Abstract
Single nucleotide polymorphisms (SNPs) are genetic variations that determine the differences between any two unrelated individuals. Various population groups can be distinguished from each other using SNPs. For instance, the HapMap dataset has four population groups with about ten million SNPs. For more insights on human evolution, ethnic variation, and population assignment, we propose to find out which SNPs are significant in determining the population groups and then to classify different populations using these relevant SNPs as input features. In this study, we developed a modified t-test ranking measure and applied it to the HapMap genotype data. Firstly, we rank all SNPs in comparison with other feature importance measures including F-statistics and the informativeness for assignment. Secondly, we select different numbers of the most highly ranked SNPs as the input to a classifier, such as the support vector machine, so as to find the best feature subset corresponding to the best classification accuracy. Experimental results showed that the proposed method is very effective in finding SNPs that are significant in determining the population groups, with reduced computational burden and better classification accuracy.
Key words: SNP, modified t-test, F-statistics, SVM
Introduction
Single nucleotide polymorphisms (SNPs) are at present the most common type of genetic variations, that is, the base variations of genetic sequences. It is believed that SNPs determine human diversities, such as different physical traits, different predispositions to diseases, and different responses to medicine. Hence we believe it is important to find out which set of SNPs differentiate individuals into different population groups as well as to be able to accurately classify individuals into different population groups using these relevant SNPs.
For different research objectives, various algorithms on selecting informative SNPs have been developed. For example, selection algorithms for informative SNPs (namely tag SNPs) in association studies 1., 2., 3. are based on a kind of correlation among SNPs, such as the linkage disequilibrium (LD) measure 4., 5., 6., and relevant evaluation measures are adopted to see how those selected tag SNPs predict or represent other SNPs. In population studies, SNPs are selected to classify different populations, therefore, tag SNP selection methods are different from those in association studies. Related researches, such as selecting genetic markers with the highest informativeness for inference of individual ancestry (7), selecting informative marker panels for population assignment (8), and detecting ethnically variant SNPs, have already been developed. In 2003, Rosenberg et al. (7) proposed to use the informativeness for assignment (In) to measure the ability of each genetic loci or marker (feature) to infer individuals’ ancestry, which is proved to be similar to the F-statistics measure (9). In 2005, Rosenberg et al. (8) proposed four algorithms, including exhaustive, univariate, greedy, and maximum algorithms, to select marker panels with performance approaching the maximum. The four algorithms were realized through a given performance function, namely the optimal rate of correct assignment, which measures the probability of correctly assigning an individual to the population from which the genotype of the individual has originated with the greatest possibility. The application of the algorithms on eight species seems effective (8).
In this study, we propose a novel computational way to find out the set of tag SNPs that should lead to the best classification accuracy. Different from previous algorithms 1., 2., 3., 7., 8., firstly we use a feature importance ranking measure, for instance a modified t-test (10) or F-statistics (9), to rank each SNP (the input feature) according to its discriminative capability. Secondly, according to the ranking list, we greedily choose different SNP subsets with different numbers of SNPs (5, 10, 50, 100, and so on), and test them on a classifier, such as the support vector machine (SVM) 11., 12.. The proper feature subset (tag SNP subset) is the one with the highest classification accuracy and the minimum size.
Since no paper has been published on population classification using the SNPs of 22 chromosomes (without considering the sex chromosomes X and Y) in the HapMap genotype data (www.hapmap.org), we cannot make a comparison between our method and other methods. Instead, we compared the two ranking measures, that is, the modified t-test and F-statistics, with different numbers of top ranking features.
Algorithms
In many existing feature selection algorithms, feature ranking is often used to show which input features are more important 13., 14., especially when datasets are very large. Here we introduce two feature importance ranking measures: t-test (10) and F-statistics (9), both of which were used in our experiment for comparisons, and we modified the t-test for our application 15., 16..
