Abstract
Muscle wasting is common in mammals during extended periods of immobility. However, many small hibernating mammals manage to avoid muscle atrophy despite remaining stationary for long periods during hibernation. Recent research has highlighted roles for short non-coding microRNAs (miRNAs) in the regulation of stress tolerance. We proposed that they could also play an important role in muscle maintenance during hibernation. To explore this possibility, a group of 10 miRNAs known to be normally expressed in skeletal muscle of non-hibernating mammals were analyzed by RT-PCR in hibernating little brown bats, Myotis lucifugus. We then compared the expression of these miRNAs in euthermic control bats and bats in torpor. Our results showed that compared to euthermic controls, significant, albeit modest (1.2–1.6 fold), increases in transcript expression were observed for eight mature miRNAs, including miR-1a-1, miR-29b, miR-181b, miR-15a, miR-20a, miR-206 and miR-128-1, in the pectoral muscle of torpid bats. Conversely, expression of miR-21 decreased by 80% during torpor, while expression of miR-107 remained unaffected. Interestingly, these miRNAs have been either validated or predicted to affect multiple muscle-specific factors, including myostatin, FoxO3a, HDAC4 and SMAD7, and are likely involved in the preservation of pectoral muscle mass and functionality during bat hibernation.
Keywords: MicroRNA, Hibernation, Metabolic rate depression, Atrophy, Dicer, Myostatin
Introduction
Skeletal muscle tissue differentiates early in vertebrate embryogenesis and requires the combinatorial actions of multiple signaling pathways. The mechanisms of maintaining and remodeling skeletal muscle are of particular interest, since skeletal muscle tissue is capable of adapting to the changing physiological and environmental conditions [1]. For example, hypertrophy of skeletal muscle is induced through increased muscle use and results in an enlarged size, augmented ability to produce force, increased resistance to fatigue and enhanced oxidative metabolism [1], [2]. However, prolonged disuse or immobility of skeletal muscle, such as during prolonged bed rest, various disease states or age-related sarcopenia, typically results in the opposite effect, i.e., atrophy of muscle [2]. Several molecular pathways involved in the development and maintenance of skeletal muscle mass have been well characterized. Major signaling pathways include the phosphoinositol 3-kinase (PI3-K)/Akt/mammalian target of rapamycin (mTOR) pathway, the enzyme histone deacetylase 4 (HDAC4), the secreted protein myostatin and its associated membrane receptor (Acvr2b), as well as muscle-specific E3 ubiquitin ligases tripartite motif-containing protein 63 (TRIM63) and muscle atrophy F-box (Atrogin-1) [3], [4], [5] (Figure S1). Transcription factors including the forkhead box O (FoxO3a), myogenin, myogenic differentiation 1 (MyoD), myogenic factor (Myf5), myogenic regulatory factor 4 (Mrf4), and myocyte enhancer factor 2 (MEF2A) are also crucial [2], [4], [6]. Development of skeletal muscle during embryogenesis, maintenance of adult skeletal muscle, and any changes leading to hypertrophy or atrophy typically fall under the control of one or more of these biological factors.
With the discovery of microRNAs (miRNAs), the small non-coding RNAs capable of regulating protein expression within a cell, additional post-transcriptional modes of regulation in skeletal muscle are being elucidated. With the aid of the miRNA-initiated silencing complex (miRISC), these 17–22 nucleotide transcripts are able to bind with full or partial complementarity usually to the 3’ UTR of mRNA targets [7], [8], [9]. Target binding either inhibits translation or leads to mRNA degradation [7], [8], [9]. This form of gene expression control at the post-transcriptional level has been shown to be of great importance in mammalian systems; to date, miRNAs are known to be critically involved in biological development, cell differentiation, apoptosis, cell-cycle control, stress response and disease pathogenesis [7], [10], [11], [12].
