Abstract
The global features of H3K4 and H3K27 trimethylations (H3K4me3 and H3K27me3) have been well studied in recent years, but most of these studies were performed in mammalian cell lines. In this work, we generated the genome-wide maps of H3K4me3 and H3K27me3 of mouse cerebrum and testis using ChIP-seq and their high-coverage transcriptomes using ribominus RNA-seq with SOLiD technology. We examined the global patterns of H3K4me3 and H3K27me3 in both tissues and found that modifications are closely-associated with tissue-specific expression, function and development. Moreover, we revealed that H3K4me3 and H3K27me3 rarely occur in silent genes, which contradicts the findings in previous studies. Finally, we observed that bivalent domains, with both H3K4me3 and H3K27me3, existed ubiquitously in both tissues and demonstrated an invariable preference for the regulation of developmentally-related genes. However, the bivalent domains tend towards a “winner-takes-all” approach to regulate the expression of associated genes. We also verified the above results in mouse ES cells. As expected, the results in ES cells are consistent with those in cerebrum and testis. In conclusion, we present two very important findings. One is that H3K4me3 and H3K27me3 rarely occur in silent genes. The other is that bivalent domains may adopt a “winner-takes-all” principle to regulate gene expression.
Keywords: H3K4me3, H3K27me3, Mouse
Introduction
The methylation of lysine 4 (H3K4) and lysine 27 (H3K27) of histone H3 attracts particular attention, since both modifications regulate gene expression and therefore play key roles in cell or tissue development [1], [2]. H3K4 trimethylation (H3K4me3) positively regulates transcription by recruiting nucleosome remodeling enzymes and histone acetylases [3], [4], [5], [6], [7], while H3K27 trimethylation (H3K27me3) negatively regulates transcription by promoting a compact chromatin structure [1], [8]. Genome-wide studies of H3K4me3 and H3K27me3 have been performed in several mammalian cell types, such as mouse embryonic stem cells (ESCs), neural progenitor cells (NPCs), and fibroblasts [9], as well as human T cells, ESCs, hematopoietic stem cells (HSC), and erythrocytes [10], [11], [12], [13], [14]. These studies revealed the general features of both modifications, and their important regulatory roles in cell differentiation and development.
Presently, very few comparative analyzes of H3K4me3 and H3K27me3 among mammalian tissues have been performed, particularly ones involving integrated analysis with RNA-seq data. In this study, we report the results of genome-wide mapping of H3K4me3 and H3K27me3 and whole transcriptome profiling in mouse cerebrum and testis, based on ChIP-seq and rmRNA-seq methods, respectively [15]. By combining analysis of both datasets, we globally described tissue-specific modifications and their relationship to tissue-specific expression, function and development. Furthermore, we also revealed several novel patterns of H3K4me3 and H3K27me3. This study would significantly advance our understanding of the biological functions of histone modifications in governing gene expression.
Results
Genome-wide maps of H3K4me3 and H3K27me3
We generated genome-wide profiles of H3K4me3 and H3K27me3 in the mouse cerebrum and testis. For each case, we prepared about 200 ng ChIP DNA samples with an average length of 300 bp for high-throughput sequencing. We obtained 321.86 million 50-bp high-quality reads with an average of 53.64 million reads for each ChIP-seq library. About 42.87% of these reads can be uniquely mapped to the mouse genome (mm9) (Table S1). These uniquely-mapped reads were used to determine the methylated H3 enrichment of ChIP fragments. H3K4me3 and H3K27me3 enriched intervals were defined as regions where number of reads exceeded a threshold estimated by randomization based on pan-H3 read distribution across genomes as described previously [16]. In the cerebrum, 82,264 H3K4me3 and 43,132 H3K27me3 intervals were identified, and in the testis, 64,110 H3K4me3 and 26,828 H3K27me3 intervals were identified (Tables S2–S6). The lengths of the intervals appear to reach a saturation point along with the growing depth of uniquely mapped reads (Figure S1A). Based on the obtained interval lengths, we estimated that H3K4me3 and H3K27me3 covered ∼6% and ∼8% of the mouse genome, respectively. At this sequencing depth, we estimated that the average read coverage (per bp) for H3K4me3 and H3K27me3 intervals is ∼3× (ncerebrum = 3.28 and ntesits = 3.24) and ∼1× (ncerebrum = 1.25 and ntesits = 0.98) in the two tissues examined, respectively. This is in contrast to the low average read coverage for non-H3K4me3-modified and non-H3K27me3-modified genomic sequences, which is ∼0.25× (ncerebrum = 0.21 and ntesits = 0.29) and ∼0.30× (ncerebrum = 0.32 and ntesits = 0.27), respectively. These results suggest that both H3K4me3 and H3K27me3 are enriched in their intervals. As an illustration, ChIP-Seq maps of H3K4me3 and H3K27me3 show significant enrichment at specific locations in the genome, whereas the pan-H3 distributions are relatively uniform (Figure S1B).
In order to correlate H3K4me3 and H3K27me3 with gene expression, we generated transcriptomic data from the two tissues using the ribo-minus RNA sequencing method (rmRNA-seq) [15]. We obtained 638 million reads, of which 33.6% was uniquely mapped onto the mouse genome (Table S1). Gene expression was initially estimated by calculating the density of uniquely-mapped reads as “reads per kilobase of exon model per million mapped reads” (RPKM) [17]. These estimates were typically performed based on publicly-available gene annotations [18]. Using a threshold RPKM value of 0.046 for the cerebrum and 0.049 for the testis as “background expression” (Figure S2), we identified 16,992 and 17,400 genes that are significantly expressed in the two tissues, respectively. Moreover, these gene numbers were consistent across different sequencing depths (Figure S1C), reflecting their adequacy for gene detection.