Modified t-test
The most common type of t-test, namely the student t-test (10), is often used to assess whether the means of two classes are statistically different from each other by calculating a ratio between the difference of two class means and the variability of the two classes. The t-test has been used to rank features (genes) for microarray data 17., 18. and for mass spectrometry data 19., 20.. These uses of t-test are limited to two-class problems. For multi-class problems, Tibshirani et al. (15) calculated a t-statistics value (Equation 1) for each gene of each class by evaluating the difference between the mean of one class and the mean of all the classes, where the difference is standardized by the within-class standard deviation.
| (1) |
| (2) |
| (3) |
Here tic is the t-statistics value for the i-th gene (feature) of the c-th class; is the mean of the i-th feature in the c-th class, and is the mean of the i-th feature for all classes; xij refers to the i-th feature of the j-th sample; N is the number of all the samples in the C classes and nc is the number of samples in class c; Si is the within-class standard deviation and S0 is set to be the median value of Si for all the features. Tibshirani et al. (15) used the t-statistics to shrink class means toward the mean of all classes to constitute a nearest shrunken centroid classifier, but did not mention how to use the t-statistics value to rank genes with regard to all the classes. In our previous study (16), we extended the t-score for feature i to be the greatest t-score for all classes for feature i:
| (4) |
The SNP data have nominal components, for example, AA, AT, and TG. However, the existing t-statistics do not handle nominal data. Therefore, we generalize the t-score of each feature as follows:
-
1.
Suppose the feature set is F = {f1, …, fi, …, fg}, and feature i has mi different nominal values represented as .
-
2.Transform each nominal feature value into a vector with the dimension mi:
- 3.
(T denotes transposition of matrix)
The ranking rule is: the greater the t-scores, the more relevant the features.
F-statistics
Another ranking measure used in our experiment is F-statistics, which was originally developed by Wright (9) and used in population genetics to describe the level of heterozygosity in a population. It is somewhat unfortunate that there are many versions of F-statistics that cause confusion in the literature. In our experiment, we adopted the Trochim’s definition (21).
Assuming there are C sub-populations for a given data and each feature contains two alleles (any two different representations in a chromosome region, such as A and T), the F-statistics value (Fst) is calculated as:
| (7) |
where p is the frequency of one allele for one population, and are the mean frequencies of the two alleles for all the populations, respectively, and V arp refers to the variance of one allele. If pc designates the frequency of one allele for the c-th population, we have the following:
| (8) |
| (9) |
Features with larger Fst values are more significant for population classification.
Classifier
SVM 11., 12. has often been applied in bioinformatics because of its attractive features, such as effectively avoiding overfitting and accomodating large feature spaces. Compared with many traditional machine learning approaches, SVM shows significantly better or at least matched performance (22). For these reasons, we used SVM in our experiment to test different feature subsets for finding the best discriminative feature subsets, that is, the one with the best classification accuracy and the minimum size.
Implementation and Results
Experimental data
We applied the method to the genotype data obtained from the HapMap database (www.hapmap.org). The data include the following four populations: CEU, YRI, CHB, and JPT. Here CEU represents Utah residents with ancestry from northern and western Europe. YRI represents Yoruba individuals from Ibadan and Nigeria in Africa. Each of the two population groups has 90 reference individuals (samples) comprised of 30 father-mother-offspring trios. CHB means Chinese Han individuals from Beijing, and JPT represents Japanese individuals from Tokyo. Each of the two population groups has 45 unrelated individuals. For CEU and YRI samples, we removed the children samples so that all the samples are unrelated. Thus the total number of samples used in our experiment is 210. First we carried out classification with the original four population groups. Then we re-did the experiment with the JPT and CHB samples combined as one Asian group, since they have many similar DNA sequence segments.
Most of the data samples are strings of bi-allelic SNPs with each SNP feature containing only two alleles. Few of the SNP features have three or more alleles at each position, which are called multi-allelic, and were omitted in our experiment according to previous studies 1., 3.. We also removed those SNP positions (features) that do not have any popolation information and finally obtained nearly four million SNPs for our experiment.
Data preparation for modified t-test ranking
For each bi-allelic SNP feature, there are at most three feature types (values). For example, if two alleles that constitute a feature are the same, such as A and A, there will be only AA for this feature, which is known as homozygous. Otherwise, two different alleles, such as A and T, will constitute three feature types: AA, AT, and TT, which is called hyterozygous. Therefore, for the three nominal values of each feature, we use three vectors with three dimensions to represent them, that is, (0, 0, 1), (0, 1, 0), and (1, 0, 0). For the feature with the homozygous type, one numeric value is enough to represent it.
