Abstract
Whole genome sequencing of buffalo is yet to be completed, and in the near future it may not be possible to identify an exome (coding region of genome) through bioinformatics for designing probes to capture it. In the present study, we employed in solution hybridization to sequence tissue specific temporal exomes (TST exome) in buffalo. We utilized cDNA prepared from buffalo muscle tissue as a probe to capture TST exomes from the buffalo genome. This resulted in a prominent reduction of repeat sequences (up to 40%) and an enrichment of coding sequences (up to 60%). Enriched targets were sequenced on a 454 pyro-sequencing platform, generating 101,244 reads containing 24,127,779 high quality bases. The data revealed 40,100 variations, of which 403 were indels and 39,218 SNPs containing 195 nonsynonymous candidate SNPs in protein-coding regions. The study has indicated that 80% of the total genes identified from capture data were expressed in muscle tissue. The present study is the first of its kind to sequence TST exomes captured by use of cDNA molecules for SNPs found in the coding region without any prior sequence information of targeted molecules.
Keywords: Hybridization; Exome; SNP, Bubalus bubalis, Temporal gene expression; 454 Sequencing
Introduction
Efficient functioning of a whole multicellular organism involves complex but specific interactions between different tissues of the body in a timely manner. A number of genes are turned off and on at particular points of time in response to various physiological processes. To fully understand how interactions between genes expressed in different tissues result in disease, increased milk or meat production or any other physiological response, it is important to study temporal gene expression in the tissues involved at different times/stages of the physiological process in question. Sequencing of the coding part of the genome (exome) helps in identifying variations which give rise to phenotypic variation. Collection of all the genes expressed in a single tissue at a specific point of time can be denoted as tissue specific temporal exome (TST exome).
Massively parallel sequencing technologies available currently are rapidly enhancing existing knowledge about the genomes of various organisms. Nonetheless, progress in genome sequencing efforts for buffalo is very limited. Buffalo is an important farm animal for dairying and agriculture particularly in Asia and contributes nearly 55% of total bovine milk production in India. The buffalo genome sequencing is expected to be completed in the near future (http://www.ncbi.nlm.nih.gov/projects/genome/guide/buffalo/). Up till now, only a total of 66,935 nucleotide sequences for the water buffalo (Bubalus bubalis) have been deposited in the GenBank database [1], out of which 64,212 genome shotgun sequences (ACZF01000001-ACZF01064212) were generated by our group. Recently, researchers from the National Bureau of Animal Genetic Resources, Karnal, India have published the first whole genome sequence assembly of water buffalo with a read depth of 17–19× and ∼91–95% coverage in comparison to the cattle assembly Btau 4.0 [2].
Buffalo whole genome sequencing and re-sequencing is almost cost prohibitive for a single institution. Therefore, targeted sequencing of selected genomic regions is the best alternative for characterization of SNPs [3], genome-wide association studies [4] and re-sequencing specific exons believed to be involved in specific disease processes and many others [5]. Of these, the re-sequencing of specific disease-associated exons seems very promising as it consists of less than 1% of total genome size [6] and is also functionally important. However, it is necessary to have knowledge of completely sequenced exomes in a species for exome re-sequencing. In the absence of complete genomic information regarding water buffalo, this approach may not be applicable. However, sequencing of the coding region of genome alone represents a possible and economically viable project to create a database containing all expressed genes in an organism that can be utilized later on for genome-wide association or mutation scanning procedures.
The coding region of a genome can be isolated and enriched by genome capture after hybridization with suitable baits. Various approaches have been reported for exome capture including microarray, in which an array containing all coding sequences is hybridized with genomic DNA to capture whole exome [7], [8]. Unfortunately, such an array is not available in buffalo at present. Another approach is to selectively amplify targeted regions with PCR for sequencing. Although a very sensitive procedure, PCR is difficult to use on a larger scale and it would be very cumbersome to amplify whole portions of exomes by PCR with uniformity. Furthermore, such multiplex amplification of exomes suffers from uneven representation of sequencing targets and poor reproducibility [5]. For these reasons this strategy cannot be applied in the case of buffalo.
In this study, we have used the cDNA prepared from buffalo muscle tissue as a bait to capture muscle TST exomes in buffalo and then sequence them with the 454 sequencer based on pyro-sequencing technology. This represents a simple method involving use of customized baits and, if optimized, can result in a robust method which can be used routinely in genome sequencing laboratories.