To verify the gene expression data, we also obtained mRNA-selected RNA-seq (mRNA-seq) data for mouse cerebrum and testis (our unpublished data). It was shown that there was a high correlation coefficient of gene expression between rmRNA-seq data and mRNA-seq data (R = 0.94, Spearman). In addition, we randomly selected 13 genes to validate the RNA-seq data in cerebrum using qRT-PCR. The correlations of gene expression among qRT-PCR, rmRNA-seq data and mRNA-seq data were also very high (R = 0.96 between qRT-PCR and rmRNA-seq data and R = 0.93 between qRT-PCR and mRNA-seq data, Spearman).
H3K4me3/H3K27me3 around promoters
We firstly analyzed H3K4me3 and H3K27me3 patterns at the known promoters, and correlated them to gene expression in the two tissues. We defined 21,215 known promoters based on RefSeq-annotated full-length transcripts. In addition, we further classified the active genes into high-, medium-, and low-expression categories according to their expression levels. To ensure a clean separation of lowly-expressed genes from silent genes, we defined a group of ambiguously-expressed genes (ncerebrum = 1905 and ntesits = 2511) that had rmRNA-seq reads detected but whose expression levels are too low to stand out from the background.
H3K4me3 in the cerebrum and testis
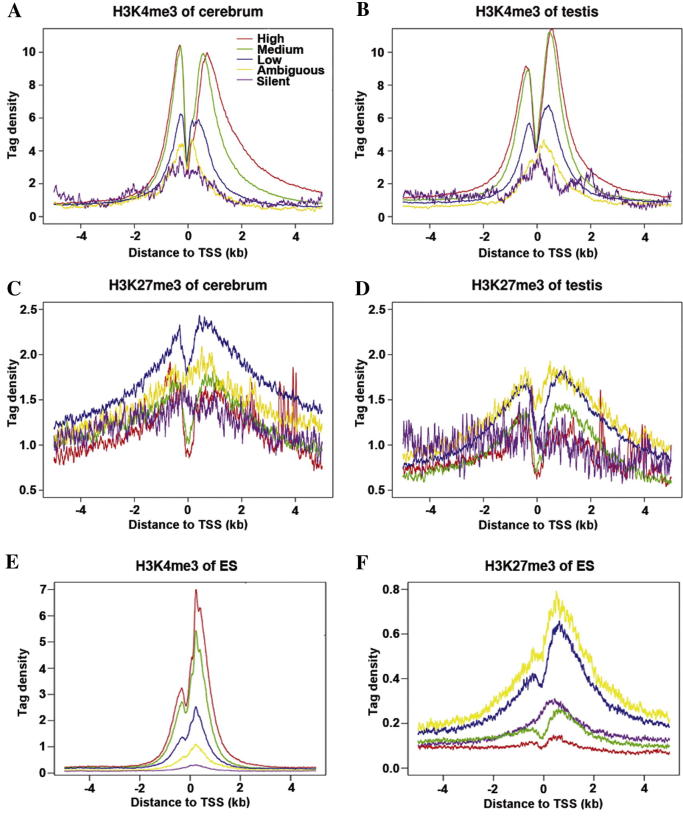
A significant fraction (pcerebrum = 68% and ptesits = 74%) of promoters are marked by H3K4me3 in the cerebrum and testis (Table S7). H3K4me3 modified regions are typically confined to a punctate interval of 1–2 kb, which is shorter than H3K27me3 modification regions (Figures S3A and B). Moreover, there is an obvious correlation between the intensity of H3K4me3 and expression level of the associated genes (Figure 1A and B, and Figure S4A). These results are in agreement with previously-reported observations [19], [20]. However, not all active genes had H3K4me3. In fact, we found that ∼15% of active genes lacked H3K4me3, which are expressed at lower levels and possess higher percentage of low-CpG promoters (LCPs) (Figure S5). Furthermore, we found that <5% of silent genes were marked by H3K4me3 in both tissues, which led us to believe that H3K4me3 is rarely associated with silent genes. This contradicts previous findings that a higher proportion of H3K4me3 was found in silent genes [10], [11], [21]. For example, Barski et al. found that H3K4me3 islands were detected in 59% of silent promoters [10].
Figure 1.
H3K4me3 and H3K27me3 modifications around TSS Profiles of H3K4me3 modifications across the TSS were shown for high, medium, low, ambiguous, and silent expression genes in cerebrum (A) and testis (B). Profiles of H3K27me3 modifications across the TSS were shown for high, medium, low, ambiguous, and silent expression genes in cerebrum (C) and testis (D). The significantly expressed genes (ncerebrum = 16,992 and ntesits = 17,400) were equally classified into three groups (high, medium and low) according to gene expression level. To ensure a clean separation between low and silent expression genes, we defined a group of ambiguous expression genes (ncerebrum = 1905 and ntesits = 2511). These genes had rmRNA-seq reads detected, but their expression level is lower than background. This group of genes displayed properties consistent with being a mixture of low and silent expression. Silent genes (ncerebrum = 2318 and ntesits = 1304) were defined as genes with no reads obtained. Shown in (E) and (F) were profiles of H3K4me3 and H3K27me3 across the TSS in ESCs, respectively.