Data preparation for F-statistics and In ranking
Instead of using feature vectors to represent the SNP features, we use 1 and 2 to represent the two different alleles for each SNP feature. For example, 1 represents A and 2 represents T if the two alleles at the position are A and T. For each allele, we can calculate its frequency and variation for each SNP feature of each population, as well as the whole population. Besides, we made special calculations for some special allele frequencies, such as for p or q equals to 0 or 100% at certain position. This means that at the SNP position the two alleles are the same for all the individuals. For example, if the SNP feature only has the value AA, then the frequency of the SNP allele A is 100%. Thus this feature has no classification capability for any populations. In this case, we set the Fst value of that feature as 0. In summary, the greater the numerator and the smaller the denominator in Equation 7, the greater the Fst value and the more important the corresponding feature for classification.
Based on this data preparation, we also adopted the measure of informativeness for assignment (In) (7) to rank SNPs and compared the classification result with those of the modified t-test and F-statistics measures.
Classification results
After feature ranking, we used the greedy selection method (23) to form different feature subsets with different sizes for classification. Since the number of features is so large that we cannot handle all the data simultaneously due to memory constraint in the computer, we dealt with one chromosome at a time. First we ranked features in each chromosome separately. Then we combined the 22 ranking lists for the 22 chromosomes together and ranked again to obtain the total ranking list, from which we selected 5, 10, 50, 100, 200, 300, 400, 500, and 1,000 top features to form 9 different feature subsets, respectively. For each feature subset, the training and testing were run 30 times by the SVM. Each time we randomly chose 140 samples as the training set and 70 as the test set.
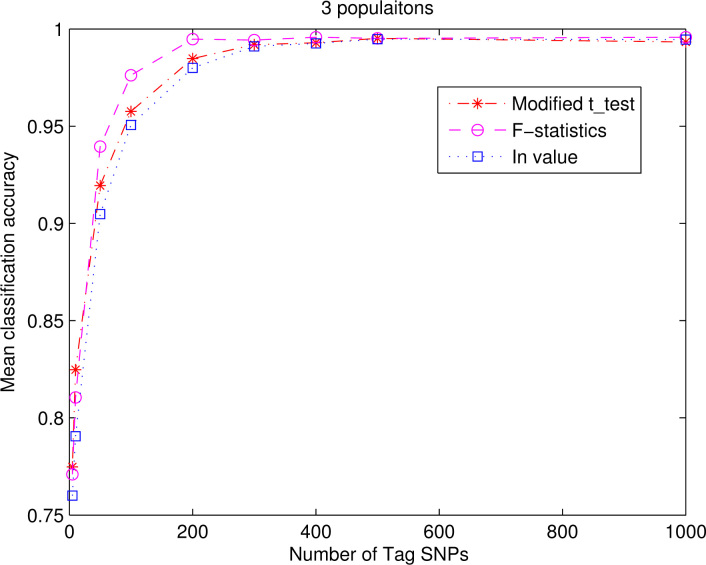
We chose the radial basis function (RBF) kernel for the SVM. The kernel parameter and the penal parameter were decided by cross-validation and grid search method (24). The classification results are shown in Fig. 1, Fig. 2 and Table 1, Table 2, Table 3, Table 4. Due to the rather long computational time required, we used the In measure on only three populations (CEU, YRI, and Asian). Figure 1 shows the classification results on four populations by the modified t-test and F-statistics, and Figure 2 shows the classification result of the In measure together with those of the modified t-test and F-statistics.
Fig. 1.
Average (mean) classification accuracy for the four populations using the modified t-test and the F-statistics.
Fig. 2.
Average (mean) classification accuracy for the three populations using the modified t-test, the F-statistics, and the In measure.
Table 1.
