any molecular clock types of methodology, are ascertained. Nevertheless, we have to move ahead and jump over theoretical hurdles.
Box 1. The common dogma.
The common dogma refers to a set of doctrines (or principles) that most scientists actually believe based on incomplete data, often followed by over-interpretations, which may not be all correct—as it may turn out in the future science—and some are certainly wrong even when the believers are actively defending it.
I used it as a rather negative sense here even though the scientific prophet Francis Crick used it in a positive sense first but it was still rejected by and large because of the discovery of the RNA world albeit himself being part of it [23].
In his autobiography, What Mad Pursuit, Francis Crick wrote about his choice of the word dogma [24]: “I called this idea the central dogma, for two reasons, I suspect. I had already used the obvious word hypothesis in the sequence hypothesis, and in addition I wanted to suggest that this new assumption was more central and more powerful. ... As it turned out, the use of the word dogma caused almost more trouble than it was worth.... Many years later Jacques Monod pointed out to me that I did not appear to understand the correct use of the word dogma, which is a belief that cannot be doubted. I did apprehend this in a vague sort of way but since I thought that all religious beliefs were without foundation, I used the word the way I myself thought about it, not as most of the world does, and simply applied it to a grand hypothesis that, however plausible, had little direct experimental support.”
References
- 1.Yang Z., Wong G.K., Eberle M.A., Kibukawa M., Passey D.A., Hughes W.R. Sampling SNPs. Nat Genet. 2000;26:13–14. doi: 10.1038/79113. [DOI] [PubMed] [Google Scholar]
- 2.Wong G.K., Passey D.A., Huang Y., Yang Z., Yu J. Is “junk” DNA mostly intron DNA? Genome Res. 2000;10:1672–1678. doi: 10.1101/gr.148900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong G.K., Passey D.A., Yu J. Most of the human genome is transcribed. Genome Res. 2001;11:1975–1977. doi: 10.1101/gr.202401. [DOI] [PubMed] [Google Scholar]
- 4.Wong G.K., Wang J., Passey D.A., Yu J. Codon-usage gradients in Gramineae genomes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Zhang J., Li R., Zheng H., Li J., Zhang Y. Evolutionary transients in the rice transcriptome. Genomics Proteomics Bioinformatics. 2010;8:223–228. doi: 10.1016/S1672-0229(10)60023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui P., Lin Q., Ding F., Hu S., Yu J. Transcript-centric mutations in human genomes. Genomics Proteomics Bioinformatics. 2012;10:11–22. doi: 10.1016/S1672-0229(11)60029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J., Yang Z., Kibukawa M., Paddock M., Passey D.A., Wong G.K. Minimal introns are not “junk”. Genome Res. 2002;12:1185–1189. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Yu J. Evaluation of six methods for estimation synonymous and nonsynonymous substitution rates. Genomics Proteomics Bioinformatics. 2006;4:173–181. doi: 10.1016/S1672-0229(06)60030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Li J., Zhao X., Wang J., Wong G.K., Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Yu J. Both size and GC-content of minimal introns are selected in human population. PLoS One. 2011;6:e17945. doi: 10.1371/journal.pone.0017945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J., He F., Wang D., Liu K., Huang D., Xiao J. A novel role for minimal introns: routing mRNAs to the cytosol. PLoS One. 2010;5:e10144. doi: 10.1371/journal.pone.0010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui P., Liu W., Zhao Y., Lin Q., Ding F., Xin C. The association between H3K4me3 and antisense transcription. Genomics Proteomics Bioinformatics. 2012;10:74–81. doi: 10.1016/j.gpb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui P., Ding F., Zhang L., Hu S., Yu J. Replication and transcription contribute differently in mutation rates of human genome. Genomics Proteomics Bioinformatics. 2012;10:4–10. doi: 10.1016/S1672-0229(11)60028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K., Meng Q., Ma L., Liu Q., Tang P., Chiu C. A novel DNA sequence periodicity decodes nucleosome positioning. Nucleic Acids Res. 2008;36:6228–6236. doi: 10.1093/nar/gkn626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Wang L., Yang M., Liu J., Xin C., Hu S. Sequence signatures of nucleosome positioning in Caenorhabditis elegans. Genomics Proteomics Bioinformatics. 2010;8:92–102. doi: 10.1016/S1672-0229(10)60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui P., Zhang L., Lin Q., Ding F., Xin C., Fang X. A novel mechanism of epigenetic regulation: nucleosome-space occupancy. Biochem Biophys Res Commun. 2010;391:884–889. doi: 10.1016/j.bbrc.2009.11.157. [DOI] [PubMed] [Google Scholar]
- 18.Cui P., Lin Q., Zhang L., Ding F., Xin C., Zhang D. The disequilibrium of nucleosomes distribution along chromosomes plays a functional and evolutionarily role in regulating gene expression. PLoS One. 2011;6:e23219. doi: 10.1371/journal.pone.0023219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L., Yu J. A comparative analysis of divergently-paired genes (DPGs) of Drosophila and vertebrate genomes. BMC Evol Biol. 2009;9:55. doi: 10.1186/1471-2148-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D., Zhang Y., Fan Z., Liu G., Yu J. LCGbase: a comprehensive database for lineage-based co-regulated genes. Evol Bioinform Online. 2012;8:39–46. doi: 10.4137/EBO.S8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W., Zhao Y., Cui P., Lin Q., Ding F., Xin C. Thousands of novel transcripts identified in mouse cerebrum, testis, and ES cells based on ribo-minus RNA sequencing. Front Genet. 2011;2:93. doi: 10.3389/fgene.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui P., Liu W., Zhao Y., Lin Q., Zhang D., Ding F. Comparative analyses of H3K4 and H3K27 trimethylations between the mouse cerebrum and testis. Genomics Proteomics Bioinformatics. 2012;10:82–93. doi: 10.1016/j.gpb.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cech TR. The RNA worlds in context. Cold Spring Harb Perspect Biol 2011. http://dx.doi.org/10.1101/cshperspect.a006742. [DOI] [PMC free article] [PubMed]
- 24.Crick F. Basic Books; New York: 1988. What mad pursuit: a personal view of scientific discovery. [Google Scholar]