A collection of miRNAs have been shown to be regulated during periods of skeletal muscle atrophy; these include miR-1a-1, miR-29b, miR-181b, miR-15a, miR-20a, miR-206, miR-128-1, miR-21, miR-23a and miR-107 [6], [13], [14]. Target validation has illustrated some ties, both direct and indirect, between these miRNAs and the host of muscle-specific effects of the aforementioned factors (Figure S1). Thus, miRNAs are critically involved in development, maintenance and adaptation of skeletal muscle [6]. In order to further characterize the roles of miRNAs in muscle metabolism, we turned to an unusual model of mammalian muscle metabolism: hibernation. Many small mammals use hibernation to survive over the winter months when food is scarce and cold environmental temperatures place high demands on thermogenesis if a high constant body temperature (Tb) is to be maintained. For example, body fat reserves of hibernating little brown bats, Myotis lucifugus, would last only about 25 days if the animals maintained euthermia but by entering prolonged periods of torpor during which Tb falls to near ambient, the bats readily endure a 7–9 month hibernation season without eating [15]. Indeed, M. lucifugus may remain in constant torpor for several weeks at a time during the hibernation season. A recent study reported torpor bout durations of 2–7 weeks for free-ranging little brown bats (interspersed with short arousals back to euthermia) [16]. Much is known about the physiology and biochemistry of hibernation in this species (e.g., [16], [17] and references therein). Most mammals that undergo long periods of immobility display significant disuse atrophy of their skeletal muscles [2]. However, hibernators, including M. lucifugus, appear to avoid this negative outcome [18] despite spending long weeks in torpor during the winter. Selected molecular mechanisms are clearly at work to preserve both skeletal muscle mass and functionality during hibernation.
The present study was undertaken to evaluate the responses of aforementioned muscle-associated miRNAs in the skeletal muscle metabolism of M. lucifugus during hibernation. RT-PCR was employed to investigate expression of these miRNAs in the pectoral muscle that powers flight in bats. The differential expression of several miRNAs was identified during hibernation and suggests an important role for these non-coding RNAs in the maintenance of skeletal muscle. The protein expression of several important miRNA targets (Myostatin, FoxO3a, HDAC4 and SMAD7) were also analyzed by immunoblotting. The data also provide intriguing leads for therapeutic targets that could be investigated to or avoid muscle disuse atrophy in humans. Furthermore, presence of all 10 miRNAs in bats, which until now had only been predicted algorithmically from genome searches, was verified by sequencing Sequences of these miRNAs were identical to those of mouse, indicating that the miRNA sequences in a member of the Chiroptera are highly conserved with those of a rodent, the mouse (Mus musculus).
Results
MicroRNA expression during hibernation
To amplify the selected miRNAs, we used a modified stem-loop procedure outlined by Biggar et al. [19]. This protocol allows both amplification and sequencing of mature miRNAs in organisms that have no previous miRNA annotation. Our results show that the uniform FO fiber composition of M. lucifugus pectoralis muscle does not change with season, consistent with previous report [20]. We therefore examined the miRNA expression in the pectoral muscle of M. lucifugus. Expression levels of miR-1a-1, miR-29b, miR-181b, miR-15a, miR-20a, miR-206, miR-128-1, miR-21, miR-23a and miR-107 in the pectoral muscle of M. lucifugus were assessed by RT-PCR and all miRNA PCR products were confirmed by sequencing. The sequences of all mature miRNAs investigated were 100% identical with the known mature miRNA sequences from the house mouse, M. musculus.
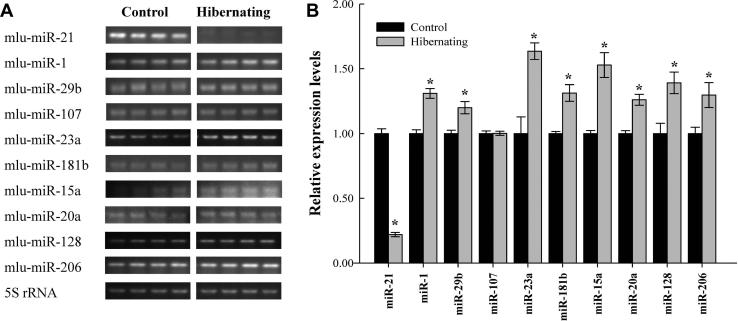
We further compared the expression of these miRNAs in hibernating bats (Tb ∼ 5–6 °C) and aroused euthermic bats (Tb ∼ 35–37 °C). Eight miRNAs were significantly upregulated in muscle from torpid bats as compared to euthermic controls (P < 0.05) (Figure 1). For example, expression of miR-29b increased by 1.2 fold (1.2 ± 0.05) and that of miR-23a increased by 1.63-fold (1.63 ± 0.07). However, expression of miR-21 decreased substantially during torpor, which corresponded to only 22% of the expression in control (22 ± 2%; P < 0.05), whereas expression levels of miR-107 (1.00 ± 0.02) didn’t change significantly with hibernation.
Figure 1.
Effect of hibernation on the expression of selected microRNAs in skeletal muscle of M. lucifugus A. Representative RT-PCR data were shown for expression of indicated miRNAs under control and hibernation conditions. B. Relative expression of miRNA-21, miR-1a-1, miR-29b, miR-107, miR-23a, miR-181b, miR-15a, miR-20a, miR-128 and miR-206. Expression of the indicated miRNAs was normalized to that of 5S rRNA from the same pectoral muscle RNA samples and shown in histogram. Data are presented as means ± S.E.M of independent samples from separate animals (n = 3–4). ∗indicated that expression of the indicated miRNA in hibernated bats was significantly different from controls (P < 0.05).
