Abstract
Bioinformatic approaches have complemented experimental efforts to inventorize plant miRNA targets. We carried out global computational analysis of rice (Oryza sativa) transcriptome to generate a comprehensive list of putative miRNA targets. Our predictions (684 unique transcripts) showed that rice miRNAs mediate regulation of diverse functions including transcription (41%), catalysis (28%), binding (18%), and transporter activity (11%). Among the predicted targets, 61.7% hits were in coding regions and nearly 72% targets had a solitary miRNA hit. The study predicted more than 70 novel targets of 34 miRNAs putatively regulating functions like stress-response, catalysis, and binding. It was observed that more than half (55%) of the targets were conserved between O. sativa indica and O. sativa japonica. Members of 31 miRNA families were found to possess conserved targets between rice and at least one of other grass family members. About 44% of the unique targets were common between two dissimilar miRNA prediction algorithms. Such an extent of cross-species conservation and algorithmic consensus confers confidence in the list of rice miRNA targets predicted in this study.
Key words: miRNA, target prediction, conservation, consensus, rice
Introduction
MicroRNAs (miRNAs), a class of ~22-nucleotide non-coding transcripts, have been shown to play a significant role in plant biology as negative regulators of gene expression 1., 2.. Understanding the functions of these miRNAs needs identification and characterization of their target sequences as well as the affected phenotype. Presently, miRNA targets are known in Arabidopsis thaliana 3., 4., 5., 6., 7., Oryza sativa 8., 9., 10., Zea mays (2), Brassica napus (11), and Populus trichocarpa (12). Experimentally, miRNA functions (not mere target sequences) are studied either in mutants or by generating knockdown lines, both of which are difficult and complicated; moreover, such phenotypes are pleiotropic and the systems are not optimized in plants except Arabidopsis. Furthermore, miRNAs and their targets do not exist as 1:1 pairs, and the pairs are not constant across tissues and cell types and along the developmental stages. Hence, computational prediction, incorporating as many factors as possible that influence miRNA–mRNA interaction, assists in generating a set of miRNA targets upon which wet experiments can be planned.
Ever since plant miRNAs and their targets were first identified and characterized, bioinformatic approaches to plant miRNA target prediction, exemplified by miR171:SCL, have been considered straightforward owing to virtually perfect base pairing between miRNA and target sequence 6., 13., 14.. As a result of the stringent base pairing and the phylogenetic conservation employed in their prediction, both of which were considered absolutely essential, most of the plant miRNA target predictions have turned out be true and have been validated experimentally (1). As a consequence, it has been deduced that nearly 70% of the plant miRNA targets are transcription factors (TFs) and most plant miRNA targets are possibly all identified 1., 15.. On the flipside, however, it is likely that we may have overlooked targets with less stringent sequence match as well as those miRNA–target pairs that are species specific. Hence, it is essential to revisit the computational methodologies employed in plant miRNA target prediction, principally to assess the implications of stringent sequence match and to analyze the influence of cross-species conservation on the target repertoire.
The challenge, therefore, is to optimize target prediction algorithms to predict plant miRNA–target pairs with less extensive sequence match without deviating from the established principles of plant miRNA–target interaction. For instance, a pattern scan for 10 miRNAs of Arabidopsis detected 23 targets (16), whereas another algorithm, miRU, predicted as many as 203 potential targets (17). The downside of predicting non-canonical plant targets is the occurrence of false hits. Under such circumstances, there can be two in silico filters for target validation. The first filter is to ensure that the algorithm is not generating either lopsided targets or false hits by comparing the results of more than one target prediction algorithms. In plants, since the focus has been on stringent sequence match between miRNA and target, the need to develop and compare different algorithms was rarely perceived. The second filter is to ensure that targets are “conserved” across taxa to increase the confidence in the predicted targets.
