Abstract
We have used a bioinformatics approach for the identification and reconstruction of metabolic pathways associated with amino acid metabolism in human mitochondria. Human mitochondrial proteins determined by experimental and computational methods have been superposed on the reference pathways from the KEGG database to identify mitochondrial pathways. Enzymes at the entry and exit points for each reconstructed pathway were identified, and mitochondrial solute carrier proteins were determined where applicable. Intermediate enzymes in the mitochondrial pathways were identified based on the annotations available from public databases, evidence in current literature, or our MITOPRED program, which predicts the mitochondrial localization of proteins. Through integration of the data derived from experimental, bibliographical, and computational sources, we reconstructed the amino acid metabolic pathways in human mitochondria, which could help better understand the mitochondrial metabolism and its role in human health.
Key words: mitochondria, metabolic pathways, amino acid metabolism, human
Introduction
Reconstruction of the metabolic maps of fully sequenced organisms is becoming a task of major importance in order to help biologists identify and analyze the maps of newly sequenced organisms and newly found pathways (1). There are several online databases that provide information on metabolic content and pathway diagrams for a majority of the fully sequenced microbes and eukaryotic species. Databases such as EcoCyc (http://ecocyc.org) describe the pathways of a single organism (Escherichia coli), while others such as KEGG (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/) and MetaCyc (Encyclopedia of Metabolic pathways; http://metacyc.org) provide pathway information on multiple organisms including prokaryotic and eukaryotic species. However, none of these resources emphasize on the subcellular localization of metabolic pathways, which is an important part of pathway analysis given that many biological pathways are distributed across more than one subcellular location. Studies of this nature hold potential value to biomedical research areas such as drug targeting, reconstruction of disease pathways, understanding protein interaction networks, and annotation of new or alternative pathways.
A number of methods have been used for pathway reconstruction by integrating a combination of data determined by experimental and computational means 2., 3., 4.. Most of these methods have been developed to understand metabolic networks in prokaryotic systems. However, reconstruction and functional characterization of ATP production, heme synthesis, and mixed phospholipid synthesis have been reported in human mitochondria (5). Mitochondria are primarily known as energy centers, while they also play vital roles in the metabolism of many other biological macromolecules including amino acids. Amino acid metabolism has been well studied in the context of cellular metabolism, but specific pathways elucidating their subcellular localization to mitochondrial organelles have not been reported before. Out of 20 amino acids, the metabolic pathways of 17 amino acids utilize mitochondrial enzymes, and dysfunction of these enzymes is the causative factor for over 40 known mitochondrial diseases or disorders in human (partly in Table 1).
Table 1.
Diseases associated with the enzymes involved in the metabolism of amino acids in human mitochondria
| Pathway | EC number | Disease or disorder | OMIM ID |
|---|---|---|---|
| Arginine degradation | 2.1.1.2 | Guanidinoacetate methyltransferase deficiency | 601240 |
| Arginine degradation | 3.5.3.1 | Argininemia | 207800 |
| Arginine metabolism | 2.1.3.3 | Ornithine transcarbamylase deficiency | 311250 |
| Arginine synthesis | 2.6.1.13 | Ornithinemia with gyrate atrophy of choroid and retina | 258870 |
| Asparatate/asparagine metabolism | 6.4.1.1 | Pyruvate carboxylase deficiency | 266150 |
| Glutamate metabolism | 1.4.1.3 | Hyperinsulinism-hyperammonemia syndrome (HHS) | 606762 |
| Glutamate/glutamine synthesis | 1.5.1.12 | Hyperprolinemia II | 239510 |
| Glutamine degradation | 1.2.1.24 | 4-hydroxybutyricaciduria | 271980 |
| Glutamine degradation | 2.6.1.19 | GABA-transaminase deficiency | 137150 |
| Glutamine degradation | 4.1.1.15 | Pyridoxine-dependent infantile convulsions | 266100 |
| Glycine degradation | 1.1.1.79 | Hyperoxularia primary type II (HP 2) | 260000 |
| Glycine degradation | 1.4.4.2 | Nonketotic hyperglycinemia (NKH) | 605899 |
| Glycine degradation | 2.1.2.10 | NKH | 605899 |
| Isoleucine degradation | 6.4.1.3 | Beta-methylcrotonylglycinuria type II | 210210 |
| Leucine degradation | 1.3.99.10 | Isovalericacidemia | 243500 |
| Leucine degradation | 2.8.3.5 | Infantile ketoacidosis | 245050 |
| Lysine degradation | 1.1.1.35 | Hydroxyl-CoA dehydrogenase deficiency | 300438 |
| Lysine degradation | 1.5.1.8 | Hyperlysinemia | 238700 |
| Lysine degradation | 2.3.1.61 | Maple syrup urine disease type II | 248610 |
