Abstract
Identification of proteins by mass spectrometry (MS) is an essential step in proteomic studies and is typically accomplished by either peptide mass fingerprinting (PMF) or amino acid sequencing of the peptide. Although sequence information from MS/MS analysis can be used to validate PMF-based protein identification, it may not be practical when analyzing a large number of proteins and when high-throughput MS/MS instrumentation is not readily available. At present, a vast majority of proteomic studies employ PMF. However, there are huge disparities in criteria used to identify proteins using PMF. Therefore, to reduce incorrect protein identification using PMF, and also to increase confidence in PMF-based protein identification without accompanying MS/MS analysis, definitive guiding principles are essential. To this end, we propose a value-based scoring system that provides guidance on evaluating when PMF-based protein identification can be deemed sufficient without accompanying amino acid sequence data from MS/MS analysis.
Key words: peptide mass fingerprinting, Mowse, Mascot, ProFound, proteomics
Introduction
Protein identification using mass spectrometry (MS) is an essential step in studies that employ proteomic methods such as two-dimensional electrophoresis (2-DE), and is typically accomplished by either peptide mass fingerprinting (PMF) or amino acid sequencing of the peptide using tandem mass spectrometry (MS/MS). In PMF analysis, proteolytic cleavage using an enzyme such as trypsin results in a collection of peptides, which serves as a unique identifier or fingerprint of the protein. The accurate determination of peptide mass-to-charge (m/z) ratio, typically using a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer, allows identification of the unknown protein by matching the resulting peptide masses with the theoretical peptide masses of proteins in a database (such as NCBI and Swiss-Prot). For peptide sequence analysis, peptides obtained by proteolytic cleavage are subjected to MS/MS to fragment the peptide along the amide backbone. The amino acid sequence of the peptide is then obtained from the differences in m/z ratios for a series of daughter ions. Subsequently, the sequence ions and the intact peptide masses are matched against protein databases to identify the unknown protein.
To determine the prevalence of PMF analysis as well as the type of search algorithms employed in protein identification, we surveyed articles published in Proteomics (August 2005 to July 2007) and Proteome Science (January 2003 to September 2007). Out of the 581 articles surveyed, approximately 35% of the studies only used PMF-based protein identification, 32% of the studies only utilized MS/MS, and 33% of the studies used both PMF and MS/MS for protein identification. Thus, 68% of the studies utilized PMF for protein identification.
Although sequence information from MS/MS analysis can be used to validate PMF-based protein identification, it may not be practical when analyzing a large number of proteins and when high-throughput MS/MS instrumentation is not readily available. Besides, in comparison to MS/MS analysis on a single peptide, identification of proteins using PMF has two significant advantages that are often ignored. Firstly, in PMF, unsuspected post-translational modifications lead to only a marginal loss in the quality of data and do not affect the outcome. In contrast, in MS/MS, unspecified post-translational modifications can adversely affect the matching and scoring process, thus precluding unambiguous protein identification. Secondly, although MS/MS analysis can be used to determine the amino acid sequence of a peptide, that peptide may be unique to a particular protein or common to a number of different proteins (for example, enzymes from the same family). Conversely, the use of multiple peptides for protein identification in PMF allows more extensive coverage of the protein, thereby increasing the confidence in a positive identification. Thus, in some cases the MS/MS analysis on a single peptide may actually be less specific than PMF.
A pertinent question when using PMF for protein identification is that: at what point does one use MS/MS analysis for protein identification? A commonly used approach is to use PMF as an initial step, and then use MS/MS to corroborate proteins that are considered ambiguous. However, this brings up an interesting question: when can one conclude protein identification using PMF to be “unambiguous”? The traditional definition of “unambiguous” (having or exhibiting no ambiguity or uncertainty) can hardly be applied to PMF-based protein identification, which is probability based and there always remains a chance of obtaining a false positive protein match. So when used in the context of protein identification, a pragmatic definition of “unambiguous” would be “having or exhibiting little ambiguity or uncertainty”. Without accompanying amino acid sequence data, can one increase the confidence in protein identification using PMF data? Western blotting can be used to confirm protein identification; however, this technique is not high-throughput, limited by the availability of antibodies, and impractical for analyzing large number of samples.