Genetic and molecular approaches for the improvement of rice have helped establish rice as a model for plant functional genomic studies. We also know that how the growth and development of rice could be influenced by miRNA-mediated regulation 8., 9., 10.. However, despite the availability of whole genome sequences of two subspecies (indica and japonica), and robust and abundant genomic resources from rice as well as a number of species belonging to the same Poaceae family, a complete repertoire of rice genes regulated by miRNA mediation is yet to be established. The objective of the present study was to generate a comprehensive list of rice miRNA targets by carrying out computational prediction, internalizing some of the above-mentioned key factors like minimum sequence match 18., 19., conservation across taxa (1), and algorithmic consensus (20) to ascertain the influence of each of these components on the number and repertoire of rice miRNA targets. Our results support the prospect of predicting additional plant miRNA targets and we report more than 70 such novel miRNA–target pairs in rice that could have been ignored by an archetypal plant miRNA target prediction algorithm.
Results and Discussion
Validation of the computational algorithm
The miRanda scanning algorithm has been successfully used earlier 21., 22., 23.. However, the suitability of this algorithm to detect miRNA targets in plants was never verified. It was, therefore, critical for the present analysis to ascertain how miRanda could be employed in plants and what kinds of modifications are necessary. Based on known principles of plant miRNA–target interactions, we arrived at a set of filters to minimize false hits (see Materials and Methods for details). The reliability of this approach was tested on Arabidopsis, which has computationally and experimentally well worked out miRNA target lists. Our analysis predicted 582 Arabidopsis miRNA targets including multiple splice forms of the target transcripts (Table S1). The hits included all the 66 known miRNA–target pairs of Arabidopsis reported by 7 different studies (Table S2). Besides, it is equally important to ascertain that a prediction algorithm does not generate redundant and false hits. Our analysis produced only 330 unique miRNA–target pairs, which is equivalent to 1.14% of the input sequences (2.8 targets/miRNA). These observations showed that the algorithm employed in the study ensured adequate stringency while additional targets to the existing ones were generated.
Prediction and analysis of rice miRNA targets
Open access rice sequence data include nucleotide sequence entries, amino acid sequences, and unigenes. Since our work was confined to computational analysis, we wanted to avoid the input that might contain predicted mRNAs and false joining of expressed sequence tags (ESTs). Hence, we opted for the experimentally derived set of rice full-length mapped and annotated cDNA sequences. Out of 242 rice miRNA sequences available in the miRBase database (24), the miRanda-based methodology predicted 228 miRNA sequences to have targets among 32,127 full-length cDNA sequences explored. The hits of the rest miRNAs did not qualify the algorithm criteria or could not get through the filters, or the target sequences could be absent in the cDNA collection, since they do not represent the entire rice transcriptome. The predicted targets comprised of 684 unique cDNA sequences (2.13% of the total sequences scanned) with an average minimum energy of the duplex structure ≤ −30 kcal/mol and an average homology ≥ 89%. A list of these miRNAs and comprehensive annotations of the corresponding targets including chromosomal locations, mRNA and protein lengths, source tissue, start–stop positions of the alignment, location of hit, hit sequence, and putative functions are given in Table S3. From this long list of targets, a set of targets attached with high probability was short listed for researchers to consider carrying out experimental validation on priority (Table 1). These top predictions exhibited extensive sequence matching of miRNA–target pairs with total mismatches (including G–U pairs) not exceeding 3. They comprised of targets of 34 miRNAs, which were earlier predicted to mostly regulate TFs. Present analysis added a set of 73 novel miRNA–target pairs putatively regulating functions like stress-response (jacalin, stress-inducible protein, heat shock protein, and NBS-LRR protein), catalysis (flavin mono-oxygenase, multi-copper oxidase, CAAX protease, and fucosyl transferase), and binding (ATP-binding protein, Ca-binding protein, and RNA-binding protein).
Table 1.