| Lysine degradation | 2.3.1.9 | Alpha-methylacetoaceticaciduria | 203750 |
| Lysine degradation | 4.2.1.17 | Trifunctional protein deficiency, type 2 | 607037 |
| Lysine degradation | 4.2.1.17 | 3-methylglutaconicaciduria | 250950 |
| Methionine degradation | 1.2.4.4 | Maple syrup urine disease | 248600 |
| Methionine degradation | 5.4.99.2 | Methylmalonicaciduria I (B12-unresponsive) | 251000 |
| Methionine degradation | 6.4.1.3 | Propionicacidemia | 606054 |
| Phenylalanine metabolism | 1.14.16.1 | Phenylketonuria | 261600 |
| Proline metabolism | 1.5.1.12 | Hyperprolinemia II | 239510 |
| Serine degradation | 2.6.1.44 & 2.6.1.51 | Oxalosis I (glycolicaciduria) | 259900 |
| Serine metabolism | 2.1.2.10 | NKH | 605899 |
| Serine metabolism | 1.1.1.79 | Hyperoxaluria primary type II | 260000 |
| Threonine degradation | 1.4.3.4 | Brunner syndrome | 309850 |
| Tyrosine metabolism | 1.11.1.8 | Defect in thyroid hormonogenesis II | 274500 |
| Valine degradation | 1.1.1.31 | 3-hydroxyisobutyricaciduria | 236795 |
| Valine degradation | 1.3.99.2 | Short-chain acyl-CoA dehydrogenase deficiency | 201470 |
| Valine degradation | 1.3.99.2 | Medium-chain acyl-CoA dehydrogenase deficiency | 201450 |
| Valine degradation | 5.4.99.2 | Methylmalonicaciduria I (B12-unresponsive) | 251000 |
| Val/Leu/Ile degradation | 1.2.1.3 | Sjoegren-Larsson syndrome (SLS) | 270200 |
| Val, Leu, Ile degradation | 1.2.4.4 | Maple syrup urine disease | 248600 |
The main goal of this study is to identify and annotate complete or partial segments of mitochondrial pathways associated with amino acid metabolism in human, and reconstruct these pathways by filling in the gaps. We use a sequence-based homology approach to identify mitochondrial pathways from KEGG, search for literature-based annotations and data from isozyme studies to systematically identify the reactions, and also employ computational prediction methods to identify proteins targeted to mitochondria. By effectively integrating all these data, we have reconstructed a majority of the pathways associated with amino acid metabolism in human mitochondria.
Results
Identification of amino acid pathways associated with human mitochondria
A non-redundant set of human mitoproteome was compiled from experimental, bibliographical, and computational sources (see Materials and Methods). KEGG database is the most comprehensive resource on metabolic pathways, which builds maps based on the Enzyme Classification (EC) numbers. In human, KEGG mapping is available only for about 3,000 enzymes, since it fails to map the enzymes lacking EC numbers. We have used a homology-based approach to find the amino acid pathways associated with mitochondria by searching the human mitoproteome set against KEGG’s human proteome set.
Reconstruction of amino acid pathways associated with human mitochondria
Plants and some bacteria are capable of synthesizing all the 20 amino acids necessary for the formation of proteins and other vital molecules; however, that is not the case in mammals including human. Human can synthesize only 11 amino acids known as “non-essential amino acids” while the other 9 (essential amino acids) need to be taken in the form of food. Hence, essential amino acids have only catabolic pathways while the non-essential ones have both anabolic and catabolic pathways. All the 20 amino acids except histidine, alanine, and cysteine have metabolic pathways associated with mitochondria. Nevertheless, these three amino acids are eventually converted into pyruvate in cytosol, which enters into mitochondria for consumption by the tricarboxylic acid (TCA) cycle. Below, we provide the reconstructed metabolic pathways and pathway maps for those amino acids associated with mitochondria. Note that some of them are grouped together because they are connected interdependently.
Lysine degradation
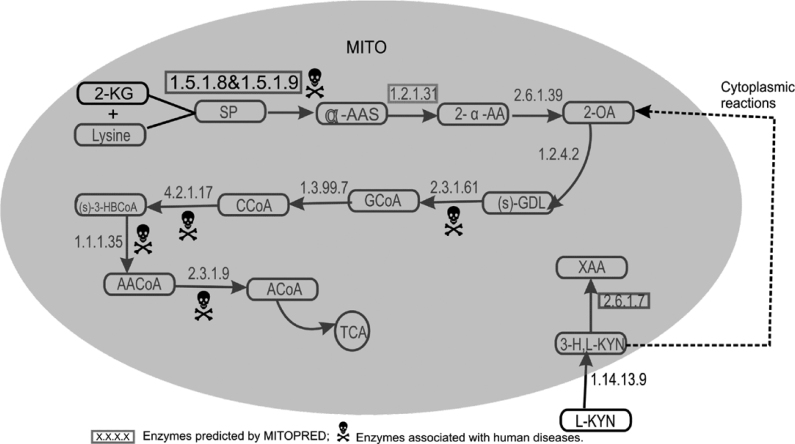
In human, lysine degradation is known to occur both in mitochondria and cytosol, and here we describe only the mitochondrial pathway (Figure 1). Lysine is transported into mitochondria by ornithine carrier that has two isoforms in human. Inside mitochondria, lysine together with 2-ketoglutarate is first reduced to saccharopine by lysine-ketoglutarate reductase (EC 1.5.1.8) and then to α-aminoadipate semialdehyde (α-AAS) by saccharopine dehydrogenase (EC 1.5.1.9). In human, there exists a bifunctional enzyme that can catalyze both reactions. This protein is predicted as mitochondrial by MITOPRED (100%) and its mouse orthologue is also believed to be mitochondrial (6).
Fig. 1.
Lysine–Tryptophan metabolism. 2-KG, 2-ketoglutarate; SP, saccharopine; α-ASS, α-aminoadipate semialdehyde; 2-α-AA, 2-α-aminoadipate; 2-OA, 2-oxoadipate; S-GDL, S-glutaryldihydrolipoamide; GCoA, glutaryl-CoA; CCoA, crotonoyl-CoA; (S)-3-HBCoA, (S)-3-hydroxybutanoyl CoA; AACoA, acetoacetyl-CoA; ACoA, acetyl-CoA; TCA, tricarboxylic acid cycle; XAA, xanthurenic acid; 3-H,L-KYN, 3-hydroxy L-kynurenine; L-KYN, L-kynurenine.