Putative miRNA targets of the differentially-expressed miRNAs
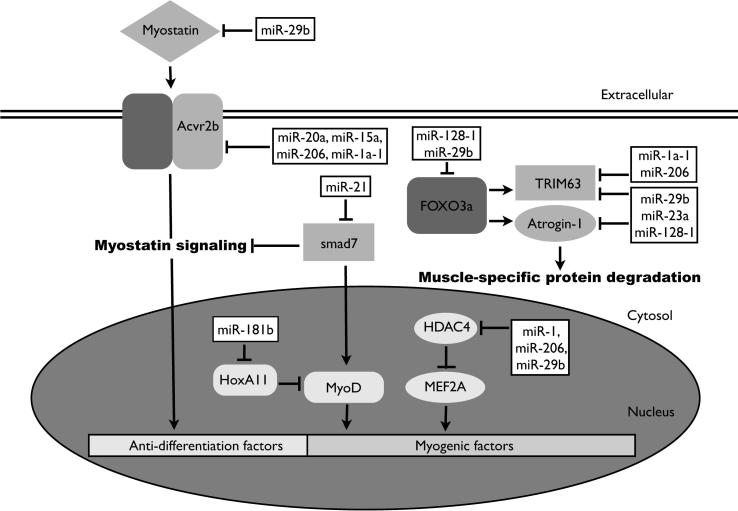
Targets of these differentially-expressed miRNAs were analyzed based on the available literature regarding these miRNAs. In addition, putative targets were also obtained from an algorithm-based miRNA target identification database (TargetScan release 4.2; http://www.targetscan.org/vert_42/). These potential targets include ACVR2b, HDAC4, myostatin, FoxO3a, MEF2A, homeobox protein HoxA11, SMAD7, MyoD, TRIM63 and Atrogin-1. A summary of the putative targets of differentially regulated miRNAs and their potential effects in skeletal muscle is provided in Table 1.
Table 1.
Differential expression of miRNAs in skeletal muscle of M. lucifugus and their putative targets
| microRNA | Expression change | Putative target(s) | Potential effects | Reference |
|---|---|---|---|---|
| miR-21 | ↓ | Smad7 | Enhance MyoD signaling; inhibit myostatin signaling | [40] |
| miR-1a-1 | ↑ | HDAC4 | Allow MEF2A signaling; inhibit TRIM63 expression | [41] |
| ACVR2b | Inhibit myostatin signaling | [37]a | ||
| miR-181b | ↑ | HoxA11 | Enhance MyoD signaling | [33] |
| miR-206 | ↑ | HDAC 4 | Allow MEF2A signaling; inhibit TRIM63 expression | [46] |
| ACVR2b | Inhibit myostatin signaling | a | ||
| miR-29b | ↑ | FOXO3a | Prevent TRIM63 and Atrogin-1 expression | a |
| Myostatin | Prevent myostatin expression | a | ||
| HDAC4 | Allow MEF2A signaling; inhibit TRIM63 expression | [42] | ||
| ACVR2b | Inhibit myostatin signaling | [42] | ||
| miR-15a | ↑ | ACVR2b | Inhibit myostatin signaling | a |
| miR-20a | ↑ | ACVR2b | Inhibit myostatin signaling | a |
| miR-128-1 | ↑ | FOXO3a | Prevent TRIM63 and Atrogin-1 expression | [14] |
| miR-23a | ↑ | TRIM63 | Prevent muscle-specific protein ubiquitination | [43], [44] |
| Atrogin1 | Prevent muscle-specific protein ubiquitination | |||
| miR-107 | NC | DMPK | Targeting of the CTG trinucleotide repeat of DMPK | [45] |
Note: No change is defined as ‘NC’.
Indicates that data was obtained from TargetScan release 4.2 (http://www.targetscan.com/vert_42/).