High-probability rice miRNA targets proposed for experimental work
| No. | miRNA family | No. of target transcripts | Predicted function (Known) | Predicted function (Novel) | Mark* |
|---|---|---|---|---|---|
| 1 | miR156 | 13 | Squamosa promoter-binding protein SPL2, SPL9, SPL10 | Jakalin homolog of barley | C, K |
| 2 | miR159 | 10 | MYB family transcription factor MYB33, MYB65 | Inositol 1,3,4-trisphosphate 5/6-kinase family protein; calcium-binding protein | C |
| 3 | miR160 | 4 | Transcriptional factor B3 family protein | Far-red impaired responsive protein | C |
| 4 | miR164 | 10 | Transcription activator NAC1-No apical meristem (NAM) | Dihydrolipoamide S-acetyltransferase | C |
| 5 | miR166 | 8 | Homeobox-leucine zipper transcription factor (HB-14); homeodomain-leucine zipper protein Revoluta (REV) | Stress-inducible protein | C |
| 6 | miR167 | 3 | Probable leucine zipper; isoleucyl-tRNA synthetase | – | |
| 7 | miR168 | 8 | Argonaute protein (AGO1) | Quinone reductase family protein DNAJ heat shock N-terminal domain; flavin-containing monooxygenase family protein | K |
| 8 | miR169 | 12 | CCAAT-binding transcription factor | Glycine-rich RNA-binding protein (GRP7); leucine-rich repeat transmembrane protein kinase; multi-zipper protein | C |
| 9 | miR171 | 5 | Scarecrow-like transcription factor 6 (SCL6) | – | C |
| 10 | miR172 | 4 | Floral homeotic protein APETALA2 (AP2) | Starch synthase-related protein | C, K |
| 11 | miR319 | 4 | MYB family transcription factor MYB33, MYB65 | – | C |
| 12 | miR390 | 3 | Leucine-rich repeat family protein | – | |
| 13 | miR395 | 7 | Sulfate transporter, sulfate adenylyl-transferase 1/ATP-sulfurylase 1 (APS1) | – | C |
| 14 | miR396 | 10 | Transcription activator GRL1, GRL2, GRL3, GRL5 | Phytochrome A-related containing 7 WD-40 repeats; ATP-binding region containing non-consensus splice site | K |
| 15 | miR397 | 4 | Laccase | Pyruvate dehydrogenase E1 beta subunit, mitochondrial; diphenol oxidase | C |
| 16 | miR398 | 2 | Copper/zinc superoxide dismutase (CSD1) | – | |
| 17 | miR399 | 9 | Phosphate transporter (PT2) | Disease resistance protein (NBS-LRR class); pentatricopeptide (PPR) repeat-containing protein; DNAJ heat shock N-terminal domain | K |
| 18 | miR408 | 6 | – | Auxin-responsive AUX/IAA7 family protein; E2F transcription factor-3; multi-copper oxidase type I family protein; plastocyanin-like domain-containing protein/plantacyanin; helicase domain-containing protein; laccase | C, K |
| 19 | miR415 | 5 | – | Auxin-responsive AUX/IAA7 family protein; leucine-rich repeat family protein; AP2 domain-containing transcription factor; viviparous-14 protein (maize) | C, K |
| 20 | miR443 | 2 | – | Beta-expansin (EXBP2) | K |
| 21 | miR444 | 1 | Expressed protein supported by MPSS (similar to AT1G54385) | – | |
| 22 | miR445 | 2 | – | Transformer serine/arginine-rich ribonucleoprotein | K |
| 23 | miR446 | 20 | – | CAAX protease (STE24); DNA repair and recombination protein PIF1, mitochondrial pre-cursor; fucosyltransferase-like protein FucT2; glutaredoxin family protein; metallo-beta-lactamase family protein; PPR repeat-containing protein; C3HC4-type zinc finger family protein | K |
| 24 | miR528 | 7 | – | F-box family protein (ORE9) E3 ubiquitin ligase SCF; L-ascorbate oxidase; uclacyanin I | K |
| 25 | miR531 | 5 | – | Nodulin family protein; cell division cycle protein 48 (CDC48) | K |
| 26 | miR806 | 2 | L1P family of ribosomal protein | ATP-dependent protease domain-containing protein; epoxidehydrolase (ATsEH) | C |
| 27 | miR808 | 4 | Helicase associated domain; cytochrome P-450; cysteine protease; plant protein family | – | C |
| 28 | miR809 | 8 | Mlo (pathogen resistance) protein; helicase associated domain; new cDNA-based gene; zinc finger protein; F-box domain protein; isoflavone reductase; cytochrome P-450; plant protein family | GTP-binding regulatory protein beta chain; exportin-related protein; glutaredoxin family protein | C |
| 29 | miR812 | 3 | Protein kinase; glycosyl hydrolases; chloroplast import receptor | – | C |
| 30 | miR814 | 1 | Peroxidase | Nucleolar protein similar to proliferating-cell nucleolar antigen p120 | |
| 31 | miR815 | 5 | – | Zinc finger (C2H2-type) family protein; dentin sialophosphoprotein-type protein; exocyst complex subunit Sec15-like family protein; disease resistance protein (CC-NBS-LRR class); protein phosphatase 2C-like protein; 5′–3′ exoribonuclease XRN4 | C |