The next step is the conversion of α-AAS into 2-α-aminoadipate (2-α-AA) by aminoadipate semialdehyde dehydrogenase (EC 1.2.1.31). The yeast orthologue of this human protein (7) is predicted by MITOPRED as mitochondrial (100%). The next enzyme 2-aminoadipate transaminase (EC 2.6.1.39) that catalyzes the conversion of 2-α-AA into 2-oxoadipate (2-OA) is localized in mitochondria. Eventually 2-OA is converted into acetyl-CoA by a series of previously well-known mitochondrial enzymes (8) as shown in Figure 1.
Tryptophan degradation
Degradation of tryptophan takes place partly in cytoplasm and partly in mitochondria. First, tryptophan is catalyzed by two cytoplasmic enzymes (EC 1.13.11.11 and EC 3.5.1.9) to produce L-kynurenine, which is further converted into 3-hydroxy L-kynurenine by kynurenine monooxygenase (EC 1.14.13.9), an outermembrane mitochondrial enzyme (8). Under normal conditions, 3-hydroxy Lkynuranine is converted into 2-OA by a series of cytoplasmic reactions, which is further transported into mitochondria by oxodicarboxylate carrier in a counter-exchange mechanism with 2-oxoglutarate. Eventually 2-OA is converted into acetyl-CoA as shown in the lysine degradation pathway (Figure 1). Under infectious conditions, 3-hydroxy L-kynurenine is converted into xanthurenic acid by kynurenine aminotransferase-II (EC 2.6.1.7). The same enzyme also catalyzes the conversion of L-kynurenine into kynurenic acid in cytoplasm for which two known isoforms have been identified in human brain. One of the isoforms is predicted as mitochondrial (99%) by MITOPRED and its mouse orthologue has a mitochondrial isoform in brain tissue (9).
Phenylalanine degradation
Phenylalanine is utilized or degraded in mitochondria by two different pathways. The first one is the formation of phenylalanyl-tRNA by phenylalanyl-tRNA synthetase (EC 6.1.1.20). In the second pathway, it is converted into tyrosine by phenylalanine-4-hydroxylase (PAH, EC 1.14.16.1) (10), which is predicted by MITOPRED as mitochondrial (100%). Gene mutation in PAH is associated with an autosomal recessive disease known as phenylketonuria (Table 1).
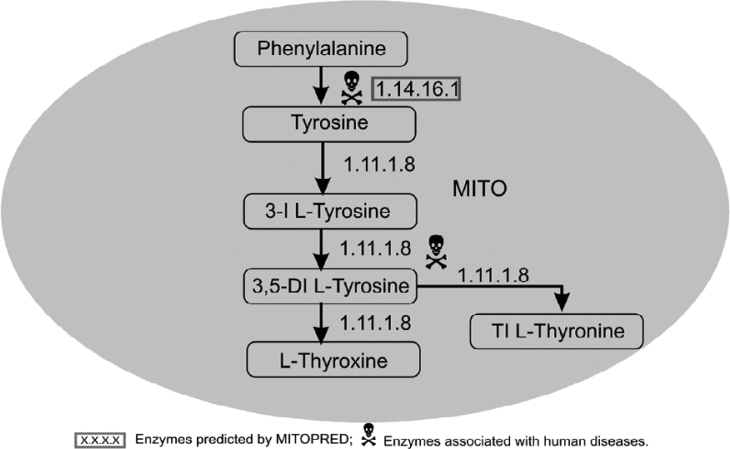
Tyrosine metabolism
Tyrosine is converted into tyrosyl-tRNA by a potential mitochondrial isoform (as per the Swiss-Prot database) of tyrosyl-tRNA synthetase (EC 6.1.1.1) that is utilized in mitochondrial protein synthesis. In another pathway (Figure 2), tyrosine is converted into thyroxine in a series of reactions, all of which are catalyzed by thyroid peroxidase (EC 1.11.1.8). The mitochondrial localization of this protein is currently unknown; however, thyroid peroxidase activity and its intermediate products have been reported in human mitochondrial fractions (11). Moreover, there are eight known isoforms to this enzyme, six of which are predicted as mitochondrial by MITOPRED at various confidence levels ranging from 62% to 77%.
Fig. 2.
Phenylalanine–Tyrosine metabolism. 3-I L-Tyrosine, 3-Ido L-Tyrosine; 3,5-DI L-Tyrosine, 3,5-Diido L-Tyrosine; TI L-Tyronine, Triido L-Tyronine.
