Abstract
We describe the GALT-Prot database and its related web-based application that have been developed to collect information about the structural and functional effects of mutations on the human enzyme galactose-1-phosphate uridyltransferase (GALT) involved in the genetic disease named galactosemia type I. Besides a list of missense mutations at gene and protein sequence levels, GALT-Prot reports the analysis results of mutant GALT structures. In addition to the structural information about the wild-type enzyme, the database also includes structures of over 100 single point mutants simulated by means of a computational procedure, and the analysis to each mutant was made with several bioinformatics programs in order to investigate the effect of the mutations. The web-based interface allows querying of the database, and several links are also provided in order to guarantee a high integration with other resources already present on the web. Moreover, the architecture of the database and the web application is flexible and can be easily adapted to store data related to other proteins with point mutations. GALT-Prot is freely available at http://bioinformatica.isa.cnr.it/GALT/.
Key words: database, mutation, homology modeling, galactosemia, GALT enzyme
Introduction
Genetic diseases are caused by single or multiple nucleotide mutations that are reflected at protein sequence level as point mutations in most cases (1). Databases for genetic pathologies are generally conceived to show mainly a list of mutations at gene level and possibly the related amino acid mutation, with additional information such as clinical features, associated literature, and so on (2). However, less attention is generally given to the impact of the mutations on the protein tertiary structure; it is assumed that a mutation will cause negative consequences, but often we do not know why. The lack of experimental information on the structural organization of mutated proteins is a relevant problem, which is due to the evident difficulty to obtain the needed amount of proteins and to perform experimental structural studies on dozens or even hundreds of different mutants. In this context, the availability of data to understand the effects of the mutations at protein structure level would be very useful to fill the gap between the experimental study and the knowledge of the molecular bases of the pathology.
In this paper, we present a database, GALT-Prot, for storing analysis results on the structure of the human enzyme galactose-1-phosphate uridyltransferase (GALT) (EC 2.7.7.12) and its single point mutants, with a web application to allow its consultation worldwide. This enzyme is associated with the genetic disorder called galactosemia type I (classical galactosemia) (OMIM: 230400), which is caused by more than 180 sequence variations in the GALT gene, of which about 150 are missense mutations 3., 4., 5., 6., 7..
The three-dimensional (3D) structure of this human enzyme has not yet been obtained by experimental methods, but it has been created by homology modeling methods (8). On the basis of this model, we have been able to investigate the position and the influence of each residue on the structure and on the dimeric assembly of the enzyme. Moreover, using a fully automated procedure, we have been able to create the structures of GALT mutants described in literature, and to analyze their structural features as well, with the aim of explaining molecular events that could be related to this pathology (Marabotti et al., manuscript in preparation).
These analyses may improve the comprehension of the structural and functional features of the wild-type enzyme, and the effects of the missense mutations on GALT structure and function. Therefore, we decided to organize them in a database and to share them with the widest possible number of people via a web-based interface, in an interactive and up-to-date way. The database is now freely available at http://bioinformatica.isa.cnr.it/GALT/. To our knowledge, this is the first database and web resource for galactosemia that is dedicated to the analysis of the protein and the effects of the mutations on the protein structure and function, since other resources [such as ARUPdb (3)] are mainly focused on the collection of GALT mutations at genetic level and on the description of their clinical outcome. The integration of information stored in “traditional” databases with those hosted by our database would give more complete and direct information, with a positive impact on the comprehension of all the elements linked to the genetic disease.
Resource Description
Application overview
GALT-Prot allows storing and disseminating information about structural and functional features of human GALT enzyme, with the possibility of constant update. Moreover, the architecture of the database and the web application is flexible and allows storing data related to other proteins with mutations, without the need of main changes.