Results
Sequences containing low quality bases were trimmed so that high quality data can be used for analysis and subsequent SNP finding. As a result, we obtained 101,244 reads containing 24,127,779 high quality bases, of which 59,211 were assembled into a total of 750 contigs. Nearly 41% of the reads mapped against the Bos taurus mRNA reference database. Table 1 presents a summary of assembly and mapping results of the sequencing data. 634 unique genes were revealed in 720 out of 750 mapped contigs. Similarly, 20 unique genes were revealed in 38 out of 230 contigs formed after assembly of unmapped and too short reads; whereas, 541 unique genes were revealed in 819 out of 29,273 singletons.
Table 1.
Summary of sequencing data obtained from Bubalus bubalis muscle TST exome capture
| gsMapper assembly (with Bos taurus reference mRNA database) | ||
| No. of reads | 101,244 | |
| No. of bases | 24,127,779 | |
| No. of mapped reads | 59,211 | 41.40% |
| No. of mapped bases | 6389,585 | 26.48% |
| No. of unmapped reads | 41,530 | 41.02% |
| No. of too short reads | 503 | 0.50% |
| No. of contigs | 750 | |
| No. of bases | 151,663 | |
| De novo assembly for unmapped and too short reads | ||
| No. of contigs | 230 | |
| No. of bases | 102,080 | |
| No. of singleton reads | 29,273 | |
| No. of singleton bases | 6503,535 | |
Note: Reads available after sequencing were mapped against the Bos taurus reference mRNA database. Unmapped reads after merging with too short reads are subjected to de novo assembly to form contigs. Assembled contigs were utilized for analysis.
Functional annotation of identified genes
Genes identified from blastn were subjected to KEGG analysis for identification of pathways. Out of 634 unique genes identified from mapped contigs, pathways could be attributed to 411 genes. Likewise, for unmapped and too short reads and singletons, pathways were identified for 15 and 395 genes, respectively. KEGG pathway analysis is summarized in Table 2. The majority of pathways identified in our experiment were involved in metabolism followed by other categories. This pathway analysis suggests that the dataset analyzed was collected from a metabolically active state. Genes identified in annotation were subjected to gene ontology (GO) to summarize the information contained in the genes and top 10 GO terms were plotted in Table 3. In our dataset, the most enriched category was molecular function (24%) followed by metabolism (17%), intracellular (16%), binding (13%) and other categories.
Table 2.
Pathway classification of genes based on KEGG analysis
| Category | Mapped contigs | Unmapped and too short contigs | Singletons | Total | % |
|---|---|---|---|---|---|
| Metabolism | 114 | 7 | 109 | 230 | 28.01 |
| Organismal systems | 90 | 3 | 89 | 182 | 22.16 |
| Human diseases | 68 | 0 | 75 | 143 | 17.41 |
| Environmental information processing | 53 | 4 | 52 | 109 | 13.27 |
| Cellular processes | 49 | 1 | 39 | 89 | 10.84 |
| Genetic information processing | 37 | 0 | 31 | 68 | 8.28 |
| Total | 411 | 15 | 395 | 821 |
Note: Percentage of enriched pathways in buffalo muscle tissue was calculated after analysis with KEGG pathway. Importantly, genes related with metabolic pathways were enriched in muscle tissue.
Table 3.
Top 10 GO terms enriched in muscle TST exome
| Go category | Proportion in muscle TST exome (%) |
|---|---|
| Molecular function | 24 |
| Metabolism | 17 |
| Intracellular | 16 |
| Binding | 13 |
| Catalytic activity | 7 |
| Cytoplasm | 5 |
| Biosynthesis | 5 |
| Development | 5 |
| Protein binding | 4 |
| Nucleic acid metabolism | 4 |
Genes involved in muscle-specific gene expression
Searching against the TiGER database demonstrated that 269 out of 634 unique genes had no records in the database. Of the remaining 365 genes, 290 (80%) genes were expressed in muscle tissue to different extents, while 75 genes are not expressed in human muscle tissue at all. The top 10 genes with preferential expression in muscle tissue are TNNI1, NRAP, NOS1, SMYD1, FBXO40, MYOM1, IKZF5, FNDC5, UBA5 and Fam13A1. Amongst these, important ones are (1) TNNI1, which encodes for Troponin-central regulatory protein of striated muscle contraction [9]; (2) NRAP, which encodes for nebulin-related anchoring protein-involved in anchoring the terminal actin filaments in the myofibril to the membrane [10]; (3) NOS1, which encodes for nitric oxide synthase which is highly expressed in skeletal muscle [11]; (4) FBXO40, which encodes for F-box protein 40 with a probable function in myogenesis [12]; (5) MYOM1, which encodes for Myomesin 1 which binds myosin, titin, and light meromyosin and interconnects the major structure of sarcomeres [13] and (6) IKZF5, which encodes for IKAROS family zinc finger 3 transcription factor that plays an important role in the regulation of lymphocyte differentiation [14].