Glycine metabolism
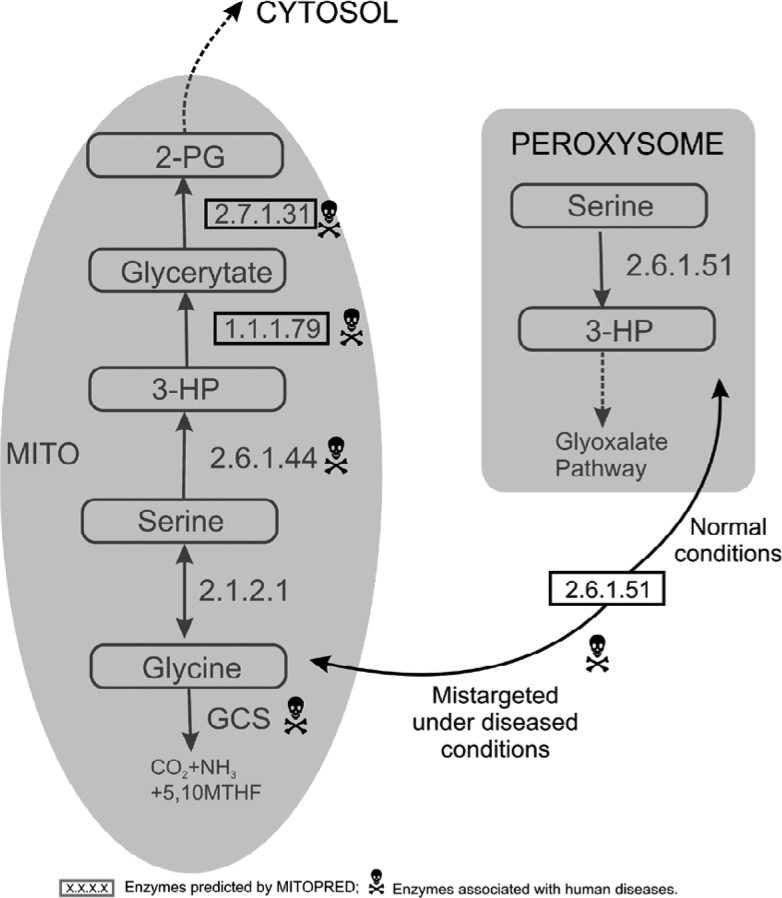
Glycine cleavage system (GCS) is a multienzyme complex comprised of glycine decarboxylase (P-protein, EC 1.4.4.2), aminomethyltransferase (T-protein, EC 2.1.2.10), lipoamide dehydrogenase (L-protein, EC 1.8.1.4), and a cosubstrate component H protein (GCSH). This complex catalyzes oxidative cleavage of glycine in a multistep reaction in almost all organisms. GCS is loosely bound to the mitochondrial inner membrane and the reaction is catalyzed in the mitochondrial matrix. Glycine degradation is catalyzed by GCS in the presence of nicotinamide adenine dinucleotide (NAD) and tetrahydrofoliate (THF), resulting in the production of CO2, ammonia, 5,10-methylene THF, and NADH (reduced form of NAD) as byproducts in multiple reactions (Figure 3). In an alternative pathway, glycine degradation and synthesis are reversibly catalyzed by serine hydroxymethyl-transferase (EC 2.1.2.1), which is a pyridoxal phosphate dependent enzyme existing as both cytosolic and mitochondrial isoforms.
Fig. 3.
Serine-Glycine metabolism. 3-HP, 3-hydroxypyruvate; 2-PG, 2-phosphoglycerate; GCS, glycine cleavage system; MTHF, methylene tetrahydrofoliate.
Serine metabolism
There are three mitochondrial pathways involved in the degradation or utilization of L-serine in mammals (Figure 3), and considerable differences exist among species in the relative contributions of these three pathways. The first pathway operates inside the mitochondrial matrix (8) in a three-step process. Serine is first converted into 3-hydroxypyruvate (3-HP) by the mitochondrial alanine-glyoxylate transaminase (AGT2, EC 2.6.1.44), followed by reduction of 3-HP to glycerate by hydroxypyruvate dehydrogenease (EC 1.1.1.79). Dysfunction of the second enzyme leads to hyperoxaluria type II disease in human (Table 1), which has been identified as mitochondrial by MITOPRED at 70% confidence. It was proposed that the intermediates of this pathway are exchanged between mitochondria and cytoplasm since the previous and the next enzymes in the pathway are known to be mitochondrial (12). Finally, glycerate is phosphorylated into 2-phosphoglycerate by mitochondrial glycerate kinase (EC 2.7.1.31) before it is transported into cytosol to take part in glycolysis. In human, two putative isoforms, glycerate kinase 1 (GLYCK1) and glycerate kinase 2 (GLYCK2), have been identified, but their subcellular locations remain unknown. However, both proteins have been predicted by MITOPRED as mitochondrial with 62% and 85% confidence, respectively. In rat liver, this enzyme is known to be localized in mitochondria and cytosol at 72% and 26% proportions, respectively (13).
In the second pathway, serine:pyruvate/alanine: glyoxylate aminotransferase (SPT/AGT, EC 2.6.1.51) converts serine into 3-HP that enters into glyoxylate pathway under normal conditions. SPT/AGT exists in the mitochondria of carnivores, the peroxisomes of herbivores and human, and both the mitochondria and peroxisomes of rodents. In human, it was reported that AGT has lost its mitochondrial targeting signal by a point mutation in the initiation codon and is localized to peroxisomes under normal conditions (14). Nevertheless, it is mistargeted to mitochondrial matrix in patients with primary hyperoxaluria type I (PH1), an inborn error of glyoxylate metabolism characterized by increased excretion of oxalate and glycolate (Table 1). The third pathway in serine metabolism is the same as the alternative metabolism of glycine synthesis and degradation.