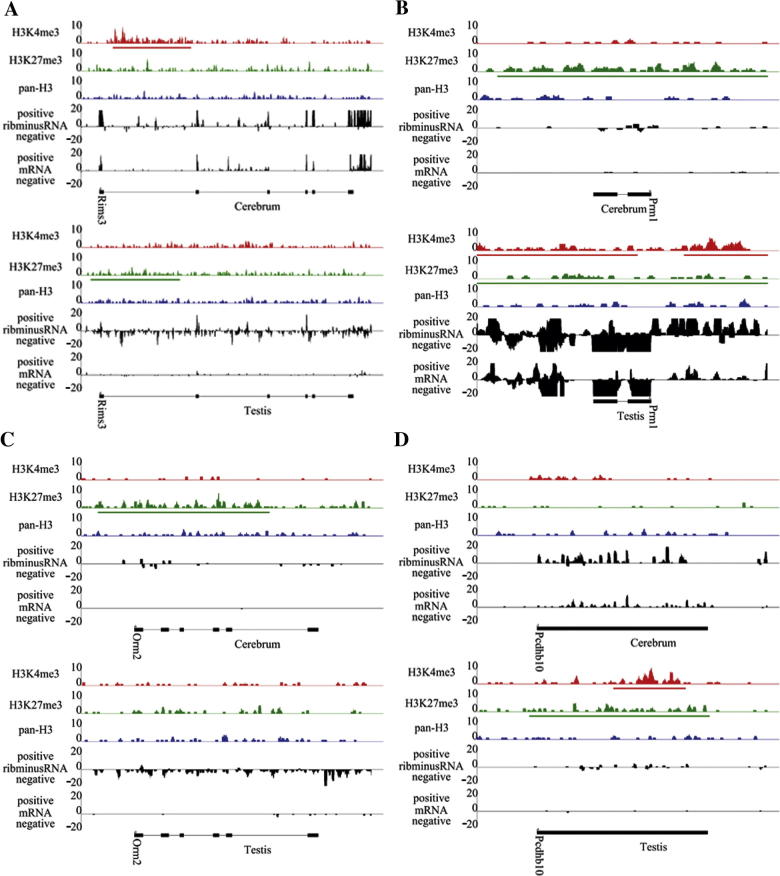
We next examined the tissue specificity of H3K4me3. Although most (96.85%) promoters marked by H3K4me3 in the cerebrum were also found in the testis, we still identified a limited, yet significant, number of the promoters which were only marked by H3K4me3 modification in either cerebrum or testis. There were 456 H3K4me3-marked promoters present in the cerebrum but absent in the testis. Most of these promoters regulate genes associated with stress responses (Table S8), such as immune, inflammatory, and defense responses. Moreover, these genes have higher transcriptional activity in the cerebrum than their counterparts in the testis (Figure S6A). For example, the gene Rims3 involved in regulating nerve signal transduction [22] appears to be cerebrum-specific in modification and expression (Figure 2A). We also identified 1611 promoters marked by H3K4me3 in the testis but not in the cerebrum. These testis-associated active genes are mostly regulatory genes with testis-specific functions (Table S9), including reproduction, spermatogenesis, and gamete generation, which are expressed at high level in the testis (Figure S6B). One of these genes, Prm1, is related to spermatogenesis [23], [24] and shows testis-specific modification and expression (Figure 2B).
Figure 2.
Cerebrum or testis specific chromatin marking In this figure, we set the maximum coverage for modification and expression as 10 and 20, respectively. In addition, the range of horizontal axes is from 1 kb upstream of the gene to 1 kb downstream of the gene in chromosome. We also displayed the SICER defined enrichment intervals for H3K4me3 and H3K27me3 under horizontal axes. (A) The gene, Rims3, involved in regulating nerve signal transduction, is marked by H3K4me3 at promoter region in cerebrum, but not in testis. Moreover, H3K4me3 modification is well associated with gene expression with higher expression level in cerebrum than in testis. (B) The gene, Prm1, related to spermatogenesis, shows testis-specific H3K4me3 modification and expression. (C) The gene, Orm2, involved in immune suppression, shows cerebrum-specific H3K27me3 modification. (D) The gene, Pcdhb10, involved in cell-cell connection shows testis-specific H3K27me3 modification and expression.
H3K27me3 in the cerebrum and testis
About 25% and 28% of promoters are marked by H3K27me3 in the cerebrum and testis, respectively (Table S7). As described previously, H3K27me3 intervals were longer compared to H3K4me3 intervals, ranging from ∼1 to 10 kb in length (Figure S3B). However, the size of the intervals appears to be tissue-specific. In the cerebrum, most modifications are limited to ∼5 kb, but those in the testis are longer (Figure S3B). In fact, the number of H3K27me3 intervals in the testis (26,828) is much lower than that in the cerebrum (43,132), so it could be that one long interval in testis is separated into two or more intervals in cerebrum. We speculated that these larger H3K27me3-modified regions could be correlated to the stronger transcriptional repression of the associated genes in the testis. In fact, we observed that H3K27me3-modified genomic regions in the testis tended to have lower transcriptional activity than those in the cerebrum (Figure S7), which supported our hypothesis that there might be tissue-specific H3K27me3 that maintained gene repression.
In addition, we found that there was a significant correlation between H3K27me3 and gene expression (Figure 1C and D, and Figure S4B). H3K27me3 was obviously less prevalent among silent promoters, covering only about 10% of them (pcerebrum = 12.2% and ptestis = 6.3%). Furthermore, even within this 10% of promoters, H3K27me3 levels were relatively low (Figure 1C and D). These results contradicted with previous observations which associated the highest levels of H3K27me3 with silent gene promoters in some cell types [10].