| 32 | miR818 | 25 | Serine threonine kinase; hydrolase; ENT domain; isoflavone reductase; leucine-rich repeat; new cDNA-basegene; pyruvate kinase | UDP-glucose:indole-3-acetate beta-D-glucosyl-transferase; suppressor of lin-12-like protein; MYB family transcription factor (MYB20); 3-hydroxy isobutyryl-coenzyme A hydrolase; 2′-hydroxy isoflavone reductase; beta-glucosidase; WRKY family transcription factor; probable DNA replication licensing factor; PPR repeat-containing protein; expressed protein similar to At1g70550; phospho inositide-specific phospholipase C | C |
| 33 | miR819 | 7 | Elongation factor; diacylglycerol kinase; strubble Ig receptor family; ABC transporter | Leucine-rich repeat family protein; probable LRR receptor-like protein kinase | |
| 34 | miR820 | 3 | DNA cytosine methyltransferase | WWE domain-containing protein |
The multiple hits conserved between indica and japonica rice subspecies are marked as “C”. Those targets that are predicted both by miRanda and miRU algorithms (consensus targets) are marked as “K”.
Location of predicted target sites on the transcript
miRNAs bind to complementary regions of mRNAs in a sequence-specific manner. In animals, almost all known miRNA target sites were found in 3′ untranslated regions (UTRs) of protein coding genes (20), whereas in plants they are only occasionally in 3′ UTRs but are predominantly in coding regions 3., 4., 5., 7., 16. and rarely reside in 5′ UTRs (16). Among rice targets, 61.7% belonged to coding regions whereas only 22% and 16.3% were in 3′ UTR and 5′ UTR regions, respectively. It was observed that miRNAs bringing about repression of multiple transcripts could target different regions of the transcripts. Among 86 such rice miRNAs that had more than one targets, 16 had targets only in one region, 31 targeted at least two regions, and the remaining 39 miRNAs could mediate regulation via binding to either of the 3′ UTR, 5′ UTR, or coding regions.
Spatial expression profile and genomic location of predicted miRNA targets
We categorized the predicted miRNA targets according to the tissues of the rice plant in which they express. It was observed that the number of miRNA targets identified in a particular tissue is directly proportional to the total number of cDNAs represented from that tissue in the analysis (Table 2). Proportion of cDNAs from each tissue source having miRNA target sites was comparable across tissues, for example, 2.6% (shoot) to 4.0% (panicle). Among all the tissues, callus shows minimum development related changes and hence we hypothesized that in order to maintain a bare minimum differentiation, callus might show conspicuous miRNA-mediated operation, since known actions of all the miRNAs are restrictive rather than amelioratory. This was indeed the case when top miRNA targets were analyzed. A total of 33.6% of such high-probability hits were expressed in callus, and not surprisingly, more than half of them were TFs such as CCAAT-binding TF, homeobox leucine zipper TF, MYB TF, and so on. Other examples of tissue-specific targets included scarecrow-like TF in nine flower and six shoot cDNAs (miR171), WD-40 repeat family protein in callus (miR396), PPR-containing protein in ABA-treated callus (miR399) and untreated callus (miR446). Information of this nature—where a relationship between known function, tissue, and possible involvement of miRNA-mediated regulation can be constructed—highlights the utility of computational target prediction in planning wet lab experiments.
Table 2.
Tissue-wise distribution of rice miRNA targets
| Source tissue | No. of miRNA targets | Fraction of miRNA targets (%) | Total cDNA sequences (bp) | Targets expressed as fraction of total cDNAs (%) |
|---|---|---|---|---|
| Shoot | 378 | 37.3 | 14,452 | 2.6 |
| Callus | 238 | 23.5 | 6,752 | 3.5 |
| Flower | 209 | 20.6 | 5,849 | 3.6 |
| Others | 98 | 9.7 | 2,750 | 3.6 |
| Panicle | 68 | 6.7 | 1,684 | 4.0 |
| Root | 22 | 2.2 | 640 | 3.4 |
| Total | 1,013 | 100 | 32,127 | 3.2 |
In addition, miRNAs and the predicted targets were mapped onto the 12 rice chromosomes using Karyoview software (http://www.gramene.org/Oryzaj_sativa/karyoview) (Figure 1). While this exercise showed that rice miRNAs and their targets are rather distributed across all the chromosomes, there are some regions that lack both miRNAs and target sequences (for example, short arms of chromosomes 4 and 9), which could be of particular interest for mining novel rice miRNAs and cognate targets.