Protein expression
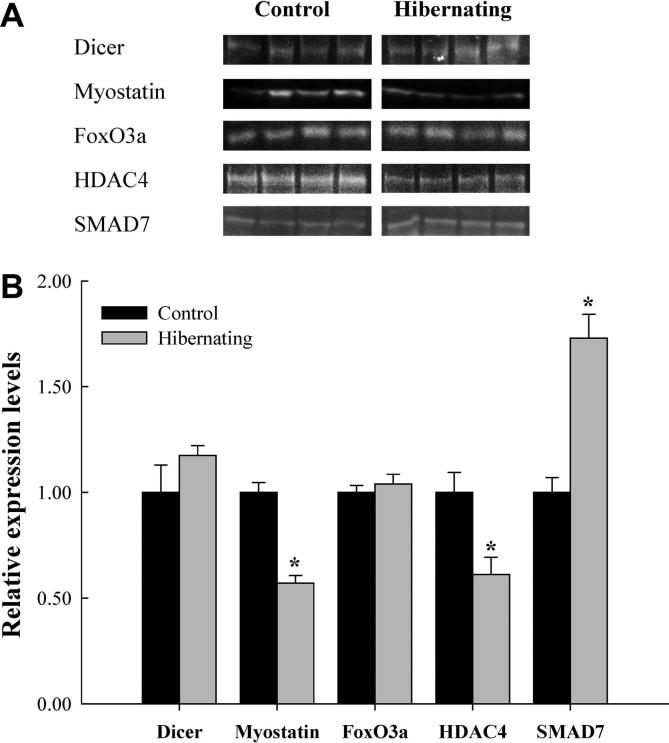
Immunoblotting was performed to examine the relative expression of several key proteins in miRNA processing and muscle maintenance/atrophy. These include Dicer, which is a major protein in processing of precursors into mature miRNAs, and several proteins involved in inducting or transducing pathways of muscle maintenance/atrophy, such as myostatin, FoxO3a, HDAC4 and SMAD7. As shown in Figure 2, the relative expression of Dicer or FoxO3a protein did not change between control and hibernating states. However, the relative expression of myostatin was found to decrease to 57 ± 4% of control in response to hibernation (P < 0.05). Similarly, relative expression of HDAC4 decreased to 61 ± 8% of that in controls. However, the relative expression of SMAD7 was found to increase 1.73 ± 0.11 fold compared to controls in response to hibernation (P < 0.05).
Figure 2.
Effect of hibernation on the protein expression of Dicer and myostatin in skeletal muscle of M. lucifugus A. Representative immunoreactive bands were shown for Dicer, myostatin and other proteins under control and hibernation conditions. B. Relative expression of Dicer, myostatin, FoxO3a, HDAC4 and SMAD7 protein between control and hibernating states. Expression of the indicated proteins was normalized and shown in histogram. Histogram shows normalized protein levels. Data are presented as means ± SEM of independent samples from separate animals (n = 4). ∗indicated that expression of the indicated protein in hibernated bats was significantly different from controls (P < 0.05).
Discussion
Since their initial discovery, miRNAs have been implicated in most major areas of biological function: development, tissue differentiation and maintenance, cell cycle control, apoptosis, and metabolism. Perhaps as a result of their clear importance in regulating biological functions, dysregulation of miRNAs has been determined to exist in various disease states such as cancer, diabetes, muscular dystrophy and heart disease [21]. These small regulatory molecules provide immense potential as applied therapies or therapeutic targets, and hence it is useful to determine how miRNAs respond in diverse physiological situations including animal survival of extreme environmental stress. An example of such a model system is the avoidance of muscle disuse atrophy in bats and other hibernating mammals over the winter months [18], [22], [23]. While expression profiling of transcription factors and various other proteins has been undertaken to investigate cellular processes that may be involved in this lack of muscle degradation [22], [23], [24], [25], our lab is the first to explore the possible contribution of miRNA regulation to create an insightful outline of the molecular mechanisms at work to minimize muscle disuse atrophy in the pectoral muscle of little brown bats during hibernation.
In M. lucifugus, differential miRNA expression in pectoral muscle appears to be a regulatory component of hibernation and perhaps atrophy resistance (Figure 1). Eight miRNAs were significantly upregulated in the muscle during torpor (including miR-1a-1, miR-9b, miR-23a, miR-181b, miR-15a, miR-20a, miR-128 and miR-206) and, in addition, miR-21 was found to be strongly downregulated. Interestingly, any changes of mature miRNA transcripts appeared to be independent of Dicer protein levels, which did not change during hibernation (Figure 2). These miRNAs are all known to have regulatory roles in muscle maintenance [6] and may aid in the atrophy resistance of differentiated muscle cells by positively influencing factors that promote muscle maintenance, while negatively targeting factors that favor atrophy. Given the nature of miRNA targeting and the translational impact of a single miRNA, even relatively small changes in miRNA expression (1.2 to 1.6-fold) as seen in this study, may facilitate rapid translational repression and/or degradation of target genes. The extent to which combinatorial mRNA-miRNA pairing occurs, may allow the expression of key genes necessary for survival (Figure S1).