We identified 1229 H3K27me3-marked cerebrum-specific promoters (not modified in the testis). Interestingly, these H3K27me3-marked promoters were similar to the cerebrum-specific H3K4me3-marked promoters in that they are also associated with responses to environmental stimuli (Table S10), including defense, immune, and inflammatory. As expected, these genes also exhibited lower transcriptional activity in the cerebrum than their counterparts in the testis (Figure S6C). This cerebrum-specific H3K27me3 modification, and the presence of cerebrum-specific H3K4me3, suggests that epigenetic mechanisms play a critical role in regulating the cerebrum-specific function of perception. In addition, we identified 1838 testis-specific H3K27me3-marked promoters, whose functions were restrained to cellular communication and homeostasis (Table S11). These genes seemed to be tightly regulated, since their expression levels were quite low in the testis compared to those in the cerebrum (Figure S6D). Two tissue-specific modified genes, Orm2 in cerebrum and Pcdhb10 in testis, are involved in immune suppression [25] and cell–cell connection [26], respectively (Figure 2C and D). Surprisingly, the expression of Orm2 in testis is very low although it does not have H3K27me3 modification.
Bivalent chromatin domains of H3K4me3 and H3K27me3
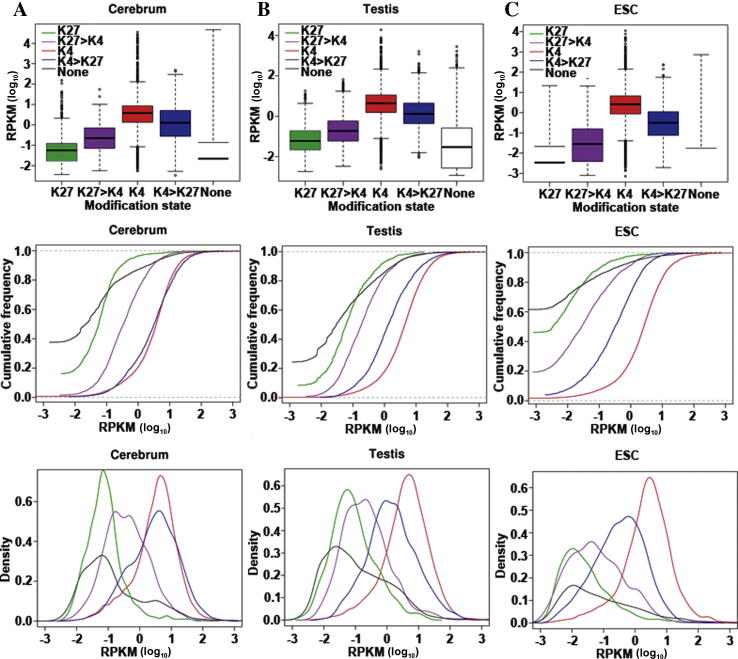
The bivalent chromatin domains, harboring both H3K4me3 and H3K27me3, were reported to be enriched in embryonic stem cells (ESCs) and proposed to regulate key genes for lineage-specific activation or repression [27]. In our data, there are 18% (3823) and 25% (5290) bivalent promoters in the cerebrum and testis (Table S12), respectively. The percentages are somewhat higher here than that in ES cells (15%) [9]. Intriguingly, genes with bivalent promoters, or bivalent-regulated genes in both tissues were found to have complex expression patterns, which are dependent on intensities of H3K4me3 and H3K27me3 as measured by number of ChIP-seq reads per kilobase of modification intervals per million mapped reads. We observed that bivalent genes which had higher intensities of H3K4me3 than H3K27me3 also had higher expression levels, similar to genes marked with H3K4me3 alone (Figure 3A and B). In contrast, the bivalent genes with higher intensities of H3K27me3 exhibited lower expression, similar to H3K27me3 only-marked genes. These results suggest that bivalent domains tend to use the “winner-takes-all” approach to regulate the expression of the associated genes although this regulatory trend is less obvious in testis than in cerebrum.
Figure 3.
Expression pattern of bivalent chromatin domains in cerebrum and testis (A) Box plot showing 25th, 50th, and 75th percentile expression levels in cerebrum (left), testis (middle) and ESC (right) for genes associated with no histone methylation (none, black), H3K4me3 only (K4, red), H3K27me3 only (K27, green), stronger H3K4me3 in the bivalent domain (K4 > K27, blue) and stronger H3K27me3 in the bivalent domain (K27 > K4, purple). Whiskers show 1.5 times of 25th and 75th percentile expression. The gene expression level is measured by RPKM values (Y axis). (B) Cumulative distribution of expression levels for genes associated with no histone methylation (none, black), H3K4me3 only (K4, red), H3K27me3 only (K27, green), stronger H3K4me3 in the bivalent domain (K4 > K27, blue) and stronger H3K27me3 in the bivalent domain (K27 > K4, purple). (C) The distribution of expression levels for genes associated with no histone methylation (none, black), H3K4me3 only (K4, red), H3K27me3 only (K27, green), stronger H3K4me3 in the bivalent domain (K4 > K27, blue) and stronger H3K27me3 in the bivalent domain (K27 > K4, purple).
Virtually 2522 bivalent promoters are shared by both the testis and the cerebrum. Using existing data on mouse ESCs [9], we calculated that 89% of ESC bivalent promoters are also present in cerebrum and testis (Figure S8). Functional analysis demonstrated that these widespread bivalent promoters regulated genes that were mostly involved in cell differentiation and development, including neuron differentiation and development, axonogenesis, cell motion, cell–cell signaling, cell adhesion, cell morphogenesis, cell fate commitment, cell migration, and neural-tube development (Figure S9A and Table S13). While most of these bivalent promoters appeared to play house-keeping roles in regulating development, a significant fraction of them might be associated with tissue-specific or cell-specific regulation. For instance, we identified 695 cerebrum-specific bivalent promoters, which were usually associated with immune response, ion transport, cell activation and inflammatory response (Figure S9B and Table S14). We also identified 2162 testis-specific bivalent promoters, most of which regulated genes that perform regionalization, adhesion and embryonic and skeletal system morphogenesis (Figure S9C and Table S15).