SNP detection
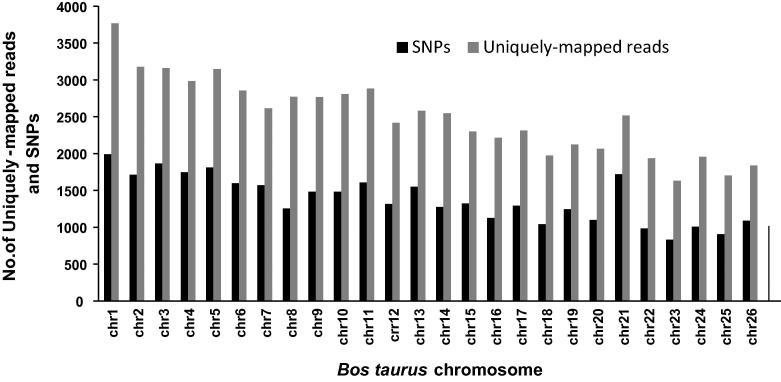
A total of 24.1 Mb of sequence was generated by TST exome capture and used for mapping with Burrows-Wheeler Aligner (BWA) algorithm. BWA is successfully used for mapping and alignment of large and complex genomes sequenced by 454 sequencing [15], repeat finding [16] as well as SNP finding [17]. Using BWA, 97,829 out of 101,244 obtained reads (96%) were successfully mapped to the reference sequence (Btau 4.0). Among them, 72,090 (71%) were uniquely mapped reads, 25,795 (25%) were mapped to multiple sites, while 3415 (4%) remained unmapped. After variant calling with SAMtools, a total of 40,100 variants were detected containing 39,697 SNPs and 403 indels. Figure 1 shows number of uniquely-mapped reads and SNPs found in our dataset after mapping against each bovine chromosome.snpEff version 1.7 was used to predict the effect of candidate SNPs using the ENSEMBL version Btau 4.0.60 database as a reference. Out of 39,697 identified SNPs, 19,642 (48.98%) were located in intergenic regions and 6184 (15.42%) were located upstream or downstream of gene bodies. Out of remaining 14,274 SNPs, 13,953 (34%) were located in intronic regions, while 218 (0.54%) encoded synonymous changes and 195 (0.48%) encoded nonsynonymous changes. Only 479 (1.2%) SNPs were reported and established in bovine according to Btau 4.0.60 and none of them were nonsynonymous.
Figure 1.
Number of uniquely-mapped reads and SNPs found in buffalo against each bovine chromosome
Results obtained from repeat masker revealed that there was a prominent reduction of repeats in captured genomic sequences. The comparison between captured genome sequence and whole genome sequences is summarized in Table 4.
Table 4.
Percentage of repeat sequences present in Bubalus bubalis muscle TST exome and in whole genome
| Repeat | In captured genomic sequencea (%) | In whole genome sequenceb (%) |
|---|---|---|
| Simple repeat | 0.09 | 4.00 |
| Satellite | 15.38 | 0.63 |
| Low complexity | 0.30 | 2.78 |
| LINE | 13.89 | 26.64 |
| SINE | 2.44 | 35.37 |
| LTR | 4.07 | 7.25 |
| DNA elements | 1.78 | 3.91 |
| RNA | 0.08 | 0.1 |
| Unknown | 0.02 | 0.06 |
| Total | 38.05 | 80.74 |
Note: Analysis of repeat masking of captured data with repeat masker software. Capture resulted in almost 40% reduction in repeat sequences. Percentage of repeats in captured genomic sequences (a) and in whole genome sequence assembly of sequencing data (b) was shown. LINE, long interspersed nuclear element; SINE, short interspersed nuclear element; LTR, long terminal repeat.