Fig. 1.
Physical map of miRNA loci (red arrow heads, left) and predicted targets (blue arrow heads, right) on 12 rice chromosomes.
Multiple hits
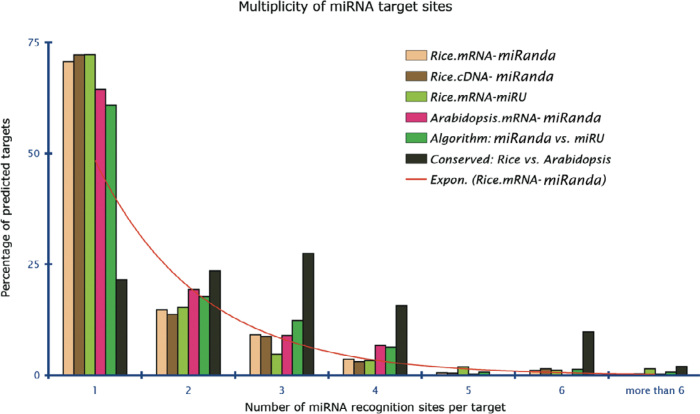
A single miRNA can regulate different mRNAs at different stages of growth or in different tissues with a common target site. In the present analysis, unique cDNA sequences of rice (684) corresponded to 6.9 targets per miRNA, allowing such a possibility. It is also known that each target, depending upon the magnitude of the downstream implication, could be targeted by multiple miRNA species to ensure stringent regulation. Nearly 72% of the hits were found to have a solitary target miRNA binding site. The fraction was more or less the same across the board. Sequences with two or three possible target sites were about 14% and 9%, respectively, and reduced further exponentially (Figure 2). In contrast, there were 21.6%, 23.5%, 27.5%, 15.7%, and 9.8% predicted targets conserved between rice and Arabidopsis that possessed 1, 2, 3, 4, and 6 recognition sites, respectively.
Fig. 2.
Multiplicity of miRNA target sites. Similar trend is observed in different sequence source–algorithm sets except for the targets conserved between rice and Arabidopsis.
Functional repertoire of rice miRNA targets
Putative functions of predicted miRNA targets were collected based on Arabidopsis homologues and PIR (Protein Information Resource) hits. The functions of the predicted miRNA targets include transcription regulator activity (MYB family TF, transcriptional factor B3 family protein, transcription activator NAC1 containing NAM domain, homeobox-leucine zipper TF HB-14, homeodomain-leucine zipper protein Revoluta, CCAAT-binding TF, scarecrow-like TF, floral homeotic protein APETALA2, GRL transcription activator, auxin-responsive AUX/IAA family protein, and C3HC4-type zinc finger family protein), catalytic activity (dihydrolipoamide S-acetyltransferase, inositol 1,3,4-trisphosphate 5/6-kinase family protein, far-red impaired responsive protein, isoleucyl-tRNA synthetase, laccase, putative/diphenol oxidase, and quinone reductase), and other activities such as structural molecule activity, ligand binding, and transporters (argonaute protein, glycine-rich RNA-binding protein, sulfate transporter, and NBS-LRR class disease resistance protein). Among those targets whose gene ontology (GO) terms for molecular function (www.geneontology.org) could be obtained, it was observed that as many as 41% exhibited catalytic function, 28% were transcription regulators, 18% performed binding activity, and 11% were transporters. On the whole, it was observed that rice also could be possessing functionally as diverse targets as those found in animal counterparts, if targets with relatively relaxed sequence match were also included. Besides categorization, the GO terms for molecular function revealed that miRNA families can often be specialized in mediating the regulation of distinct class of function. For instance, miR162, miR398, miR419, miR439, and miR535 were found to exclusively target catalytic activity whereas miR156, miR159, miR171, miR398, miR441, and miR445 were mainly involved in transcription regulation.