Classification accuracy (standard deviation) for the original four populations using the modified t-test ranking measure
| Number of SNPs | CEU | CHB | JPT | YRI |
|---|---|---|---|---|
| 5 | 66.68% (20.20%) | 39.51% (36.77%) | 21.06% (28.70%) | 88.20% (7.78%) |
| 10 | 72.14% (17.42%) | 33.86% (22.64%) | 39.34% (27.85%) | 93.17% (5.28%) |
| 50 | 82.67% (12.64%) | 43.74% (23.14%) | 55.95% (19.71%) | 99.85% (0.82%) |
| 100 | 88.57% (8.50%) | 54.59% (21.72%) | 49.10% (20.00%) | 100% (0) |
| 200 | 95.98% (5.54%) | 56.88% (20.25%) | 54.95% (19.78%) | 100% (0) |
| 300 | 98.76% (3.21%) | 59.76% (24.17%) | 55.18% (24.59%) | 100% (0) |
| 400 | 99.67% (1.22%) | 58.42% (23.61%) | 55.84% (18.78%) | 100% (0) |
| 500 | 99.86% (0.75%) | 59.09% (21.52%) | 50.06% (21.59%) | 100% (0) |
| 1,000 | 99.40% (1.87%) | 58.34% (23.37%) | 51.40% (26.76%) | 100% (0) |
Table 2.
Classification accuracy (standard deviation) for the original four populations using the F-statistics ranking measure
| Number of SNPs | CEU | CHB | JPT | YRI |
|---|---|---|---|---|
| 5 | 68.55% (23.70%) | 37.63% (32.49%) | 32.12% (27.27%) | 90.11% (9.85%) |
| 10 | 73.27% (12.19%) | 42.12% (30.44%) | 31.02% (22.85%) | 90.65% (9.52%) |
| 50 | 93.42% (7.08%) | 57.17% (33.00%) | 36.39% (32.70%) | 99.61% (1.48%) |
| 100 | 98.79% (2.01%) | 65.00% (33.58%) | 30.81% (33.61%) | 99.85% (0.82%) |
| 200 | 99.62% (1.45%) | 55.80% (39.66%) | 41.50% (40.45%) | 100% (0) |
| 300 | 99.78% (1.20%) | 61.97% (39.59%) | 32.75% (39.26%) | 100% (0) |
| 400 | 99.29% (1.83%) | 64.99% (39.94%) | 32.49% (40.31%) | 100% (0) |
| 500 | 99.78% (1.20%) | 64.33% (39.88%) | 31.95% (39.30%) | 100% (0) |
| 1,000 | 99.47% (1.63%) | 61.18% (41.72%) | 36.55% (39.80%) | 100% (0) |
Table 3.
Classification accuracy (standard deviation) for the three populations using the modified t-test ranking measure
| Number of SNPs | CEU | Asian (CHB and JPT) | YRI |
|---|---|---|---|
| 5 | 48.06% (34.28%) | 91.98% (8.69%) | 87.34% (9.34%) |
| 10 | 62.38% (29.83%) | 90.44% (10.59%) | 92.61% (9.13%) |
| 50 | 82.07% (13.34%) | 94.27% (5.22%) | 99.08% (2.68%) |
| 100 | 90.45% (8.94%) | 97.02% (3.37%) | 100% (0) |
| 200 | 96.96% (3.64%) | 98.61% (2.12%) | 100% (0) |
| 300 | 98.42% (3.21%) | 99.13% (1.45%) | 100% (0) |
| 400 | 98.94% (1.93%) | 99.13% (1.45%) | 100% (0) |
| 500 | 99.67% (1.25%) | 99.13% (1.45%) | 100% (0) |
| 1,000 | 99.22% (2.05%) | 99.03% (1.49%) | 100% (0) |
Table 4.