Validation of the relationship between H3K4me3/H3K27me3 and gene expression and the “winner-takes-all” principle for bivalent domains
To further validate our result, we downloaded the publicly-available ChIP-seq [9] and RNA-seq data [28] for mouse ESCs and performed similar analysis. It was shown that, 67% genes are marked by H3K4me3 in ESCs. The correlation between the intensity of H3K4me3 and gene expression is also obvious (Figure 1E). Moreover, we also noticed that 17% of silent genes have H3K4me3 in ESCs, which is higher than those observed in the mouse cerebrum and testis (Table S7). We proposed that this is due to the insufficient depth of sequencing in ESCs such that many lowly-expressed genes are classified as silent genes. However, this percentage is significantly lower than that reported by Barski et al., which was 59% [10]. It further suggested that H3K4me3 is rarely associated with silent genes. In addition, about 29% of promoters are marked by H3K27me3 in ESCs. Like in cerebrum and testis, H3K27me3 are also less prevalent among silent genes, covering only about 23%, compared to lowly-expressed genes which covered 54%. Meanwhile, the modification extent of H3K27me3 was lower in silent genes, compared to lowly-expressed genes (Figure 1F). These results were consistent with our findings in cerebrum and testis.
In ESCs, genes with bivalent modification (both H3K4me3 and H3K27me3) also tend to be regulated following the “winner-takes-all” principle (Figure 3C). This indicates that “winner-takes-all” may be a universal regulatory mechanism in tissues or cells. This winner-takes-all pattern appears to be different from the behavior previously described in ESCs [9] where H3K27me3 plays a dominant role in repressing transcriptional activities of the bivalent genes.
Moreover, we did differential gene expression analysis among mouse cerebrum, testis and ESCs (Tables S16–S18). We found that most of genes are expressed significantly differentially among three samples. However, the transcriptome profiles that differ significantly in the three samples have common characteristics of association with H3K4me3 and H3K27me3. This suggests that our findings are common in tissues or cells.
Special patterns of H3K4me3/H3K27me3 in silent genes
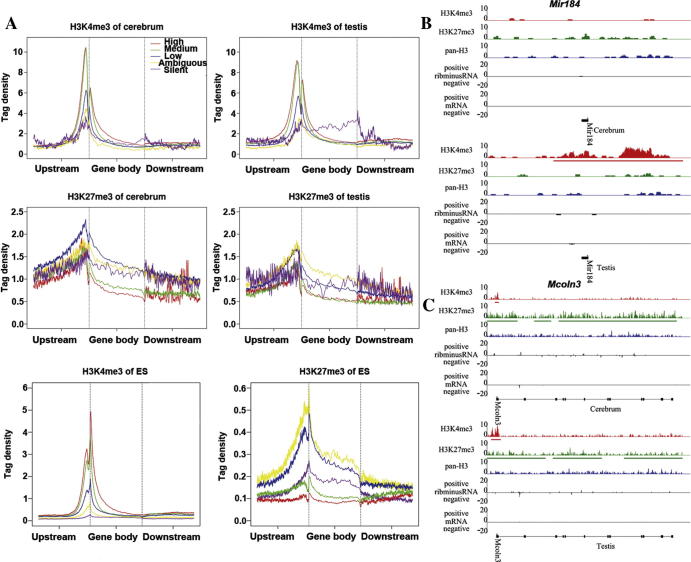
We found only a few silent genes that were marked by H3K4me3/H3K27me3. Interestingly, these silent genes tended to be uniformly modified across whole gene region, rather than enriched at the active promoters (Figure 4A). As expected, the silent genes in ESCs were also modified in entire gene regions (Figure 4A). For example, Mir184 and Mcoln3 are two such genes shown in Figure 4B. Moreover, these genes appear to be enriched in several specific functional pathways. In the cerebrum, silent H3K4me3-marked genes are associated with protein-DNA complex, nucleosome, chromatin, and cellular macromolecular complex assemblies (Table S19). In the testis, these genes are involved in sensory perception, cognition, and cell surface receptor linked signal transduction (Table S20). In contrast, silent genes marked by H3K27me3 are related to sensory perception, defense response, and cognition; all of these functions are shared by both tissues (Tables S21 and S22). This modification pattern could reflect a special regulation of H3K4me3 and H3K27me3 for silent genes.
Figure 4.
Even distribution of H3K4me3/H3K27me3 along silent genes (A) The silent genes tend to be equally modified across the whole gene body in cerebrum, testis and ES cells. (B) The gene, Mir184, show equably modified by H3K4me3 across the gene body in cerebrum and testis. (C) The gene, Mcoln3, show equably modified by H3K27me3 across the gene body in cerebrum and testis.