Cross-species conservation of miRNA–target pairs
Conservation of target sequences between rice subspecies
The cultivated rice (O. sativa) is classified into two primary subspecies, indica and japonica, based on the morphological and biochemical characters, hybrid sterility, and molecular analyses 25., 26., 27., 28.. Both subspecies are the products of separate domestication events from the ancestral species, O. rufipogon, and have evolved considerable genetic variation over the period of time 29., 30. in addition to differential genome sizes (indica 466 Mb and japonica 389 Mb). Indica (tropical) and japonica (temperate) have adapted to contrastingly different eco-geography experiencing independent genetic variation for ~0.44 million years, requiring extensive readjustments in genetic regulatory make-up 31., 32.. For instance, characteristics like photosensitivity, period of cultivation, and grain features greatly differ between indica and japonica rice cultivars. These differences are expected to be reflected in the variations in regulatory circuit including the miRNA-mediated genes. Hence, indica and japonica subspecies provide an excellent platform to assess the conservation of the miRNA–target pairs in rice. In this study, homologous indica sequences of every japonica rice miRNA target sequence were obtained by BLAST analysis. We found that out of 684 putative unique targets predicted in japonica rice, 339 (54.9%) miRNA–target pairs possessed homologues in indica rice (Table S4). Among the conserved targets for which GO terms for molecular function were known, it was observed that 49% of the targets were involved in catalytic functions, 23.2% were in the transcription regulation circuitry, whereas 15.5% and 9% were involved in binding and transporter activities, respectively.
Conservation of target sequences among members of the grass family
Rice belongs to the cereal and grass family (Poaceae). Considerable genomic resources of many other members of Poaceae are available including those of bread-wheat and maize. Assessment of conservation of miRNA–target combinations among the members of Poaceae could be highly informative towards understanding the nature of conserved miRNA targets. The miRanda algorithm was run individually on transcript sequences of each of the following species: maize (Zea mays), barley (Hordeum vulgare), oat (Avena sativa), bread-wheat (Triticum aestivum), sugarcane (Saccharum officinarum), sorghum (Sorghum bicolor), and 33 other grass species. Members of 31 miRNA families were found to possess conserved targets between rice and at least one of other grass members (Table S5). The conserved targets include regulatory proteins (SBP, GAMYB, heat shock protein, rolled leaf1, CCAAT-binding TF, floral homeotic APETALA, glossy15, indeterminate spikelet 1, and NB-ARC protein) as well as enzymes (glycosyltransferase, glutathione peroxidase, pyruvate dehydrogenase complex, beta-2-xylosyl transferase, calpain, DNA-directed RNA polymerase of chloroplast, superoxide dismutase, alcohol dehydrogenase, DNA-directed RNA polymerase-II, protein kinase, glucanase, and laccase).
Conservation of target sequences between rice and Arabidopsis
miRNAs and their target sequences are well worked out in Arabidopsis, both computationally and experimentally. Examination for conserved target sequences between rice and Arabidopsis yielded 146 miRNA–target combinations, of which 44 were proteins involved in transcription regulation. The results indicated that conservation of the miRNA–target pairs between rice and Arabidopsis was rather very low (7.5%; Table S6) compared with as many as 371 reported earlier (7). However, comparing the targets exclusively involved in transcription regulation, our results (126) match the 129 targets reported earlier (7) in terms of conserved targets between rice and Arabidopsis.
To ensure a high signal-to-noise ratio, bioinformatic approaches employed evolutionary conservation of the targets as one of the filters 33., 34.. In the present analysis, we computed the target conservation using different options. Since most of the efforts in plants were concentrated upon Arabidopsis, predicted targets were tested for cross-species conservation in rice 3., 5., 7.. Although such corroboration has helped in a way that most of the early predicted targets with high miRNA–mRNA sequence match and presence of a homologue in rice have been experimentally characterized 1., 6., it was contended that rice–Arabidopsis comparison may not be always feasible and would miss rice-specific miRNAs and targets (9). This was evident by the fact that in rice–Arabidopsis comparison, only 7.5% of the targets were conserved with putative functions as almost exclusively transcription regulation.