Presently, there are huge disparities in criteria used to identify proteins using PMF. Plomion et al. (1) selected a molecular weight search (Mowse) score of 71 or more as significant for a protein match, while Hoffrogge et al. (2) used a Mowse score of greater than 69 as significant. On the other hand, it has been suggested that the Mowse score should be 50 more than the significant threshold level for protein identification (3). Because the Mowse score depends on the number of sequences in a database, for species with small database sizes this approach would not be appropriate. Naranjo et al. (4) categorized a match as successful if a protein had a score greater than the significant threshold of the Mascot algorithm (5., 6.; www.matrixscience.com) and was the top match in both Mascot and ProFound 7., 8. algorithms. Whereas proteins with a Mascot score of at least 95 or a ProFound score of 2.2 have been considered significant by others (9). Also, Guipaud et al. (10) and Sinclair et al. (11), in addition to the significant scores of protein algorithms, incorporated sequence coverage, number of peptides matched, and the congruence of isoelectric point (pI) and molecular mass of the protein with the sequence database in the evaluation of PMF data. Consequently, definitive guidelines for what constitutes a positive match are lacking.
To this end, we propose a value-based scoring system that provides guidance on evaluating when PMF-based protein identification can be deemed sufficient without accompanying amino acid sequence data from MS/MS analysis. To construct this value-based scoring system, we have used parameters that are considered important in substantiating protein identification, such as congruence of the observed pI and molecular mass with the protein sequence database, percent sequence coverage (percentage of the theoretical protein that is covered by the experimental peptide masses), number of peptides matched, and the matching scores from two different protein search engines.
Protein search algorithms
Since PMF-based protein identification is probability-based, the choice of protein search algorithms is of paramount importance. To date, thirteen different PMF search algorithms have been reported (12). There are several ways that these protein search engines differ from one another, but the fundamental difference is in the scoring algorithm that is employed. Some of the protein search engines calculate the probability of obtaining an incorrect match and report significance threshold scores to increase the confidence in a protein match, whereas others just rank possible protein matches without reporting a significance level. In our sampling of the literature, Mascot was the most commonly used protein search engine (67%), followed by MS-Fit (13) (14%) and ProFound (12%). Mascot and MS-Fit implement a probability-based scoring approach using Mowse; however, while Mascot reports scores that correspond to a 5% significance level, MS-Fit does not provide a significance level for its protein matches. ProFound, on the other hand, uses the Bayesian probability and reports a significance threshold score (Z score). Incidentally, most studies that employed Ms-Fit or ProFound also used Mascot for database searching.