MEF2A, a myogenic transcription factor, is known to be epigenetically repressed by HDAC4 [26]. Interestingly, the expression of HDAC4 protein was found to decrease during hibernation while its mRNA is negatively regulated by the miRNAs miR-1a-1, miR-206 and miR-29b, all of which show increased expression in torpid bat skeletal muscle (Figure 1, Figure 2). The increased expression of HDAC4 has also been linked to positive regulation of the pro-atrophy factor, muscle-specific E3 ubiquitin ligase TRIM63, in denervation atrophy models [27]. The ubiquitin proteasome system (including TRIM63 and Atrogin-1) has been estimated to account for up to 80% of protein degradation during muscle wasting [28], [29]. Expression of both TRIM63 and Atrogin-1 are negatively regulated by miR-23a, a miRNA shown to be upregulated during hibernation, and is positively regulated by transcription factor FoxO3a [28]. FoxO3a is also a predicted target of both miR-29b and miR-128, two miRNAs that are also upregulated in skeletal muscle during hibernation. Interestingly, similar to muscle atrophy studies in the plantaris muscle tissue of 13-lined ground squirrels, an organism that is also able to undergo winter torpor, this study found no significant change in FoxO3a protein expression during torpor [30]. These results suggest that FoxO3a protein may undergo additional modes of regulation including either reduced rates of protein degradation or post-translational modification.
As one of the myogenic regulator factors, MyoD is a key regulator of muscle differentiation and maintenance as shown through knockout experiments in mice [31]. Additionally, HoxA11 is a known repressor of MyoD transcriptional activity [32]. Interestingly, it has been shown that HoxA11 is a target of miR-181b, a miRNA that we have shown to increase expression in skeletal muscle during hibernation [33]. It is possible that the increased expression of miR-181b may indirectly promote MyoD regulation of myogenesis. In addition to the possible contribution of miR-181b, a significant downregulation of miR-21 (to just 20% of control values) may also indirectly contribute to promote MyoD signaling. miR-21 has been shown to target SMAD7 and a decreased expression of miR-21 may act to influence the increase of SMAD7 protein expression as seen in this study (Figure 2). Interestingly, the function of SMAD7 in myocytes is to promote MyoD-dependent myogenesis, via a strong positive feedback loop, and also represses pro-atrophic myostatin signaling [34], [35]. To this regard, Myostatin, a protein shown in this study to decrease during hibernation in pectoral muscle, is a strong negative regulator of muscle differentiation and a predicted target of miR-29b (Figure 2). Myostatin is a secreted protein that exerts its function onto myocytes by interacting with cell surface activin type II receptors [36]. Control of myostatin signaling in hibernating M. lucifugus may not only be control via increased SMAD7 protein expression, but though the repression of activin type II receptor, ACVR2b. This receptor is a known target of miR-1a-1 and a predicted target of miR-206, miR-15a and miR-20a, all miRNAs with upregulated expression in skeletal muscle of hibernating bats [37].
Previous research has shown that little or no atrophy occurs during hibernation in several species of bats [18], [22], [23], [24], [25]. The common expression patterns among the miRNAs examined in the present study suggest the promotion of skeletal muscle maintenance and the prevention of myostatin and ubiquitin ligase directed atrophy. Studies of muscle atrophy in small mammalian hibernators, such as the little brown bat, may be useful in deducing natural pathways of preventing muscle atrophy during extended period of disuse and disease states such as amyotrophic lateral sclerosis and cachexia.
Materials and methods
Animals
Collection and treatment of little brown bats was as described previously in Eddy and Storey [38]. Briefly, bats each weighing 7−8 g were collected from caves near Sherbrooke, Quebec, at an air temperature of 5 °C. The bats were awakened by collection and remained euthermic during transport to Université de Sherbrooke. Upon arrival, half the animals were allowed to re-enter torpor in a cold room at 5 °C; full torpor was reached again in 10–12 h. Other bats were kept under euthermic conditions at an environmental temperature of 23–24 °C. Pre-calibrated miniature temperature-sensitive radio transmitters (SRX-400; LOTEK Engineering) were attached to the bats in order to monitor Tb at 5 min intervals. To do this, transmitters were attached in the interscapular region with Skin-Bond surgical adhesive (Allegro Medical Supplies, Bolingbrook, IL, USA) and skin temperature was used to approximate Tb. Immediately before sampling, euthermic bats showed Tb of 35–37 °C, whereas torpid bats showed Tb of 5–6 °C. Euthermic bats were sacrificed 2 days post collection, whereas torpid bats were sacrificed 36–38 h after re-establishment of full torpor. Animals were killed by cervical dislocation and then tissues were quickly excised, stored in liquid nitrogen, and sent to Carleton University. Tissues were kept at −80 °C until used.