Algorithmic consensus
Diverse algorithms have been employed in target prediction; however, it is impractical to determine which one, if any, is the most reliable and sensitive target prediction method (20). A comparison of target predictions in animals concluded that those prediction methods with similar algorithms produced overlapping results whereas other algorithms generated entirely different sets of targets (20). This calls for employment of more than one algorithm in plant miRNA–target matches to ensure reliability of the target prediction. There is no instance of comparison of different miRNA target prediction algorithms for plants that can establish guidelines for rejection or selection of computationally predicted plant miRNA targets in the absence of experimental information. To determine consensus, we compared the targets generated by miRanda-based algorithm with the targets generated by miRU, a web server developed specifically to predict plant miRNA targets (17).
Although we had used rice full-length cDNA sequences for the miRNA target predictions, to compare the performance of miRanda-based method with that of miRU, we had to repeat the target prediction using miRanda on TIGR rice genome mRNA (OSA1 release 3, December 28, 2004) because miRU uses only this predefined set of mRNA sequences. There were 539 targets of 81 miRNAs common between the two algorithms (Table S7). We found that 43.7% of the unique targets to be common between the two algorithms. Additionally, unlike the conserved targets between rice and Arabidopsis where 86% of the sequences were involved in transcription regulation, the targets conserved between the two algorithms exhibited putative functions of transcription regulation (57%), catalysis (34%), transporters (7%), and binding (2%). These observations imparted confidence in the additional targets predicted in this study.
Conclusion
Plants, sessile creatures, need to deal with a variety of stimuli, particularly stress, from the biotic and abiotic environments, often in a tissue- or stage-specific fashion. These responses are complex but are under stringent regulation 35., 36., 37.. It is therefore plausible that many more hitherto unknown traits are regulated by miRNAs albeit with an effect not as dramatic as observed in the case of transcription factors. Since most of the targets exhibiting nearly perfect sequence complementarity to miRNAs are identified, additional targets, if any, are expected to be those with relatively more number of mismatches with miRNAs.
Our efforts to employ miRanda-based approach resulted in predicting additional rice miRNA targets involved in diverse functions, many of which are species-specific. Conservation filter narrowed down the number of targets (to <10% of the targets between rice and Arabidopsis and half between rice subspecies). On the other hand, we observed that the signal-to-noise ratio could also be effectively improved by computing consensus between algorithms. Our analysis resulted in the prediction of more than 70 novel miRNA–target pairs for immediate experimental validation.
Materials and Methods
Dataset
The miRNA sequences of O. sativa (242) and A. thaliana (117) were downloaded from miRBase database (http://microrna.sanger.ac.uk) (24). Full-length cDNA sequences (32,127) of O. sativa japonica were accessed at KOME database (http://cdna01.dna.affrc.go.jp/cDNA) (38). TIGR O. sativa japonica mRNA sequences (62,827) were downloaded from TIGR (http://www.tigr.org/tdb/e2k1/osa1/data_download.shtml), whereas O. sativa indica mRNA sequences (149,955) were downloaded from GenBank database (http://www.ncbi.nlm.nih.gov/entrez) by choosing filtering options as Taxonomy ID: 39946 and Molecule: mRNA. Transcript sequences of Z. mays (14,480), S. bicolor (110), T. aestivum (2,341), A. sativa (66), H. vulgare (1,157), S. officinarum (322), and various grass species (245) were downloaded from GenBank excluding genome survey sequences, EST sequences, sequence-tagged sites, third-party annotation sequences, working drafts, and patents. The cDNA sequences of A. thaliana (28,952) were downloaded from TIGR (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/sequences).
miRNA target prediction algorithms
The miRanda scanning algorithm (21), which utilizes dynamic-programming alignment and thermodynamics to predict miRNA targets, was employed in a stand-alone version 1.9 (http://www.microrna.org/miranda_new.html). The thresholds used for hit detection were: scaling factor set at 2.0 to ensure stringent complementarity at the first 11 positions (from 5′ end of miRNA) of the miRNA–mRNA duplexes; initial Smith-Waterman hybridization alignment with S > 95; and the minimum energy of the duplex structure ∆G ≤ −20 kcal/mol. Previous reports 9., 18., 19. have observed that pairing to the 5′ half of the miRNA (approximately positions 2 to 12, all nucleotide positions counted from 5′ end of miRNA) is vital, since this region exhibits nearly perfect complementarity and seldom more than one mismatch. Furthermore, mismatches, if exist, are typically absent at the putative cleavage site (positions 10 and 11) in almost all confirmed targets. Therefore, we introduced a condition that the hits possess at least 19 bp in length (allowing mismatches at the extremes) of sequence match starting at least from position 2, if not from the first, and with compulsory miRNA–mRNA matches at positions 2, 3, 4, 10, and 11. It was also ensured that the hits do not possess more than three mismatches in the miRNA–mRNA pair (excluding G–U pairs), and specifically, hits with either two consecutive mismatches or with two mismatches separated by just one match or with gaps (indels) in the sequence match are shifted out.