Discussion
In our study, TST exomes were captured using mRNA from muscle tissue. It is hence, imperative that whatever genes have been identified should be expressed in muscle tissue. To validate this the genes were searched against the “TiGER” database which contains information on tissue-specific combinational gene regulation, based on transcription factor binding sites, enabling us to perform a large-scale analysis of tissue-specific gene regulation in human tissues. Though, the data generated in this experiment was from buffalo muscle tissue exome, this analysis would give a fair idea about tissue specific gene expression and efficiency of TST exome capture.
The results revealed that genes captured in this experiment were expressed in muscle tissue and thus show reliability of this approach for TST exome capture. Not only that, several genes represented by only one or two reads with very low expression in muscle tissue were successfully captured using mRNA molecules as bait. This shows the usefulness of this approach in obtaining a complete gene expression profile for the tissue in question.
Comparison against the Btau 4.0.60 database revealed that only 479 (1.2%) SNPs were reported in the reference database while 39,218 (98.8%) candidate SNPs were novel. Previous studies involving novel candidate SNP finding in B. taurus revealed around 87% of novel candidate SNPs [17], [18]. As we have used the B. taurus reference for genomic sequence from buffalo, a comparatively larger number of novel candidate SNPs were reported in our analysis.
All 195 nonsynonymous candidate SNPs were homozygous, located in 37 different genes with the highest number found on chromosome (Chr) 11 followed by Chr 19, 21, 29, 30 and 6. Our analysis showed 34 candidate SNPs (17.5%) in the CRB2 gene alone. No reports are available for candidate SNPs in the buffalo CRB2 gene, however, human mutation analysis of the CRB2 gene revealed 11 sequence variants leading to an amino acid substitution causing autosomal recessive disease [19]. CRB2 was followed by PPFIA1 with 10 candidate SNPs. Though PPFIA1 has not been studied for candidate SNP finding, the gene is associated with squamous cell carcinoma in human [20], [21].
It is evident from Table 4 that there was significant reduction in the percentage of repeats in captured sequences versus whole genome sequences [22] for all classes of repeats, except satellite repeats. It is surprising that an increase in satellite repeat sequences occurs in this situation. Satellite repeats are present with more than one copy and they flank the coding sequences in most of the cases. In exons, tri-nucleotide repeats are invariably the most abundant in all taxa, with hexa-nucleotide repeats being the second most common. Inter-genic regions and introns contain more hexa-nucleotide repeats than exons do, but the tri-nucleotide repeats are less common [23]. Hence, it is logical to assume an increase in satellite sequences in TST exome capture data as satellite repeats contain tri-nucleotide repeats and hexa-nucleotide repeats, of which the former repeats are more common in exons. In our experiment, we have not utilized any carrier to block the capturing of repeat sequencing. Nonetheless the decrease in repeat content in enriched sequencing data is significant (47.12%). In future studies, use of some blocking reagent/sequences for repeats may result in further reduction in repeat sequences and in turn capture of highly enriched sequences for the coding region.
Several strategies have been tried to reduce genome complexity which include EST sequencing, methyl filtration, and high-Cot DNA selection [24]. These approaches may decrease genome complexity to some extent but it is not possible to have targeted capture as by solution hybridization. In our experiment we noticed that approximately 2/3rd of the captured reads are on the targets, i.e., regions intended to be coding, but a still larger proportion, approximately 1/3rd of captured sequences were off the target. Blocking with repetitive DNA can be employed to increase on-target capture and to reduce off target capture. Other approaches include reducing fragment size of the library thereby reducing chances of secondary capture of off target sequences [25]. However, it is invariably necessary to develop whole genome sequence of buffalo so that high density capture arrays can be prepared for efficient and specific capture of buffalo coding regions.
Currently kits for exome capture/target enrichment are commercially available. However, these arrays are only available for humans and mice but not applicable in farm animals. This study shows use of solution hybridization to capture whole TST exome without using any array. Our method can be applied to less explored species or farm animals for which the complete genome sequence is unavailable for examining SNPs in coding regions and tissue specific gene expression. In addition, it is possible to scale up to work with large numbers of samples simultaneously using this method although it needs to be improved for blocking repeat sequences.