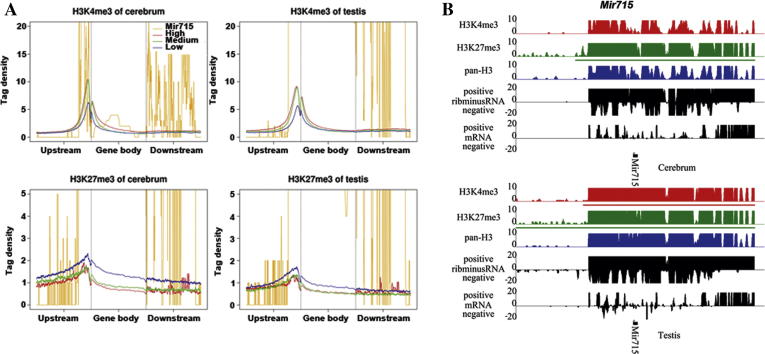
A special modification pattern at the Mir715 locus
We found that the Mir715 locus, encoding a microRNA, is regulated by bivalent domains, showing an impressive surge of H3K27me3 and H3K4me3 across the region from 1 kb upstream to 5 kb downstream of Mir715 (Figure 5). The modification levels are highest among all modified genes and the reason is unknown. In the testis, Mir715 is heavily H3K4me3-modified and has a higher expression level than in the cerebrum, which reflected the H3K4me3-driven expression for a bivalently-regulated gene. Furthermore, based on functional classifications of the 794 Mir715 targets documented in miRBase [29], we found that Mir715 regulated genes were associated with germline-specific functions, such as cell differentiation and development (Table S23). This is consistent with a recent study on this gene’s role in the testis [30]. These results suggest that H3K4me3 and H3K27me3 have important functional roles in regulating microRNA expression. In addition, we also found the same strong expression and modification pattern in ESCs.
Figure 5.
Enrichment of H3K27me3 and H3K4me3 modifications across Mir715(A) The profiles of H3K27me3/H3K4me3 were plotted across genes with different expression levels and Mir715. The H3K27me3/H3K4me3 was elevated at Mir715. (B) The modification and expression patterns of Mir715 were indicated.
Genome-wide annotation of promoters and novel transcripts
It is widely accepted that H3K4me3/H3K27me3 intervals are highly associated with known genes, but there are still numerous modification intervals that fall into either intronic or intergenic regions (69.1% H3K4me3 and 75.5% H3K27me3 in the cerebrum; 59.1% H3K4me3 and 64.9% H3K27me3 in the testis). Further inspection revealed that about 90% of these intervals had RNA-seq reads and these modifications might be associated with transcriptional activities. We estimated that about 7% of these intervals lie in an area between 10 kb upstream of transcriptional start sites (TSS) and 10 kb downstream of transcriptional terminal sites (TTS) in known genes, representing alternative promoters or exons. Nearly 75% of them are in intergenic sequences and are likely to be new transcripts. An example is shown in Figure S10. The above results suggest that there may be thousands of non-coding or even coding RNAs expressed in the mouse cerebrum and testis, which have yet to be characterized. Actually, Liu et al. have discovered thousands of novel transcripts (mostly non-coding RNAs) in intronic and intergenic regions of mouse cerebrum, testis, and ESCs through an in-depth analysis of rmRNA-seq data [31].
Discussion
General features of H3K4me3/H3K27me3 in the tissues
In our analyzes, we examined general characteristics of H3K4me3 and H3K27me3 in the mouse cerebrum and testis tissues. Consistent with previous studies, H3K4me3 was found to be enriched in promoter regions and typically confined to a genomic locus of 1–2 kb in size [19], [20]. Moreover, this modification exhibited a “twin-peak” profile around TSS, as well as a positive correlation with gene expression [32]. H3K27me3 was also enriched around promoters, and extended over a broader range of 2–10 kb in length [33]. However, it is worthy of note that the size distribution of the H3K27me3-modified intervals was different in both tissues. We believe that the larger lengths of H3K27me3-modified regions may be associated with the repression of gene expression.
We believe that tissue-specific epigenetic signals regulate tissue-specific functions by controlling gene expression. We found that in the cerebrum, genes involved in sensing and responding to environment stimuli were specifically modified by H3K4me3 or H3K27me3, and in the testis, genes related to reproduction and spermatogenesis were specifically modified in the similar way. Therefore, broadly surveying chromatin states in tissues and organs would uncover valuable information that will help us better understand the role that epigenetic mechanisms play in tissue function and development.
H3K4me3/H3K27me3 in silent genes
Another important finding was that H3K4me3 and H3K27me3 were rarely found among silent promoters in either tissue. In other words, almost all promoters marked by H3K4me3 and H3K27me3 appeared to be active. Our data indicated that <10% of the silent genes were marked by H3K4me3 and H3K27me3 in both tissues, and these marked genes were also much less modified than active genes. This feature appeared to contradict previous reports stating that H3K4me3 and H3K27me3 had been detected among silent genes in ESCs or differentiated cells and that H3K27me3 exhibited higher levels among silent genes [9], [10], [27], [34], [35], [36], [37], [38]. For example, one study found that 59% of silent genes in human T cells were modified by H3K4me3 [10]. Another study in yeast suggested that the presence of H3K4me3 among silent genes was a remnant of past transcriptional activity [21]. A third study performed on HSCs/HPCs posited that the presence of H3K4me3 was related to an epigenetic state that maintained the activating potential of genes [11]. We speculate that the inconsistencies in these findings could be caused by the inaccurate definition of silent genes. We found that previous studies mostly relied on microarray-based gene expression profiling, which was known to have problems in detecting the expression of lowly-expressed genes [39]. As a result, most lowly-expressed genes are often defined as silent genes due to the high false negatives in microarray data. This leads to flawed descriptions of H3K4me3 and H3K27me3 among silent genes. In contrast, our result is derived from recently-developed high-throughput RNA-seq data. The RNA-seq method is based on next-generation sequencing technology and is highly successful in detecting low-expression genes [40]. In addition, we also obtained the same result in cerebrum and testis as seen in ESCs using publicly-available ChIP-seq and RNA-seq data that most silent genes are not modified by H3K4me3 or H3K27me3.
The lack of H3K27me3 in silent genes suggests that silent genes could be free from the regulation of H3K27me3. Therefore, there should be another mechanism for repressing the expression of silent genes. Recent studies suggested that silent genes tend to be located at chromatin regions that are associated with nuclear lamina [41]. These lamina-associated chromatin regions show closed status and repress gene transcriptional activities. Therefore, lamina-associated chromatin structure could be one kind of regulation model for repressing gene expression.