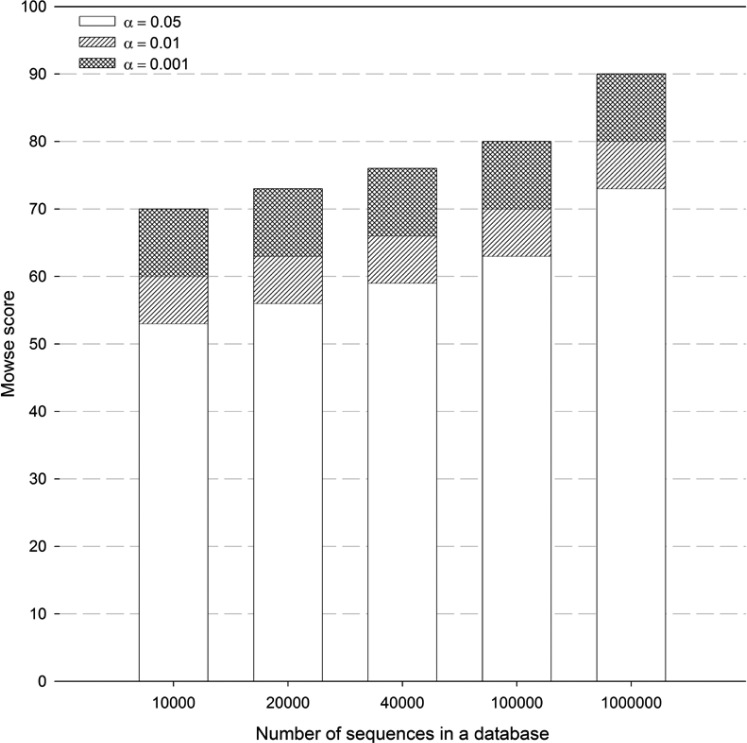
The threshold Mowse score in Mascot is reported as —10 lg(P), where P (α value/number of database sequences) is the probability that the observed match is a random event 5., 6.. Accordingly, the threshold Mowse score depends on two parameters: the number of sequences in a database and the α value used for searches. Typically, protein searches are carried out with an α value of 0.05. We plotted the Mowse scores for a number of different theoretical database sequences, ranging from 10,000 to 1,000,000 at various α values (0.05, 0.01, and 0.001) (Figure 1). Since the significance threshold score depends on the number of sequences in the database, as the number of protein sequences increase, the threshold would also increase. This issue is of greater relevance when it comes to the identification of proteins from organisms whose sequences have not been completely determined. Also, post-translational processing of proteins would increase the number of protein sequences in the databases, even for completely sequenced genomes. For instance, if the searched database contains 20,000 sequences, a protein match would be considered positive at the default α value of 0.05 when the Mowse score exceeds the threshold of 56. However, this protein match would no longer be positive if the number of sequences in the database were to increase. On the other hand, if the search was performed at a lower α value (for example 0.01), the protein match would still retain its significance (at an α of 0.05) even if there was a five-fold increase in the number of sequences in the future. Therefore, based on the distribution profile of threshold Mowse scores (Figure 1), we suggest performing PMF searches in Mascot using an α value of 0.01, as opposed to the default α value of 0.05. For organisms whose complete sequence information is not available, it would be better to carry out and report protein searches at an α value of 0.001. This ensures that the matching would still be significant at an α value of 0.05, even if there was a vast increase in the number of sequences in the future (Figure 1). Furthermore, a decrease in α value decreases the risk of Type 1 error (α error) associated with the search, and thus decreases the incidence of false protein identification with PMF. Although this would increase the Type 2 error (β error) resulting in an increase of false negatives (number of proteins that cannot be identified), we feel it is preferable to be highly averse to making a Type 1 error as the goal is to correctly identify proteins.
Fig. 1.
Effect of α value and the number of sequences in a database on the distribution of threshold Mowse scores. The threshold Mowse score was calculated as —10 lg(P), where P is α value/number of sequences in the database. A protein match above a particular Mowse threshold score indicates that the match is less likely to be random and is likely to be significant. The bars represent the threshold Mowse scores for theoretical database sequences at different α values (0.05, 0.01, and 0.001).
Unlike the Mowse score used in Mascot, the significance threshold of ProFound, Z score, is constant and is independent of the number of sequences available in the database. The Z score is used as a measure of probability of a random protein match and represents the distance of a sample from the mean in units of standard deviation. A Z score of 1.65 (α = 0.05, one tail) or lower signifies that the protein match is likely to be random and a score greater than 1.65 indicates that the protein match is significant 7., 8.. We recommend using two different protein search engines that employ dissimilar algorithms for protein identification as this should reduce the likelihood of false protein identification. As reporting of significance threshold scores help to assess the quality of a protein match, we suggest the use of Mascot and ProFound for PMF searches.
In addition to the scoring of protein matching with algorithms such as Mascot and Profound, other factors that assist in substantiating protein identification include the percent sequence coverage and the number of peptides matched. Despite the expected variation in percent coverage based on the size of the protein (the sequence coverage for large proteins tends to be less and vice versa), studies have suggested sequence coverage of at least 20% 14., 15.. Based on our observation as well as prior reports 8., 16., 17., we suggest a minimum of four peptides to be matched for positive protein identification with PMF.