Classification accuracy (standard deviation) for the three populations using the F-statistics ranking measure
| Number of SNPs | CEU | Asian (CHB and JPT) | YRI |
|---|---|---|---|
| 5 | 60.25% (25.57%) | 84.65% (11.23%) | 88.29% (12.96%) |
| 10 | 67.06% (17.97%) | 84.81% (11.01%) | 89.84% (8.43%) |
| 50 | 87.84% (8.09%) | 95.30% (4.17%) | 98.65% (3.34%) |
| 100 | 99.48% (4.62%) | 98.46% (1.66%) | 99.86% (0.78%) |
| 200 | 99.36% (1.66%) | 99.23% (1.39%) | 100% (0) |
| 300 | 99.34% (1.69%) | 99.13% (1.45%) | 100% (0) |
| 400 | 99.84% (0.85%) | 99.13% (1.45%) | 100% (0) |
| 500 | 99.69% (1.16%) | 99.13% (1.45%) | 100% (0) |
| 1,000 | 99.84% (0.85%) | 99.13% (1.45%) | 100% (0) |
Discussion
From Fig. 1, Fig. 2, we can see that when the number of features increases, the average classification accuracy gradually increases, and at certain number of input features the accuracy reaches the highest. For the original four populations (Figure 1), with the top 400 features obtained from the modified t-test and F-statistics, the accuracy is on average 81.00% and 77.43%, respectively. For the three ethnic populations (classes) (Figure 2), the accuracy with the top 400 features is on average 99.29% for the modified t-test, 99.57% for the F-statistics, and 99.27% for the In measure. Therefore, we can see that among the original nearly four million SNPs, only a minority of them, like 400 or so, are very important for differentiating the populations, while most of the four million SNPs may be redundant or irrelevant.
Classification on the three ethnic populations achieved much better results than that on the four populations, since JPT and CHB have many similar SNP features and are therefore difficult to discriminate. Table 1, Table 2, Table 3, Table 4 present the classification accuracy of each class for the previous two situations, that is, classification on the four original populations and the three ethnic populations, respectively. Comparing between Table 1, Table 3, and between Table 2, Table 4, we can see that the major difference is the accuracy between the Asian group (CHB and JPT combined) and the separate CHB and JPT groups. For example, in Table 3, with the top 300 features, the Asian group’s classification accuracy is on average 99.13% with a standard deviation 1.45%, while in Table 1, with the top 300 features, CHB as one single population has an average accuracy of 59.76% with a greater standard deviation 24.17%, and JPT as one single population has an average accuracy of 55.18% with a standard deviation 24.59%. In Table 4, with the top 200 features, the Asian group reaches the highest classification accuracy of 99.23% with a standard deviation 1.39%. While in Table 2, with the top 200 features, CHB has an average accuracy of 55.80% with a standard deviation 39.66%, and JPT has an average accuracy of 41.50% with a standard deviation 40.45%. For the YRI populations, it can be completely classified (100% accuracy) with the top 100 features from the modified t-test ranking measure, while with the F-statistics ranking measure, the average accuracy is 99.85% with the top 100 features.
The top ranked tag SNPs are not equally distributed for all chromosomes; however, the distribution does not vary significantly in chromosomes. For example, when classifying the three populations, there are at least 1 and at most 10 SNPs from each chromosome among the top 100 discriminative tag SNPs.
As to the two ranking measures, the modified t-test and the F-statistics, the classification results they brought about do not differ very much. As to the In measure (7), the results in Figure 2 show that the In measure produces results similar to those of the modified t-test and the F-statistics when we gradually add the number of tag SNPs. This is consistent with Rosenberg’s conclusions (7) on the relationship between the In measure and the F-statistics.
Conclusion
In this study, we proposed to find out which SNPs are significant in determining the population groups and then to classify different populations using these relevant SNPs as the input features. We proposed a modified t-test ranking measure based on those discussed in previous studies 15., 16., applied it to the problem of classifying populations from the HapMap genotype data, and compared the results with those obtained using the F-statistics measure (21) and the In measure (7). The results showed that the performance of the modified t-test is comparable with those of the F-statistics and the In measure.
It is very important to realize population classification with few SNPs. The significance of this work can be viewed from two aspects. From a computational point of view, it would be much cheaper to handle several hundred SNPs rather than the original ten million common SNPs directly. Some of the SNPs may be irrelevant and therefore act as “noise” to tasks of classification and clustering. From a biological point of view, reducing the number of irrelevant SNPs can facilitate geneticists to focus on fewer SNPs, so as to reduce genotyping cost and increase efficiency of association studies and population studies.
At the same time, we should notice that for this application we only did a coarse feature selection (greedy selection of feature subsets after feature ranking). We did not detect feature correlations in order to remove those redundant features. In our future work, we will deal with those redundant features by calculating correlations among features or by clustering. Furthermore, we will also try to form novel feature combinations, in which selected features need not be the most highly ranked and the size of the feature subset can be further reduced 16., 25.. Besides, comparisons with other methods were not carried out in this study for the same data because of the unavailability of results for classifying the populations.