Bivalent domains in two tissues
Our results demonstrated that bivalent domains were enriched in mouse cerebrum and testis tissues. We propose a novel principle (“winner-takes-all”) that explains the function of bivalent domains in regulating gene expression. We believe that bivalent “switching” behavior can be sensitive and rapid, through weighing the proportion of modified H3K4me3 vs H3K27me3 sites within the bivalent domains. This principle contradicts the previously-held notion that H3K27me3 plays a dominant role in bivalent domains [27]. In addition, our results suggest that bivalent domains prefer to be in the vicinity of developmentally-associated genes, which exist in very large numbers in all three tissues and cell type we analyzed (mouse cerebrum and testis, and ESCs).
Materials and methods
Ethics statement
The handling of mice and experimental procedures were guided and approved by Beijing Municipal Science & Technology Commission with SYXK2009-0022.
ChIP-seq experiment
We collected tissue samples (pooled from three individuals) of cerebrum and testis from 10-week old male BALB/c mice. We carried out ChIP-seq experiments according to the published procedure [19] (http://www.upstate.com). Briefly, the tissue samples from the mouse cerebrum and testis were homogenized and fixed with 1% formaldehyde. Chromatins were fragmented in a size range of 200–1000 bp and incubated with antibodies (against trimethyl Lys4, Abcam #8580 and against trimethyl Lys27) at 4 °C overnight. After cross-linking reversal and proteinase K treatment, DNA samples were precipitated and treated with RNase and calf intestinal alkaline phosphatase (CIAP). The DNA samples were purified with MinElute Kit (Qiagen) before library construction. About 10 ng DNA was used for adaptor ligation, gel purification and PCR with 15 cycles. Sequencing was done by using SOLiD system (Applied Biosystems).
rmRNA-seq experiment
We performed rmRNA-seq experiments as described previously [15]. Briefly, total RNAs from tissues were isolated using Trizol, and then ribosomal RNA was depleted with Ribo-minus Eukaryote kit (for RNA-seq, Invitrogen, cat.10837-08). RNA-seq library was constructed using the protocol from SOLiD™ Small RNA Expression Kit (#4397682). We put together the following mixture on ice in order: 8 μl RNA (1 μg), 1 μl 10 × RNase buffer and 1 μl RNase (#AM2290; Applied Biosystems). The mixture was incubated at 37 °C for 10 min followed by incubation for 20 min at 65 °C. We use FlashPAGE™ to collect fragmented RNA in a length range of 50–150 bp and to purify the RNA using FlashPAGE Reaction Clean-Up Kit (#AM12200; Applied Biosystems). We resuspended the air-dried RNA in 3 μl nuclease-free water and the ligation mixture was put together, which contains the RNA, Adaptor Mix and Hybridization Solution. The ligation was started by adding ligase to each sample and the mixture was incubated at 16 °C for 16 h. cDNA was synthesized by adding 20 μl RT master mix to each sample and incubating at 42 °C for 30 min. The residual RNA was removed with RNase H (10 U in 10 μl cDNA mixture) at 37 °C for 30 min. The cDNA library was amplified, cleaned with Qiagen MinElute PCR purification Kit (#28004, 28006; Qiagen) and purified on a native 6% polyacrylamide gel. Usually 400 μl reaction product is enough for sequencing and a fraction of the library in a size range of 140–200 bp (DNA ladder, #10821-015; Invitrogen) is usually selected for SOLiD sequencing.
Sequencing read mapping
Sequencing reads were mapped to the mouse genome (mm9) using a custom-designed SOLiD read mapping pipeline. The full length 50-bp or 35-bp reads were firstly aligned onto the genome by allowing up to five mismatches out of 50-bp reads, or three mismatches out of 35-bp reads. Afterwards, mapping was repeated for the first 45 or 40-bp and 30 or 25-bp truncated tags of unmapped reads with reduced mismatches (4, 4, 3 and 2 mismatches for 45 bp, 40 bp, 30 bp and 25 bp length reads). The uniquely-mapped reads were used for identifying enriched intervals and calculating gene expression level. To evaluate the mapping quality, we re-mapped the transcriptome data for mouse cerebrum and testis by Bowtie 0.12.7 using default parameters [42]. The mapping result is similar (143 M mapped reads before vs. 136 M mapped reads now in 498 M of raw reads in mouse testis, Pearson value is 0.137 and P value is 0.711 using Chi squares test). We also calculated the correlation coefficient of gene expression using two mapping methods (R = 0.94, Spearman). For replication of gene expression, we also obtained the mRNA-selected RNA-seq data for mouse cerebrum and testis (data not published) and mapped them by Bowtie. In addition, public collections of ChIP-seq data (including H3K4me3 and H3K27me3) and RNA-seq data of mouse ESCs were obtained [28], [33].
Identification of enriched intervals
We defined H3K4me3 and H3K27me3 intervals using the SICER program (v1.03). The parameters used were: (1) 200-bp window, 200-bp gap, and 0.001 for False Discovery Rate (FDR) and (2) 200-bp window, 600-bp gap, and 0.001 for FDR, were used for identifying H3K4me3 and H3K27me3 intervals, respectively. The sequencing reads from a pan-H3 experiment was used as background control. We defined the promoter regions based on inferred TSSs from full length transcripts deposited in RefSeq, as 2 kb from upstream and downstream of the TSS. The classification list of HCPs, ICPs and LCPs was obtained from previous literature [9]. Chromatin states among promoters were determined by overlapping with H3K4me3- and H3K27me3-enriched intervals. The modification levels were calculated from the mean fragment density over each promoter or transcripts.