Value-based scoring system
We have transformed the commonly used parameters (pI, molecular mass, percent sequence coverage, number of peptides matched, and significance scores of Mascot and ProFound) in PMF into a value-based scoring system for an objective evaluation of protein identification using PMF (Table 1). With this scoring system, a combined score of 15 or more can be considered sufficient for the protein match using PMF to be unequivocal without the need for subsequent MS/MS analysis. The threshold of 15 was chosen by taking the following into account: congruence of pI and molecular mass (4 points), sequence coverage of 20% or more (3 points), minimum of four peptides matched (2 points), and protein match with algorithms Mascot and ProFound at an α value of 0.01 (6 points). We have gone through an iterative procedure to score the parameters according to their degree of relevance in PMF-based protein identification. The value-based system was derived based on the criteria that are commonly used in the literature and also on our personal experience in protein identification. Though protein algorithms Mascot and ProFound were used to construct the scoring system, any comparable protein search algorithms can be substituted.
Table 1.
Value-based scoring for PMF
| Parameter | Score | |
|---|---|---|
| pI | 2 | |
| Molecular mass | 2 | |
| Sequence Coverage (%) | ||
| 10%–19% | 2 | |
| 20%–29% | 3 | |
| 30%–39% | 4 | |
| 40%–49% | 5 | |
| 50% or more | 6 | |
| Number of peptides matched | ||
| 4–7 | 2 | |
| 8–11 | 3 | |
| 12–15 | 4 | |
| 16–19 | 5 | |
| 20 or more | 6 | |
| Matching of protein algorithms | Mascot | ProFound |
| Top match (not significant) | 1 | 1 |
| Significant match (α = 0.05) | 2 | 2 |
| Significant match (α = 0.01) | 3 | 3 |
| Significant match (α = 0.001) | 4 | 4 |
While being conservative, the scoring system is also objective and flexible. For instance, protein modifications can alter the observed pI (for example, deamidation) as well as the molecular mass. However, this would not preclude an unequivocal protein determination, but rather the absence of information on pI or molecular weight can be offset by an increase in the percent sequence coverage, number of peptides matched, or use of a more stringent threshold score for Mascot and ProFound. It should be noted that this scoring system cannot substitute for the care undertaken in sample preparation and 2-DE analysis. Rather, it provides guidance on evaluating the need for a subsequent MS/MS analysis when using PMF.
One of the possible drawbacks of this value-based scoring system is that it is conservative. Therefore, while decreasing the number of false positives, it would also increase the number of false negatives (proteins that are changed but not identified). That is, with this scoring system, a score of 15 or more would be considered positive. However, in reality a protein match with a score of 14 or less can still be the correct match. Nevertheless, as the goal is to increase confidence in PMF-based protein identification without the accompanying need for sequence analysis, we feel it is preferable to be Type 1 error averse. Besides, the number of peptides matched (as well as sequence coverage) would vary depending on protein size. Since the scoring system recommends matching a minimum of four peptides, it can affect the identification of small sized proteins. However, the lack of a recommended minimum number of peptides can be offset by other parameters (pH, MW, and the significant scores of protein search engines), and thus should not affect the identification of most proteins.
In addition to the factors discussed, for reliable protein identification using PMF, the search parameters (such as error tolerance and the number of missed cleavages) that are employed should also be optimized; however, an elaborate discussion on these parameters is beyond the scope of the present study and readers are referred to germane articles 8., 18.. Though the levels of error tolerance vary across studies, a value of 100 or less is desirable. For the number of missed cleavages, a value of 1 should be optimal, although a lot of studies have used 2 missed cleavages. With regard to protein modifications, most studies employ carbamidomethylation of cysteine as a fixed modification and oxidation of methionine as a partial modification 2., 16., 17., 19..