Protein isolation
Soluble protein samples of frozen pectoral muscle (0.5 g) from individual bats were prepared in a similar manner to Biggar and Storey [39]. Protein concentrations were measured using the BioRad protein assay (Cat# 500-0006) and adjusted to 10 μg/μL in homogenizing buffer for all samples. The samples were denatured by mixing with 2× SDS loading buffer (100 mM Tris-base, 4% w/v SDS, 20% v/v glycerol, 0.2% w/v bromophenol blue and 10% v/v 2-mercaptoethanol) and incubating in boiling water for 5 min. Samples were then stored at -80 °C until use.
Immunoblotting
Aliquots containing 20 μg protein were separated and were then electroblotted onto polyvinylidenedifluoride (PVDF) membrane (Cat# IPVH00010, Millipore) using a BioRad mini Trans-Blot cell. Immunobotting was carried out as described previously [39]. The primary antibodies used in this study include rabbit anti-Dicer polyclonal antibody (Cat# sc-30226, Santa Cruz), rabbit anti-myostatin polyclonal antibody (Cat# ab3239, Millipore), goat anti-FoxO3a polyclonal antibody (Cat# sc-9813, Santa Cruz), rabbit anti-HDAC4 polyclonal antibody (Cat# A00429, GenScript) and rabbit anti-SMAD7 polyclonal antibody (Cat# sc-11392, Santa Cruz). All primary antibodies were diluted 1:1000. Signals were revealed by using HRP-conjugated secondary antibodies (1:1000) for development using enhanced chemiluminescence. The Dicer band was found at ∼200 kDa, myostatin at ∼25 kDa, FoxO3a at ∼90 kDa, HDAC4 at ∼120 kDa and SMAD7 at ∼45 kDa.
RNA isolation
Approximately 50 mg of frozen skeletal muscle from individual animals was used for each sample. Total RNAs were extracted from frozen tissue homogenized in 1 mL of Trizol® reagent (Invitrogen) as described previously [39]. RNA quality was examined by measuring the ratio of OD260/OD280; only samples with OD260/OD280 >1.8 were used for further experiments. Further quality validation was done by running 2 μg of total RNA on a 1% agarose gel to ensure the presence of normally abundant 18S and 28S rRNAs.
Reverse transcription
RT-PCR of miRNAs was conducted according to Biggar et al. [19]. An aliquot of 5 μL total RNA (0.2 μg/μL) was added to 5 μL of stem-loop primer (250 nM). Sequences for stem-loop primers are listed in Table S1. To anneal the stem-loop primer to the target miRNA, a reaction was carried out as follows: 5 min at 95 °C, followed by 5 min at 60 °C. Reactions were then centrifuged and held on ice for 1 min before starting the reverse transcription. The amplification reaction was then carried out as follows: 16 °C for 30 min followed by 60 cycles of 20 °C for 30 s, 42 °C for 30 s, then 50 °C for 1 s and a final single cycle of 85 °C for 5 min.
PCR amplification
PCR was performed to amplify the mature miRNA targets. Forward primers for all miRNAs were designed based on conserved mature miRNA sequences and listed in Table S1. A universal primer (5′-CTCACAGTACGTTGGTATCCTTGTG-3′), complementary to the stem-loop sequence, was used as the reverse primer in all amplifications. Amplification was carried out using miRNA-specific cDNA template, miRNA-specific forward primer and universal reverse primer with the following program: 95 °C for 10 min, followed by 30 cycles of 95 °C for 15 s and 60 °C for 1 min, and then a final hold at 4 °C.
The amplified products were then mixed with SYBR® Green I nucleic acid gel stain (Invitrogen) and loaded onto 2.5% agarose for electrophoresis. The bands from the most dilute cDNA sample that yielded a visible product were used for quantification, ensuring that PCR products did not reach amplification saturation. All PCR products were sent to BioBasic (Markham, ON, Canada) for sequencing using a shortened universal reverse primer (5′-CTCACAGTACGTTGG-3′) to increase sequence coverage.