Valine, leucine, and isoleucine degradation
Valine, leucine, and isoleucine are branched-chain amino acids (BCAAs) and their degradation pathways are predominantly localized in mitochondria except the first transamination step, which occurs in cytoplasm (8). Cytoplasmic transamination steps on BCCAs result in the production of corresponding α-keto acids that are transported into mitochondria by mitochondrial carnitine shuttle mechanism. Inside mitochondria, the first step is the irreversible oxidative decarboxylation of α-keto acids by a branched-chain α-keto acid dehydrogenase complex (EC 1.2.4.4 and EC 2.3.1.–), yielding three different CoA derivatives, that is, 3-methylbutanoyl-CoA, isobutyryl-CoA, and 2-methylbutanoyl-CoA for leucine, valine, and isoleucine keto acids. In the case of leucine, 3-methylbutanoyl-CoA is converted into 3-methylbut-2-enoyl-CoA by isovaleryl-CoA dehydrogenase (EC 1.3.99.10), while the acyl-CoAs of valine and isoleucine are converted into methacrylyl-CoA and trans-2-methylbut-2-enoyl-CoA respectively by butyryl-CoA dehydrogenase (EC 1.3.99.2). Another mitochondrial enzyme, medium chain acyl-CoA dehydrogenase (EC 1.3.99.3), also catalyzes the above reactions for all the three acyl-CoAs, which is the first reaction in the β-oxidation of mitochondrial fatty acids. Enzymes involved in the rest of these pathways resulting in the production of acetyl-CoA in leucine and isoleucine degradation as well as succinyl-CoA in valine degradation have been thoroughly illustrated in the literature (8), and hence are not discussed here in detail.
Threonine degradation
In mammals, there are two major L-threonine degradation pathways. L-threonine is catabolized either by L-serine/threonine dehydratase (SDH, EC 4.2.1.16) in cytoplasm or through the L-threonine 3-dehydrogenase (TDH, EC 1.1.1.103) pathway in cytoplasm or mitochondria, independently. In the second pathway, L-threonine is cleaved by TDH to produce 2-amino-3-ketobutyrate, which is further converted into glycine and acetyl-CoA by aminoacetone synthetase (AAS, EC 2.3.1.29). However, in human, only AAS is found to contain mitochondrial import sequence and TDH turns out to be a pseudogene due to one or more mutations that disrupt RNA splicing (15), indicating that this pathway is non-functional in human. In an alternative pathway, 2-amino-3-ketobutyrate produced in cytoplasm is spontaneously decarboxylated to aminoacetone (MetaCyc) and transported across mitochondrial membranes, where it is converted into 2-oxopropanol by monoamine oxidase (EC 1.4.3.4), an outer mitochondrial membrane enzyme (16). 2-oxopropanol is further converted into pyruvate by aldehyde dehydrogenase (EC 1.2.1.3) in mitochondria and is consumed by the TCA cycle.
Methionine degradation
This pathway spans across cytoplasm and mitochondria. In cytoplasm, methionine is activated in an ATP-dependent reaction to form S-adenosyl methionine (SAM), which is the major donor of methyl groups in biosynthetic methylations. In a series of cytoplasmic reactions, SAM is converted into α-ketobutyrate, which enters into mitochondria (MetaCyc). In mitochondria, α-ketobutyrate is converted into propionyl-CoA by the mitochondrial α-ketobutyrate dehydrogenase (EC 1.2.4.4) (17). Propionyl-CoA is converted into D-methylmalonyl-CoA by propionyl-CoA carboxylase (EC 6.4.1.3), then into L-methylmalonyl-CoA by methylmalonyl-CoA racemase (EC 5.1.99.1), and finally into succinyl-CoA by methylmalonyl-CoA mutase (EC 5.4.99.2) that enters into the TCA cycle.
Glutamate and glutamine metabolism
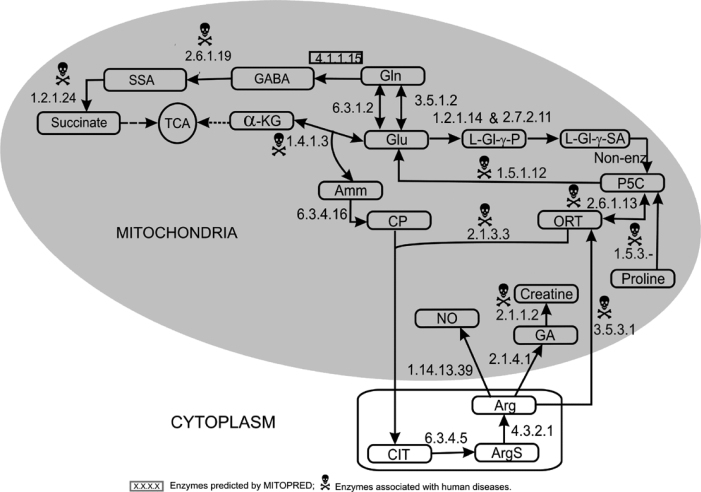
Glutamate/glutamine (Glu/Gln) metabolism is carried out in different subcellular locations depending on the tissue type. In mitochondria, Glu/Gln is synthesized in four different ways as described in Figure 4: (1) Glutamate is formed by the reductive amination of α-ketoglutarate (from the TCA cycle); (2) Glutamine and glutamate are interconverted in an ATP-requiring reaction catalyzed by glutamine synthetase (EC 6.3.1.2); (3) The previous reaction with ammonia as a byproduct is also catalyzed in a non-ATP requiring reaction by glutaminase (EC 3.5.1.2) in human brain, liver, and kidney tissues; (4) The fourth precursor (an intermediate from proline metabolism) for the synthesis of Glu/Gln is 1-pyrroline-5-carboxylate (P5C), which is catalyzed by 1-pyrroline-5-carboxylate dehydrogenase (P5CDh) (EC 1.5.1.12) in mitochondrial matrix.
Fig. 4.