Authors’ contributions
NZ collected the datasets, conducted data analysis, and prepared the manuscript. LW proposed the idea and assisted with manuscript revision. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank anonymous reviewers for their careful scrutiny of the paper and suggestions that help to improve the paper.
References
- 1.Halperin E. Tag SNP selection in genotype data for maximizing SNP prediction accuracy. Bioinformatics. 2005;21:i195–i203. doi: 10.1093/bioinformatics/bti1021. [DOI] [PubMed] [Google Scholar]
- 2.Liu T.F. Effective algorithms for tag SNP selection. J. Bioinform. Comput. Biol. 2005;3:1089–1106. doi: 10.1142/s0219720005001521. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Altman R.B. Finding haplotype tagging SNPs by use of principal components analysis. Am. J. Hum. Genet. 2004;75:850–861. doi: 10.1086/425587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phuong T.M. Choosing SNPs using feature selection. Proc. IEEE Comput. Syst. Bioinform. Conf. 2005:301–309. doi: 10.1109/csb.2005.22. [DOI] [PubMed] [Google Scholar]
- 5.Devlin B., Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard J.K., Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg N.A. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg N.A. Algorithms for selecting informative marker panels for population assignment. J. Comput. Biol. 2005;12:1183–1201. doi: 10.1089/cmb.2005.12.1183. [DOI] [PubMed] [Google Scholar]
- 9.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
- 10.Devore J., Peck R. third edition. Duxbury Press; Pacific Grove, USA: 1997. Statistics: The Exploration and Analysis of Data. [Google Scholar]
- 11.Vapnik V.N. John Wiley & Sons, Inc.; New York, USA: 1998. Statistical Learning Theory. [Google Scholar]
- 12.Wang L. Springer; Berlin, Germany: 2005. Support Vector Machines: Theory and Applications. [Google Scholar]
- 13.Guyon I., Elisseeff A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003;3:1157–1182. [Google Scholar]
- 14.Wang L., Fu X. Springer; Berlin, Germany: 2005. Data Mining with Computational Intelligence. [Google Scholar]
- 15.Tibshirani R. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L. Accurate cancer classification using expressions of very few genes. IEEE/ACM Trans. Comput. Biol. Bioinform. 2007;4:40–53. doi: 10.1109/TCBB.2007.1006. [DOI] [PubMed] [Google Scholar]
- 17.Jaeger J. Improved gene selection for classification of microarrays. Pac. Symp. Biocomput. 2003:53–64. doi: 10.1142/9789812776303_0006. [DOI] [PubMed] [Google Scholar]
- 18.Su Y. RankGene: identification of diagnostic genes based on expression data. Bioinformatics. 2003;19:1578–1579. doi: 10.1093/bioinformatics/btg179. [DOI] [PubMed] [Google Scholar]
- 19.Wu B. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics. 2003;19:1636–1643. doi: 10.1093/bioinformatics/btg210. [DOI] [PubMed] [Google Scholar]
- 20.Levner I. Feature selection and nearest centroid classification for protein mass spectrometry. BMC Bioinformatics. 2005;6:68. doi: 10.1186/1471-2105-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trochim W.M. second edition. Atomic Dog Publishing; Mason, USA: 2001. The Research Methods Knowledge Base. [Google Scholar]
- 22.Hua S., Sun Z. A novel method of protein secondary structure prediction with high segment overlap measure: support vector machine approach. J. Mol. Biol. 2001;308:397–407. doi: 10.1006/jmbi.2001.4580. [DOI] [PubMed] [Google Scholar]
- 23.Kwak N., Choi C.H. Input feature selection by mutual information based on Parzen window. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24:1667–1671. [Google Scholar]
- 24.Hsu C.W. Department of Computer Science, National Taiwan University; 2003. A practical guide to support vector classification. Technical report. [Google Scholar]
- 25.Liu B. An efficient semi-unsupervised gene selection method via spectral biclustering. IEEE Trans. Nanobioscience. 2006;5:110–114. doi: 10.1109/tnb.2006.875040. [DOI] [PubMed] [Google Scholar]