The present study reports the first instance of utilizing the simple strategy of solution hybridization for capture of exons from buffalo muscle tissue with cDNA prepared from muscle tissue as bait molecules. We have successfully enriched coding sequences present in the buffalo genome and the generated data was used for SNP and indel detection. This approach can be used for TST exome capture in farm animals, which would help to evolve a strategy for selection of animals with better production potential. Moreover, this approach provides flexibility of use with any species irrespective of genome size and prior sequence information and can be applied to the capture and sequencing of any type of variation, either SNPs or indels. With some modification and optimization of hybridizing conditions the method could be potentially applied to any organism. For the species without complete whole genome assembly, we have to develop alternate strategies to study variations in coding sequences and this study will serve as an important step in that direction.
Materials and methods
Sample collection
The thigh muscle tissue samples for the study were collected from a slaughtered male Surti breed of buffalo (B. bubalis) immediately after slaughter. The muscle tissues were collected following aseptic procedures in RNAlater® and transported to the laboratory on ice and stored at −196 °C in liquid nitrogen. Blood was collected from a male Surti buffalo in Vacuette (Greiner Bio-One, NC) containing EDTA and transported to laboratory on ice.
mRNA and cDNA preparation
Total RNA was isolated from both tissues using a standard TRIZOL protocol (Invitrogen™ Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. Contaminating genomic DNA was removed from total RNA by DNAse treatment using DNAseI, RNAse-free (Fermentas, CA) following the manufacturer’s instructions. Total RNA was assayed and quality and quantity was verified on Nanodrop ND1000 (Thermo Fischer Scientific, Wilmington, USA) and Bioanalyzer 2100 (Agilent Technologies, Mississauga, ON). A sample containing total RNA from muscle was used for cDNA preparation using biotinylated OligodT.
Genomic library preparation
Genomic DNA was extracted from a blood sample collected from a mature buffalo bull of the Jaffrabadi breed using and standard phenol/chloroform extraction methods [26]. To prepare a single stranded (sst) library, genomic DNA was fragmented and adapters from GS-FLX Titanium were ligated following the manufacturer’s protocol.
TST exome capture
The sst genomic DNA library prepared was hybridized (1:10) with biotinylated cDNA from muscle tissue. Here, muscle cDNA was used in excess so that exons encoding rarely expressed transcripts can also be captured. Hybridization was performed in 2× hybridization buffer containing 10 × SSPE, 10 × Denhardt’s, 10 mM EDTA and 0.2% SDS [27]. Hybridization was carried out for 24 h at 66 °C and captured genomic regions were recovered using NaOH. The recovered genomic DNA library was purified with MinElute columns (Quiagen, DE) and subjected to emulsion PCR.
Sequencing
DNA-positive beads from emulsion PCR were recovered, enriched and subjected to 454 sequencing standard protocols developed by Roche Diagnostics (Roche Diagnostics, Switzerland). 1/4th region on PicoTitrePlate was utilized for sequencing of capture sequences from buffalo genomic DNA on GS FLX.
Sequence analysis
Among reads generated after sequencing, only reads that contained the correct base key at the start (a portion of the 454 primer used to differentiate reads from internal quality control sequences), including at least 84 flows and less than 5% of flow cycles resulting in an ambiguous base call (N), were retained for analysis. The 3′ ends of the reads were trimmed such that <3% of the remaining flows have ambiguous signal intensity for incorporation. Each base sequenced was also annotated with a Phred equivalent quality score from Roche diagnostics and the average base score for all the reads was found to be 31 indicating the accuracy of base calling to be 99.99%.
Available reads were assembled using gsMapper provided by Roche to map obtained reads against the B. taurus reference mRNA database. For efficient utilization of obtained sequences, remaining sequence data of too short and unmapped reads after reference database mapping were again subjected to de novo assembly using inbuilt software tool gsAssembler provided by Roche using default parameters, i.e., overlap length 40 bp, seed step 12 bp and minimum sequence identity 90%.
Sequence annotation
For annotation of available sequence data, all the contigs and unassembled sequences obtained after mapping and de novo assembly were subjected to NCBI Blastn against the B. taurus mRNA reference database using default parameters. Genes found with Blastn were used in the KEGG Mapper for identification of respective pathways [28]. All genes identified were searched against Tissue-specific Gene Expression and Regulation (TiGER) for validation of muscle tissue specific gene expression [29]. All the genes found with Blast search are used to download respective GO terms from http://www.ebi.ac.uk/GOA/cow_release.html of the cow genome. Available GO terms were categorized using CateGOrizer [30] GO_Slim classification system.