Presently, the majority of proteomic studies employ PMF. Therefore, to reduce incorrect protein identification using PMF, and also to increase confidence in PMF-based protein identification without accompanying MS/MS analysis, definitive guiding principles are essential. The value-based scoring system proposed in this study is objective, flexible, and provides preliminary direction on evaluating when PMF-based protein identification can be considered sufficient without a subsequent need for amino acid sequence analysis.
References
- 1.Plomion C. Mapping the proteome of poplar and application to the discovery of droughtstress responsive proteins. Proteomics. 2006;6:6509–6527. doi: 10.1002/pmic.200600362. [DOI] [PubMed] [Google Scholar]
- 2.Hoffrogge R. 2-DE profiling of GDNF overexpression-related proteome changes in differentiating ST14A rat progenitor cells. Proteomics. 2007;7:33–46. doi: 10.1002/pmic.200600614. [DOI] [PubMed] [Google Scholar]
- 3.Li Z. Proteomic analysis of the E2F1 response in p53-negative cancer cells: new aspects in the regulation of cell survival and death. Proteomics. 2006;6:5735–5745. doi: 10.1002/pmic.200600290. [DOI] [PubMed] [Google Scholar]
- 4.Naranjo V. Proteomic and transcriptomic analyses of differential stress/inflammatory responses in mandibular lymph nodes and oropharyngeal tonsils of European wild boars naturally infected with Mycobacterium bovis. Proteomics. 2007;7:220–231. doi: 10.1002/pmic.200600527. [DOI] [PubMed] [Google Scholar]
- 5.Pappin D.J. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 6.Perkins D.N. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Chait B.T. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 8.Chamrad D.C. Evaluation of algorithms for protein identification from sequence databases using mass spectrometry data. Proteomics. 2004;4:619–628. doi: 10.1002/pmic.200300612. [DOI] [PubMed] [Google Scholar]
- 9.Wöhlbrand L. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics. 2007;7:2222–2239. doi: 10.1002/pmic.200600987. [DOI] [PubMed] [Google Scholar]
- 10.Guipaud O. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair J. Proteomic response of Schizosaccharomyces pombe to static and oscillating extremely low-frequency electromagnetic fields. Proteomics. 2006;6:4755–4764. doi: 10.1002/pmic.200500861. [DOI] [PubMed] [Google Scholar]
- 12.Shadforth I. Protein and peptide identification algorithms using MS for use in high-throughput, automated pipelines. Proteomics. 2005;5:4082–4095. doi: 10.1002/pmic.200402091. [DOI] [PubMed] [Google Scholar]
- 13.Clauser K.R. Role of accurate mass measurement (± 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 14.Biron D.G. The pitfalls of proteomics experiments without the correct use of bioinformatics tools. Proteomics. 2006;6:5577–5596. doi: 10.1002/pmic.200600223. [DOI] [PubMed] [Google Scholar]
- 15.Barrett J. Analysing proteomic data. Int. J. Parasitol. 2005;35:543–553. doi: 10.1016/j.ijpara.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S.K. Proteomic approach for identification and characterization of novel immunostimulatory proteins from soluble antigens of Leishmania donovani promastigotes. Proteomics. 2007;7:816–823. doi: 10.1002/pmic.200600725. [DOI] [PubMed] [Google Scholar]
- 17.Wu M. Proteome analysis of human androgen-independent prostate cancer cell lines: variable metastatic potentials correlated with vimentin expression. Proteomics. 2007;7:1973–1983. doi: 10.1002/pmic.200600643. [DOI] [PubMed] [Google Scholar]
- 18.Ossipova E. Optimizing search conditions for the mass fingerprint-based identification of proteins. Proteomics. 2006;6:2079–2085. doi: 10.1002/pmic.200500484. [DOI] [PubMed] [Google Scholar]
- 19.Inberg A. Cellular processes underlying maturation of P19 neurons: changes in protein folding regimen and cytoskeleton organization. Proteomics. 2007;7:910–920. doi: 10.1002/pmic.200600547. [DOI] [PubMed] [Google Scholar]