Glutamate–Glutamine–Arginine–Proline metabolism. Glu, glutamate; Gln, glutamine; GABA, gamma-aminobutyric acid; SSA, succinate semialdehyde; α-KG, α-ketoglutarate; L-Gl-γ-P, L-glutamyl-γ-phosphate; L-Gl-γ-SA, L-glutamyl-γ-semialdehyde; P5C, pyrroline-5-carboxylate; ORT, ornithine; Amm, ammonia; CP, carbomyl phosphate; CIT, citrulline; ArgS, argininosuccinate; Arg, arginine; NO, nitric oxide; GA, guanidinoacetate.
Degradation of Glu/Gln takes place via two different pathways associated with mitochondria. The first one is the conversion of glutamate into α-ketoglutarate and ammonia by glutamate dehydrogenase (EC 1.4.1.3) that enters into the TCA cycle. In the second pathway, glutamine is converted into succinate in a three-step reaction, resulting in the production of 4-aminobutanoate (GABA) and succinate semialdehyde as intermediaries. This is a tissue-specific pathway that occurs in liver, brain, and pancreatic islets (18). The enzymes involved in these three successive reactions are glutamate decarboxylase (GAD, EC 4.1.1.15), 4-aminobutyrate aminotransferase (GABAT, EC 2.6.1.19), and succinate semialdehyde dehydrogenase (SSADH, EC 1.2.1.24), respectively. In human, GAD has two isoforms (GAD65 and GAD67) encoded by individual genes, and GAD67 is predicted as mitochondrial by MITOPRED (70%). In rat, GAD activity has been reported in mitochondria (19). GABAT and SSADH are known as mitochondrial enzymes.
Proline metabolism
Proline oxidase (EC 1.5.3.–) catalyzes the first step in the degradation of proline and converts proline into P5C in mitochondria, which is further degraded into glutamate and explained in Glu/Gln metabolism (Figure 4). P5CDh deficiency is associated with Type II hyperprolinemia, an autosomal recessive disorder characterized by accumulation of P5C and proline (Table 1).
Arginine metabolism
Synthesis of arginine is carried out in cytoplasm. However, the precursors required for this process originate in mitochondria by the degradation of three other amino acids, that is, glutamine, glutamate, and proline (Figure 4). Glutamate is converted into L-glutamyl-γ-phosphate followed by L-glutamyl-γ-semialdehyde by a bifunctional enzyme, P5C synthetase (EC 1.2.1.41 and EC 2.7.2.11). A spontaneous non-enzymatic reaction converts L-glutamyl-γ-semialdehyde into Δ-P5C, which is also produced by proline oxidase in proline degradation. P5C is converted into ornithine by ornithine aminotransferase (EC 2.6.1.13). Ornithine together with carbomyl phosphate (CP) produces citrulline, and CP is produced in mitochondria by the degradation of glutamate in a two-step process. The first reaction is catalyzed by glutamate dehydrogenase (EC 1.4.1.3) that produces α-ketoglutarate and ammonia, then ammonia is utilized to produce CP in the second reaction catalyzed by carbamoyl phosphate synthase (CPS1, EC 6.3.4.16). CP and ornithine react to produce citrulline, which is catalyzed by ornithine carbomyl-transferase (EC 2.1.3.3). Citrulline enters into cytoplasm, and the following two steps are carried out by two cytosolic enzymes associated with the outer mitochondrial membrane. Citrulline is converted into argininosuccinate and then into arginine by the action of arginosuccinate synthetase (EC 6.3.4.5) and arginosuccinate lyase (EC 4.3.2.1) respectively in cytoplasm.
Arginine degradation is accomplished in mitochondria by three different pathways, resulting in the production of three compounds, including Δ-P5C, nitric oxide (NO), and creatine. In the first pathway, arginine is converted into ornithine by the mitochondrial arginase II (EC 3.5.3.1) and then into P5C. The second pathway leads to the production of NO, catalyzed by NO synthase (EC 1.14.13.39). The third pathway results in the formation of guanidinoacetate and then creatine by the actions of glycine amidinotransferase (EC 2.1.4.1) and guanidine acetate-N-methyltransferase (EC 2.1.1.2), respectively.
Aspartate/aspargine metabolism
Aspartate synthesis in mitochondria is initiated by the pyruvate carboxylase (EC 6.4.1.1) that converts pyruvate into oxaloacetate. Oxaloacetate is then converted into aspartate by aspartate aminotransferase (EC 2.6.1.1) and then into aspargine by aspargine synthase (EC 6.3.5.4). The first two enzymes are mitochondrial while the last enzyme has two cytosolic isoforms, indicating that the last reaction is carried out in cytosol.