We employed miRU (17), a plant microRNA potential target finder to compute the algorithmic consensus. Since miRU was not available as a stand-alone version, target predictions were carried out using the web interface (http://bioinfo3.noble.org/miRNA/miRU.htm). miRU provides options of minimum alignment score, maximum number of G–U wobble pairs, maximum number of indels, maximum number of mismatches, and length of miRNA (19–28 bases). All the options were maintained at the lowest stringency levels to get maximum possible hits. The input options were: score for each 20 nt = 3; G–U wobble pairs = 6; indels = 1; other mismatches = 3. The dataset is TIGR rice genome mRNA (OSA1 release 3, December 28, 2004). As the program was run on the web interface, each miRNA was input one by one to get online results in html files. These html files were converted to text files using web2text program (http://www.jetman.dircon.co.uk/software/web2text.html).
Sequence processing and analysis
All the computational analyses were carried out on UNIX-based Darwin terminal of a 1.67 GHz PowerPC G4 running on Mac OS X (version 10.4.6), and accordingly the compatible algorithms and software were utilized. Homology detection using BLASTN was carried out on a stand-alone version of the NCBI BLAST package on a Sun grid engine (LINUX platform). Certain specific text editing operations were carried out on Solaris 8.0 platform.
Authors’ contributions
SA conceived the study and carried out the computational analysis. JN provided the guidance. SA and JN prepared the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
JN was funded by the Department of Biotechnology, Government of India under Centre of Excellence programme grant. SA was supported by the Indian Council of Agricultural Research in the form of study leave.
Supporting Online Material
Tables S1–S7
References
- 1.Jones-Rhoades M.W. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B. Computational identification of microRNAs and their targets. Comput. Biol. Chem. 2006;30:395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Adai A. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet E. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Rhoades M.W. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 7.Wang X.J. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y.C. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006;580:5111–5116. doi: 10.1016/j.febslet.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Sunkar R. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J.F. Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 2004;32:1688–1695. doi: 10.1093/nar/gkh332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie F.L. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 12.Lu S. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llave C. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart B.J. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai E.C. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallory A.C. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. Embo J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwab R. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Rajewsky N. MicroRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 21.Enright A.J. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraldez A.J. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 23.John B. Human microRNA targets. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 25.Second G. Origin of the genic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isoenzyme loci. Jpn. J. Genet. 1982;57:25–57. [Google Scholar]
- 26.Harushima Y. Diverse variation of reproductive barriers in three intraspecific rice crosses. Genetics. 2002;160:313–322. doi: 10.1093/genetics/160.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J., Bennetzen J.L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang T. Genomic variation in rice: genesis of highly polymorphic linkage blocks during domestication. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng C. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- 30.Londo J.P. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao L.Z. Microsatellite diversity within Oryza sativa with emphasis on indica-japonica divergence. Genet. Res. 2005;85:1–14. doi: 10.1017/s0016672304007293. [DOI] [PubMed] [Google Scholar]
- 32.Morishima H., Oka H.I. Phylogenetic differentiation of cultivated rice. XXII. Numerical evaluation of the indica-japonica differentiation. Japan. J. Breed. 1981;31:402–413. [Google Scholar]
- 33.Grün D. MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1 doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lall S. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 35.Conrath U. Priming: getting ready for battle. Mol. Plant Microbe Interact. 2006;19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- 36.Lipka V., Panstruga R. Dynamic cellular responses in plant-microbe interactions. Curr. Opin. Plant Biol. 2005;8:625–631. doi: 10.1016/j.pbi.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Kikuchi S. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S7