Quantification and statistics
Bands of proteins, miRNAs and 5S rRNA were visualized and analyzed using the ChemiGenius BioImaging System and the GeneTools software (Syngene, MD, USA). To correct any minor variations in sample loading, immunoblot band intensity in each lane was normalized against a strong band in the same lane stained by Coomassie blue, which showed constant intensity across all samples and was well-separated from the area of the gel containing the immunoreactive protein. For RT-PCR, all miRNA bands were standardized against the intensity of the corresponding 5S rRNA band from the same RNA sample. Statistical analysis was performed using the Students t-test with a sample size of n = 3–4. Expression differences were considered significant based on P < 0.05. Data are reported as mean standardized band densities ± SEM for skeletal muscle samples from euthermic control vs. torpid bats.
Authors’ contributions
KBS conceived the project, designed and coordinated the study. SFK carried out molecular genetic studies, participated in the sequence alignment. KKB carried out molecular genetic studies, immunoassays and data analysis. KKB drafted and revised the manuscript with the help of KBS. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgments
Thanks go to Dr. Don Thomas (Université de Sherbrooke) for providing bat tissues, Dr. Angela. Beye (St. Francis Xavier University) for insightful conversation regarding the physiological parameters of muscle atrophy in hibernators, and to Janet M. Storey for editorial review of the manuscript. This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (Grant No. 6793). KB Storey holds the Canada Research Chair in Molecular Physiology, KK Biggar held an NSERC postgraduate fellowship, and SF Kornfeld was supported by an NSERC undergraduate summer research award.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.09.001.
Appendix A. Supplementary material
Supplementary Fig. 1.
References
- 1.Bassel-Duby R., Olson E. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 2.Glass D. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Saccone V., Puri P. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010;6:48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochi E., Hirose T., Hiranuma K., Min S.K., Ishii N., Nakazato K. Elevation of myostatin and FOXOs in prolonged muscular impairment induced by eccentric contractions in rat medial gastrocnemius muscle. J Appl Physiol. 2009;108:306–313. doi: 10.1152/japplphysiol.00278.2009. [DOI] [PubMed] [Google Scholar]
- 5.Yang W., Zhang Y., Li Y., Wu Z., Zhu D. Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/Akt/GSK-3β pathway and is antagonized by insulin-like growth factor 1. J Biol Chem. 2007;282:3799–3808. doi: 10.1074/jbc.M610185200. [DOI] [PubMed] [Google Scholar]
- 6.Guller I., Russell A. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol. 2010;588:4075–4087. doi: 10.1113/jphysiol.2010.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung K., Sharp P. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schier A., Giraldez A. MicroRNA function and mechanism: insights from zebrafish. Cold Spring Harb Symp Quant Biol. 2006;121:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y., Jin Y. MicroRNA in cell differentiation and development. Sci China C Life Sci. 2010;52:205–211. doi: 10.1007/s11427-009-0040-5. [DOI] [PubMed] [Google Scholar]
- 11.Chan J., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 12.Ivey K., Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Biggar K.K., Storey K.B. The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol. 2011;3:167–175. doi: 10.1093/jmcb/mjq045. [DOI] [PubMed] [Google Scholar]
- 14.Guttilla I., White B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphries M.M., Thomas D.W., Kramer D.L. The role of energy availability in mammalian hibernation: a cost-benefit approach. Physiol Biochem Zool. 2003;76:165–179. doi: 10.1086/367950. [DOI] [PubMed] [Google Scholar]
- 16.Jonasson K.A., Willis C.K.R. Hibernation energetics of free-ranging little brown bats. J Exp Biol. 2012;215:2141–2149. doi: 10.1242/jeb.066514. [DOI] [PubMed] [Google Scholar]
- 17.Storey KB. Biochemical regulation of carbohydrate metabolism in hibernating bats. In Ruf T, Bieber C, Arnold W, Millesi E, editors. Living in a seasonal world: thermoregulatory and metabolic adaptations. Heidelberg: Springer 2012; p. 411–21.