Discussion
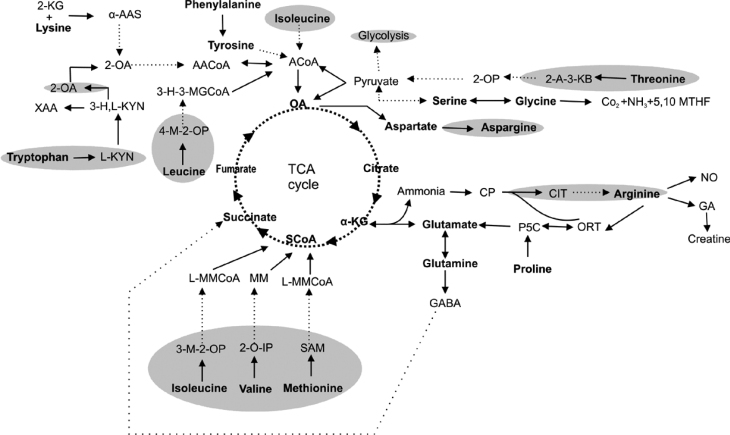
In this study, we have presented complete or partial pathways associated with the metabolism of 17 amino acids in human mitochondria (Figure 5). Pathways for six of these amino acids (leucine, isoleucine, valine, methionine, aspartate, and aspargine) have been well characterized in the literature and hence were not thoroughly discussed in this paper. For the remaining 11 amino acids, pathways were reconstructed by filling in the gaps in the existing pathways that are primarily based on KEGG’s human pathways. MITOPRED predictions have been used to obtain subcellular localization information for the enzymes lacking such information. For most of the predictions, literature-based evidence from human proteins has been provided (see Supporting Online Material). If such information is not available, then evidence from other mammalian species or eukaryotic species is used. Since most of these metabolic pathways span across multiple subcellular locations, several transport mechanisms exist to import or export the metabolites involved in these pathways into or out of mitochondria. The outer mitochondrial membrane (OMM) is freely permeable to small molecules less than 6,000 Da. In contrast, the inner mitochondrial membrane (IMM) regulates the flux of a large variety of solutes between cytosol and mitochondrial matrix using about 20 known mitochondrial carrier systems encoded by the SLC25 family of genes in human (20). In this study, we have provided known mitochondrial solute carriers involved in the transport of metabolites in appropriate pathways.
Fig. 5.
Overview of the amino acid metabolic network in human mitochondria. Shaded areas represent the cytoplasmic segments of the pathways. 2-KG, 2-ketoglutarate; α-AAS, α-aminoadipate semialdehyde; 2-OA, 2-oxoadipate; XAA, xanthurenic acid; 3-H,L-KYN, 3-hydroxy L-kynurenine; L-KYN, L-kynurenine; AACoA, acetoacetyl-CoA; ACoA, acetyl-CoA; OA, oxaloacetate; 3-H-3-MGCoA, 3-hydroxy-3-methylglutaryl-CoA; 4-M-2-OP, 4-methyl-2-oxopentanoate; SCoA, succinyl-CoA; α-KG, α-ketoglutarate; 3-M-2-OP, 3-methyl-2-oxopentanoate; 2-O-IP, 2-oxo-isopentanoate; MM, methylmalonate; L-MMCoA, L-methylmalonyl-CoA; 2-OP, 2-oxopropanol; 2-A-3-KB, 2-amino-3-ketobutyrate; 5,10 MTHF, 5,10 methylene tetrahydrofliate; CP, carbomyl phosphate; CIT, citrulline; NO, nitric oxide; ORT, ornithine; GA, guanidinoacetate; P5C, pyrroline-5-carboxylate; GABA, gamma-aminobutyric acid; SAM, S-adenosylmethionine.
Most of the amino acid metabolic pathways span across cytoplasmic and mitochondrial locations. Nevertheless, other subcellular locations such as peroxisomes are also often associated. In the case of methionine, valine, leucine, isoleucine, threonine, tryptophan, arginine, and aspargine, the metabolic pathways are executed partly in cytoplasm and partly in mitochondria. On the other hand, for glutamate and glutamic acid, the pathways are localized in a tissue-specific manner as described before. In some cases, parallel alternative pathways exist in multiple locations. In the case of glutamate metabolism, the “transamination route” is in cytoplasm while the “transdeamination route” takes place in mitochondria (8). The phosphorylated pathway of serine metabolism occurs in cytoplasm while the nonphosphorylated pathway is found in mitochondria (8). Glycine can be formed by two routes, both of which involve serine. Glycine is formed from serine by serine hydroxylmethyltransferase, which has both cytosolic and mitochondrial isoforms, confirming that this reaction occurs in both locations. The second route is catalyzed by a mitochondrial enzyme, namely glycine synthase (also known as the glycine cleavage system). For a number of cases, location-specific isoforms exist for catalyzing the same reaction in different subcellular locations.
It has been known that deficient or dysfunctional mitochondrial proteins are causative factors for over 100 human diseases (http://www.neuro.wustl.edu/neuromuscular/mitosyn.html), including Alzheimer’s disease, Type II diabetes, Parkinson’s disease, cancer, apoptosis, and so on. We have identified all the known human diseases associated with amino acid metabolism in human mitochondria from the OMIM database (Table 1). Experimental data on the enzymes, substrates, products, and their carriers have been collected and integrated from various sources, including literature-based evidence and database sources such as KEGG, ENZYME, and Swiss-Prot. Studies of this nature pave the conceptual basis for many experimental investigations to study the system-wide implications of altering individual components in metabolic pathways, which could help better understand mitochondrial metabolism and its role in human health.
Materials and Methods
Collection of human mitochondrial protein sequences
Human mitochondrial protein sequences (nucleus-encoded and targeted to mitochondria) were collected from three different sources. The first set of 615 proteins was identified from the human heart muscle by mass spectroscopy combined with rigorous bioinformatics analysis (21). This set includes a significant number of potentially new mitochondrial proteins, the biochemical functions of which are yet to be determined. A second set (366 proteins) was generated based on the annotations from public data repositories (22). The third set of 810 proteins was generated by a computational prediction method, MITOPRED, which was developed by us for the prediction of nucleus-encoded mitochondrial proteins (23). MITOPRED predictions are based on the differences in functional domains and amino acid compositions between mitochondrial and nonmitochondrial proteins. Redundant protein sequences with 100% identity were removed and a final set of 1,439 human mitochondrial proteins (mitoproteome) was used in this study. We used the KEGG database (http://www.genome.ad.jp/kegg/) to get our reference set of metabolic pathways associated with amino acid metabolism, and these pathways are merely used as starting points.