SNP finding
For alignment and annotation of the sequence reads, we used the bovine genome chromosome Btau 4.0 (http://www.hgdownload.cse.ucsc.edu/goldenPath/bosTau4/chromosomes/) as a reference source.
For long read alignment, we used BWA-SW ver 0.5.9 [31]. The BWA default values for mapping were as follow: Score of a match = 1 (matchScore), Mismatch penalty = 3 (mmPen), Gap open penalty = 5 (gapOpenPen), Gap extension penalty = 2 (gapExtPen), Number of threads in the multi-threading mode = 1 (thresh), Band width in the banded alignment = 33 (zBest), Minimum score threshold divided by a = 37 (nHspRev) and Coefficient for threshold adjustment according to query length = 5.5. After read mapping we discarded repeat sequences and unmapped reads. Only reads mapped to a unique position on the reference genome sequence were used for SNP calling.
To call SNPs, we used SAMtools ver 1.1.12a [32] and applied additional filters as follows: reads mapped with minimum mapping quality of 20, minimum read depth = 3. After SNP calling, we annotated the candidate SNPs using SNPeff (http://www.snpeff.sourceforge.net/) software.
Repeat masker analysis
Available sequencing data were subjected to Repeat Masker for repeat prediction, with the DNA source parameter set to cow, comparison species set to human with weak lineage specific masking, advance alignment option to be in repeat orientation and by masking repeats with lower case. The remaining parameters were set to default. The results were obtained in three different text files containing results of repeat data, masked sequences and alignment.
Authors’ contributions
CG conceived the whole experiment, monitored the progress and helped draft manuscript. DN helped in manuscript preparation and made advisory comments. SJ prepared the manuscript and performed the experiment with VB. PG assisted in sequencing of captured library with SJ and VB. KB performed SNP finding. All authors read and approved the final manuscript.
Competing interests
The authors have no competing interests to declare.
Acknowledgements
Authors are thankful to Government of India for providing financial assistance under Rashtriya Krishi Vikas Yojana (RKVY) scheme to carry out research work mentioned in the manuscript.
References
- 1.Michelizzi V.N., Dodson M.V., Pan Z., Amaral M.E., Michal J.J., McLean D.J. Water buffalo genome science comes of age. Int J Biol Sci. 2010;6:333–349. doi: 10.7150/ijbs.6.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tantia M.S., Vijh R.K., Bhasin V., Sikka P., Vij P.K., Kataria R.S. Whole-genome sequence assembly of the water buffalo (Bubalus bubalis) Indian J Anim Sci. 2011;81:38–46. [Google Scholar]
- 3.Dapprich J., Ferriola D., Magira E.E., Kunkel M., Monos D. SNP-specific extraction of haplotype-resolved targeted genomic regions. Nucleic Acids Res. 2008;36:e94. doi: 10.1093/nar/gkn345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter S.W., Davey J.W., Johnston J.S., Shelton A.M., Heckel D.G., Jiggins C.D. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One. 2011;6:e19315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porreca G.J., Zhang K., Li J.B., Xie B., Austin D., Vassallo S.L. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 6.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 8.Okou D.T., Steinberg K.M., Middle C., Cutler D.J., Albert T.J., Zwick M.E. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 9.Wade R., Eddy R., Shows T.B., Kedes L. cDNA sequence, tissue-specific expression, and chromosomal mapping of the human slow-twitch skeletal muscle isoform of troponin I. Genomics. 1990;7:346–357. doi: 10.1016/0888-7543(90)90168-t. [DOI] [PubMed] [Google Scholar]
- 10.Mohiddin S.A., Lu S., Cardoso J.-P., Carroll S., Jha S., Horowits R. Genomic organization, alternative splicing, and expression of human and mouse N-RAP, a nebulin-related LIM protein of striated muscle. Cell Motil Cytoskeleton. 2003;55:200–212. doi: 10.1002/cm.10123. [DOI] [PubMed] [Google Scholar]