- 18.Brigham R.M., Ianuzzo C.D., Hamilton N., Fenton M.B. Histochemical and biochemical plasticity of muscle fibers in the little brown bat (Myotis Iucifugus) J Comp Physiol B. 1990;160:183–186. doi: 10.1007/BF00300951. [DOI] [PubMed] [Google Scholar]
- 19.Biggar K.K., Kornfeld S.F., Storey K.B. Amplification and sequencing of mature microRNAs in uncharacterized animals models using stem-loop reverse transcription-polymerase chain reaction. Anal Biochem. 2011;416:231–233. doi: 10.1016/j.ab.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong R.B., Ianuzzo C.D., Kunz T.H. Histochemical and biochemical properties of flight muscle fibers in the little brown bar Myotis lucifugus. J Comp Phys B. 1977;119:141–154. [Google Scholar]
- 21.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessier S.N., Storey K.B. Expression of myocyte enhancer factor-2 and downstream genes in ground squirrel skeletal muscle during hibernation. Mol Cell Biochem. 2010;344:151–162. doi: 10.1007/s11010-010-0538-y. [DOI] [PubMed] [Google Scholar]
- 23.Brooks N.E., Myburgh K.H., Storey K.B. Myostatin levels in skeletal muscle of hibernating ground squirrels. J Exp Biol. 2011;214:2522–2527. doi: 10.1242/jeb.055764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K., So H., Gwag T., Ju H., Lee J.W., Yamashita M. Molecular mechanisms underlying muscle mass retention in hibernating bats: role of periodic arousal. J Cell Physiol. 2010;222:313–319. doi: 10.1002/jcp.21952. [DOI] [PubMed] [Google Scholar]
- 25.Lee K., Park J.Y., Yoo W., Gwag T., Lee J.W., Byun M.W. Overcoming muscle atrophy in a hibernating mammal despite prolonged disuse in dormancy: proteomic and molecular assessment. J Cell Biochem. 2008;104:642–656. doi: 10.1002/jcb.21653. [DOI] [PubMed] [Google Scholar]
- 26.Cohen T., Barrientos T., Hartman Z.C., Garvey S.M., Cox G.A., Tao T.P. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J. 2008;23:99–106. doi: 10.1096/fj.08-115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moresi V., Williams A.H., Meadows E., Flynn J.M., Potthoff M.J., McAnally J. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senf S.M., Dodd S.L., Judge A.R. FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. Am J Physiol Cell Physiol. 2010;298:C38–C45. doi: 10.1152/ajpcell.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2010;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 30.Choi H., Selpides P.J., Nowell M.M., Rourke B.C. Functional overload in ground squirrel plantaris muscle fails to induce myosin isoform shifts. Am J Physiol Regul Integr Comp Physiol. 2009;297:R19090–R19095. doi: 10.1152/ajpregu.00236.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews J., Zhang X., McCarthy J.J., McDearmon E.L., Hornberger T.A., Russell B. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok G., Sweetman D. Many routes to the same destination: lessons from skeletal muscle development. Reproduction. 2011;141:301–312. doi: 10.1530/REP-10-0394. [DOI] [PubMed] [Google Scholar]
- 33.Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M. The microRNA miR-181 targets the homeobox protein HOX-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 34.Kollias H.D., Perry R.L., Miyake T., Aziz A., McDermott J.C. SMAD7 promotes and enhances skeletal muscle differentiation. Mol Cell Biol. 2006;26:6248–6260. doi: 10.1128/MCB.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquez R.T., Bandyopadhyay S., Wendlandt E.B., Keck K., Hoffer B.A., Icardi M.S. Correlation between microRNA expression levels and clinical parameters associated with chornic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 36.Lee S.J., Reed L.A., Davies M.V., Girgenrath S., Goad M.E., Tomkinson K.N. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci U S A. 2005;102:18117–18122. doi: 10.1073/pnas.0505996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T., Wu R., Zhang Y., Zhu D. A systematic analysis of the skeletal muscle microRNA transciptome of chicken varieties with divergent skeletal muscle growth identifies novel microRNAs and differentially expressed microRNAs. BMC Genomics. 2011;12:186. doi: 10.1186/1471-2164-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy S.E., Storey K.B. P38 MAPK regulation of transcription factor targets in muscle and heart of hibernating bats Myotis lucifugus. Cell Biochem Funct. 2007;25:759–765. doi: 10.1002/cbf.1416. [DOI] [PubMed] [Google Scholar]
- 39.Biggar K.K., Storey K.B. Evidence for cell cycle suppression and microRNA regulation of cyclin D1 during anoxia exposure in turtles. Cell cycle. 2012;11:1705–1713. doi: 10.4161/cc.19790. [DOI] [PubMed] [Google Scholar]
- 40.Lui G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V.J. MiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2005;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Hassan M.Q., Jafferji M., Ageilan R.I., Garzon R., Croce C.M. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taipaleenmaki H., Bjerre Hokland L., Chen L., Kauppinen S., Kassem M. Mechanisms in endocrinology: microRNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–371. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- 44.Wada S., Kato Y., Okutsu M., Miyaki S., Suzuki K., Yan Z. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286:38456–38465. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safdar A., Abadi A., Aktar M., Hettinga B.P., Tarnopolsky M.A. MiRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57BI/6J male mice. PLoS One. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gambardella S., Rinaldi F., Lepore S.M., Viola A., Loro E., Angelini C. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. doi: 10.1186/1479-5876-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.