Identification and reconstruction of amino acid pathways in mitochondria
The mitoproteome dataset was compared against the entire human proteome set in the KEGG database and all matching sequences at 100% identity were selected. Using this list, we have traced all the KEGG’s pathways related to amino acid metabolism containing at least one mitochondrial protein sequence, and finally identified 17 pathways. For each pathway, we superposed the initial set of known mitochondrial enzymes on corresponding enzymes in the KEGG reference pathways to obtain a backbone map for reconstruction. Using available literature, the start and end points of each pathway invoked inside human mitochondria were determined. Intermediate enzymes that were not found in the backbone map were identified based on computational predictions from the MITOPRED program, mostly validated by published literature. Each predicted protein has been given a prediction confidence value, which is the ratio of calculated score to the total required score (for being a mitochondrial sequence) and is expressed as a percentage. Isozyme data were also used to determine mitochondrial associations in the case of ambiguous annotations or proteins present in multiple pathways belonging to different subcellular locations. Diseases associated with the dysfunctions or deficiencies of mitochondrial enzymes were identified from the OMIM database (http://www.ncbi.nlm.nih.gov/omim).
Authors’ contributions
PG collected the datasets, conducted data analyses, and prepared the manuscript. CG supervised the project and co-wrote the manuscript. SS conceived the idea of using this approach for pathway analysis and assisted with manuscript preparation. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by the UC Life Sciences Informatics Program and Mitokor Corporation (L99-10077) to SS and the start-up funds to CG from the State University of New York at Albany.
Supporting Online Material
References
- 1.Feist A.M. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Förster J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer F., Viari A. Ab initio reconstruction of metabolic pathway. Bioinformatics. 2003;19:ii26–ii34. doi: 10.1093/bioinformatics/btg1055. [DOI] [PubMed] [Google Scholar]
- 4.Herrgård M.J. Identification of genome-scale metabolic network models using experimentally measured flux profiles. PLoS. Comput. Biol. 2006;2 doi: 10.1371/journal.pcbi.0020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vo T.D. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J. Biol. Chem. 2004;279:39532–39540. doi: 10.1074/jbc.M403782200. [DOI] [PubMed] [Google Scholar]
- 6.Fabio P. Lysine degradation through the saccharopine pathway in mammals: involvement of both bifunctional and monofunctional lysine-degrading enzymes in mouse. Biochem. J. 1999;344:555–563. [PMC free article] [PubMed] [Google Scholar]
- 7.Praphanphoj V. Identification of the alpha-aminoadipic semialdehyde dehydrogenase-phosphopantetheinyl transferase gene, the human ortholog of the yeast LYS5 gene. Mol. Genet. Metab. 2001;72:336–342. doi: 10.1006/mgme.2000.3138. [DOI] [PubMed] [Google Scholar]
- 8.Salway J.G. third edition. Blackwell Publishing Inc.; Malden, USA: 2003. Metabolism at a Glance. [Google Scholar]
- 9.Malherbe P. Identification of a mitochondrial form of kynurenine aminotransferase/glutamine transaminase K form rat brain. FEBS Lett. 1995;367:141–144. doi: 10.1016/0014-5793(95)00546-l. [DOI] [PubMed] [Google Scholar]
- 10.Kwok S.C. Nucleotide sequence of a full-length complementary DNA clone and amino acid sequence of human phenylalanine hydroxylase. Biochemistry. 1985;24:556–561. doi: 10.1021/bi00324a002. [DOI] [PubMed] [Google Scholar]
- 11.Vashkevich I.I. Autoantibodies to human thyroid peroxidase in immunoassay and immunoaffinity chromatography. Prikl. Biokhim. Mikrobiol. 2002;38:96–102. [PubMed] [Google Scholar]
- 12.Kitagawa Y., Sugimoto E. Possibility of mitochondrial-cytosolic cooperation in gluconeogenesis from serine via hydroxypyruvate. Biochim. Biophys. Acta. 1979;582:276–282. doi: 10.1016/0304-4165(79)90390-8. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa Y. Identity of mitochondrial and cytosolic glycerate kinases in rat liver and regulation of their intracellular localization by dietary protein. Biochim. Biophys. Acta. 1979;582:260–275. doi: 10.1016/0304-4165(79)90389-1. [DOI] [PubMed] [Google Scholar]
- 14.Takada Y. Human peroxisomal L-alanine: glyoxylate aminotransferase. Evolutionary loss of a mitochondrial targeting signal by point mutation of the initiation codon. Biochem. J. 1990;268:517–520. doi: 10.1042/bj2680517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar A.J. Molecular cloning and tissue distribution of mammalian L-threonine 3-dehydrogenases. BMC Biochem. 2002;3:19. doi: 10.1186/1471-2091-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binda C. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dariush N. Structure of the gene encoding the entire mature E1 alpha subunit of human branched-chain alpha-keto acid dehydrogenase complex. FEBS Lett. 1991;284:34–38. doi: 10.1016/0014-5793(91)80755-r. [DOI] [PubMed] [Google Scholar]
- 18.Jeremiah S., Povey S. The biochemical genetics of human gamma-aminobutyric acid transaminase. Ann. Hum. Genet. 1981;45:231–236. doi: 10.1111/j.1469-1809.1981.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilkin G.P. Subcellular fractionation of rat cerebellum: separation of synaptosomal populations and heterogeneity of mitochondria. Brain Res. 1979;164:153–163. doi: 10.1016/0006-8993(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 20.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 21.Taylor S.W. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 22.Cotter D. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–D467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guda C. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]