- 11.McConell G.K., Bradley S.J., Stephens T.J., Canny B.J., Kingwell B.A., Lee-Young R.S. Skeletal muscle nNOSμ protein content is increased by exercise training in humans. Am J Physiol Regul Integr Comp Physiol. 2007;293:R821–R828. doi: 10.1152/ajpregu.00796.2006. [DOI] [PubMed] [Google Scholar]
- 12.Ye J., Zhang Y., Xu J., Zhang Q., Zhu D. FBXO40, a gene encoding a novel muscle-specific F-box protein, is upregulated in denervation-related muscle atrophy. Gene. 2007;404:53–60. doi: 10.1016/j.gene.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Li T.-B., Liu X.-H., Feng S., Hu Y., Yang W.-X., Han Y. Characterization of MR-1, a novel myofibrillogenesis regulator in human muscle. Acta Biochim Biophys Sin. 2004;36:412–418. doi: 10.1093/abbs/36.6.412. [DOI] [PubMed] [Google Scholar]
- 14.Perdomo J., Holmes M., Chong B., Crossley M. Eos and Pegasus, two members of the IKAROs family of proteins with distinct DNA binding activities. J Biol Chem. 2000;275:38347–38354. doi: 10.1074/jbc.M005457200. [DOI] [PubMed] [Google Scholar]
- 15.You F.M., Huo N., Deal K.R., Gu Y.Q., Luo M.C., McGuire P.E. Annotation-based genome-wide SNP discovery in the large and complex Aegilops tauschii genome using next-generation sequencing without a reference genome sequence. BMC Genomics. 2011;12:59. doi: 10.1186/1471-2164-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croucher N.J., Vernikos G.S., Parkhill J., Bentley S.D. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics. 2011;12:120. doi: 10.1186/1471-2164-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawahara-Miki R., Tsuda K., Shiwa Y., Arai-Kichise Y., Matsumoto T., Kanesaki Y. Whole-genome resequencing shows numerous genes with nonsynonymous SNPs in the Japanese native cattle Kuchinoshima-Ushi. BMC Genomics. 2011;12:103. doi: 10.1186/1471-2164-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eck S.H., Benet-Pages A., Flisikowski K., Meitinger T., Fries R., Strom T.M. Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biol. 2009;10:R82. doi: 10.1186/gb-2009-10-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hurk J.A., Rashbass P., Roepman R., Davis J., Voesenek K.E., Arends M.L. Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol Vis. 2005;11:263–273. [PubMed] [Google Scholar]
- 20.Dancau A.M., Wuth L., Waschow M., Holst F., Krohn A., Choschzick M. PPFIA1 and CCND1 are frequently coamplified in breast cancer. Genes Chromosomes Cancer. 2010;49:1–8. doi: 10.1002/gcc.20713. [DOI] [PubMed] [Google Scholar]
- 21.Tan K.D., Zhu Y., Tan H.K., Rajasegaran V., Aggarwal A., Wu J. Amplification and overexpression of PPFIA1, a putative 11q13 invasion suppressor gene, in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2008;47:353–362. doi: 10.1002/gcc.20539. [DOI] [PubMed] [Google Scholar]
- 22.Vaidya M.B., Sajnani M.R., Ramani U.V., Tripathi A.K., Bhatt V.D., Patel J.S. A preliminary analysis of repeat organisation in Bubalus bubalis genome. Indian J Anim Biotechnol. 2012;11:62–66. [Google Scholar]
- 23.Toth G., Gaspari Z., Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbazuk W.B., Bedell J.A., Rabinowicz P.D. Reduced representation sequencing: a success in maize and a promise for other plant genomes. Bioessays. 2005;27:839–848. doi: 10.1002/bies.20262. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y., Springer N.M., Gerhardt D.J., Ying K., Yeh C.T., Wu W. Repeat subtraction-mediated sequence capture from a complex genome. Plant J. 2010;62:898–909. doi: 10.1111/j.1365-313X.2010.04196.x. [DOI] [PubMed] [Google Scholar]
- 26.John S.W., Weitzner G., Rozen R., Scriver C.R. A rapid procedure for extracting genomic DNA from leukocytes. Nucleic Acids Res. 1991;19:408. doi: 10.1093/nar/19.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X., Zack D.J., Zhu H., Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi-Liang H., Jie B., James M.R. “CateGOrizer: a web-based program to batch analyze gene ontology classification categories”. Online J Bioinform. 2008;25:2078–2079. [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]