Defining background expression
We measured expression levels of genes by calculating the density of uniquely-mapped reads as “reads per kilobase of exon model per million mapped reads” (RPKM) [17]. These estimates are typically performed based on publicly-available RefSeq gene annotations [18]. We estimated background expression based on a published procedure [43]. Briefly, we compared gene expressions between the gene expression (exons) and the expression in the intergenic region (defined based on RefSeq gene annotation) to find a threshold for detectable expression above background. We defined the intergenic sequences by matching the length of genes (only known exons). We binned expression of all genes and intergenic regions between 0.01 and 10 RPKM for our analyzes. We counted the cumulative number of expressed regions above the expression levels for gene (genes_a) and intergenic (inters_a) sequences as well as below the expression levels for gene (genes_b) and intergenic (inters_b) sequences. A false positive rate (FPR) was calculated at each expression level as FPR = inters_a/inters_a + inters_b + genes_a + genes_b) and false negative rate (FNR) as FNR = genes_b/(inters_a + inters_b + genes_a + genes_b) (Figure S2). We used a threshold RPKM value of 0.046 and 0.049 for cerebrum and testis to balance FPR and FNR. Since it is difficult to identify un-transcribed DNA sequences with confidence, the background is often overestimated. To ensure a clean separation between lowly-expressed and silent genes, we defined a unique group of genes as ambiguously-expressed genes (ncerebrum = 1905 and ntesits = 2511), which contain rmRNA-seq reads but their expression level is lower than the background expression mathematically.
Validation of RNA-seq data by qRT-PCR
We randomly selected 13 genes to validate the RNA-seq data using qRT-PCR method in cerebrum. Specific primer pairs for 13 genes were downloaded from PrimerBank (http://www.pga.mgh.harvard.edu/primerbank/) (Table S24). The GAPDH gene served as internal standard. The relative expression of target mRNAs was determined using Maxima® SYBR Green/ROX qPCR Master Mix (Fermentas) following the manufacturer’s instructions on RNA of mouse cerebrum (Table S25). The correlation of gene expression among qRT-PCR, rmRNA-seq data and mRNA-seq data are very high (R = 0.96 between qRT-PCR and rmRNA-seq data and R = 0.93 between qRT-PCR and mRNA-seq data, Spearman).
Function analysis
We used DAVID (web version, http://www.david.abcc.ncifcrf.gov/) for functional gene classification and the raw results were summarized in Supplementary tables. The sequencing data has been deposited in the SRA database of NCBI (SRA accession Nos. SRA039962 and SRX005943).
Authors’ contributions
SH, JY and FS designed the research. DZ performed the ChIP-Seq experiments. CX performed the rmRNA-Seq experiments. PC, WL, YZ and QL analyzed the ChIP-Seq data. PC, WL, FD, ZZ and SS analyzed the rmRNA-Seq data. PC and WL wrote the manuscript. JY and SH revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This study is supported by Grants from Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-01-04), National Science and Technology Key Project (2008ZX1004-013), 863 Program (2009AA01A130), Special Foundation Work Program (2009FY120100), National Key Technology R&D Program (2008BA164B02) and 973 Program (2011CB944100, 2011CB965300 and 2007CB948101) from the Ministry of Science and Technology of the People’s Republic of China. The authors thank the anonymous reviewers for critical comments and helpful suggestions, and also thank Jeo Yu for editing the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.05.007.
Contributor Information
Jun Yu, Email: junyu@big.ac.cn.
Songnian Hu, Email: husn@big.ac.cn.
Supplementary material
References
- 1.Ringrose L. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 3.Li B. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 4.Pray-Grant M.G. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 5.Santos-Rosa H. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 6.Sims R.J., 3rd Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wysocka J. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Francis N.J. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen T.S. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barski A. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Cui K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz Y.B., Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Wei G. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X.D. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Cui P. A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. Genomics. 2010;96:259–265. doi: 10.1016/j.ygeno.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C. Methods for labeling error detection in microarrays based on the effect of data perturbation on the regression model. Bioinformatics. 2009;25:2708–2714. doi: 10.1093/bioinformatics/btp478. [DOI] [PubMed] [Google Scholar]
- 17.Mortazavi A. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 18.Pruitt K.D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein B.E. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Kim T.H. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng H.H. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y. The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem. 2000;275:20033–20044. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- 23.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr J.A., Silverman N. The heparin–protamine interaction. A review. J Cardiovasc Surg (Torino) 1999;40:659–666. [PubMed] [Google Scholar]
- 25.Depke M. Altered hepatic mRNA expression of immune response and apoptosis-associated genes after acute and chronic psychological stress in mice. Mol Immunol. 2009;46:3018–3028. doi: 10.1016/j.molimm.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Nollet F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein B.E. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Cui P. Hydroxyurea-induced global transcriptional suppression in mouse ES cells. Carcinogenesis. 2010;31:1661–1668. doi: 10.1093/carcin/bgq106. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S. MiRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan N. Microarray profiling of microRNAs expressed in testis tissues of developing primates. J Assist Reprod Genet. 2009;26:179–186. doi: 10.1007/s10815-009-9305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W., Zhao Y., Cui P., Lin Q., Ding F., Xin C. Thousands of novel transcripts identified in mouse cerebrum, testis, and ES cells by ribo-minus RNA sequencing method. Front Genet. 2011;2:93. doi: 10.3389/fgene.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman N.D. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 33.Mikkelsen T.S. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenther M.G. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner A. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roh T.Y. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber M. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guelen L. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramskold D. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.