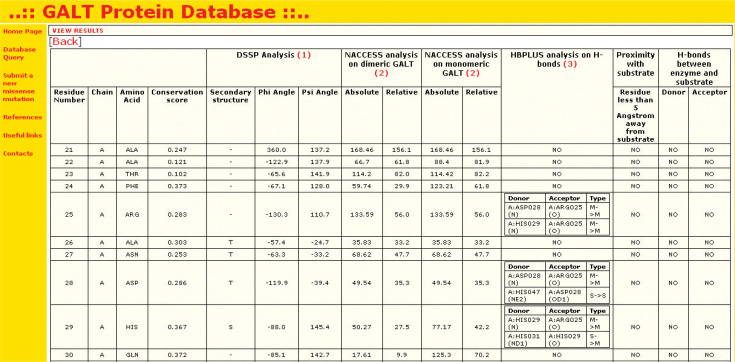
The web application is composed of two main sections, one for the wild-type protein and the other for mutants. In the first section, users can retrieve the information about wild-type GALT stored in the database, all together or focusing the research to one or more kinds of structural and functional information. Filters can be applied to retrieve information related to one residue by using its sequence number, or related to a residue type. Information available on the wild-type protein (Figure 1) includes: the conservation score of each residue in the 3D model of the protein (which starts from residue 21 of the human GALT sequence), the local secondary structure context attributed by DSSP software (9) in terms of secondary structure code, φ and ϕ angles, the solvent accessible surface area computed with the aid of NACCESS software (10), both in the monomer and in the dimeric assembly (a difference between the values suggests the involvement of the residue in the dimer interface), the involvement of the residue in H-bonds detected by means of HBPLUS software (11), and the analysis of enzyme-substrate interactions obtained by visual inspection of the 3D model of the wild-type enzyme bound to the substrate. These kinds of information can help in understanding the role of each residue in structure, activity and dimeric assembly of the protein, and, indirectly, what kind of molecular features would be affected if the selected residue(s) would be involved in mutations.
Figure 1.
Example of results of a search for the information about the wild-type protein. The table contains the analysis results of the protein structure using the tools DSSP, NACCESS and HBPLUS.
In the second section, users can indicate the sequence number of a residue involved in mutations, or select a particular sequence mutation (for example, from Ala to Ser), provided that it exists. The application outputs a table (Figure 2A) that contains information on original and mutant codons and amino acids as reported by literature, the conservation score of the corresponding residue, and the primary literature reference associated with each mutation. These references are reported in a page that allows direct link, when available, to PubMed abstracts. Moreover, for each mutation a linked web page hosts all information obtained on that mutation, in order to ensure a high flexibility of the application (Figure 2B). In each web page, it is possible to find an overview of the mutation with a description of its features derived from a cross-link to the SwissProt/UniProt database (12). A link allows people to download the PDB file of the mutant protein, obtained using a Python script implemented in the MODELLER program (13) as described in Materials and Methods. We also report the results of structural analyses performed on the mutant, with the aim to highlight some drawbacks of the structure following the introduction of these mutations and to help people in detecting which could be the most significant impairments introduced by each mutation in the structure and function of GALT enzyme. These analyses are shown together with the corresponding results obtained for the wild-type residue in the 3D model of GALT protein, to help the comparison between the different features.
Figure 2.
Example of results of a search for the information about the mutant protein. A. The table contains the name of the mutant, the original and mutant codon and amino acid, and conservation score. B. An example of static page in which the information about general and structural features and the prediction of stability of the mutant is shown.
Additional sections of the web application include several links to external web sites that provide general information on classical galactosemia and on GALT gene or protein stored in scientific or in general resources (including links to GALT gene databases), and to web sites of patient associations and non-profit organizations.
Data submission and management
Users can submit information about newly detected mutations by means of a form. The administrator receives the information and performs the validation of the submission, then the structure of the new mutant is modeled, analyzed and the database is updated by means of a stand-alone application. When the data are added, information linked to the particular residue is retrieved and visualized in the database. At this point, people submitting the mutations are alerted that their model is available for analysis. Another form is provided to contact the database administrator without submitting mutations. At present, it is not possible to model and analyze the structure in an interactive way, but we are planning to allow it in the future.
Materials and Methods
Creation and analysis of mutants
The 3D model of human GALT enzyme (8) was used as a starting point to analyze structural features of the wild-type enzyme and to create 107 single point mutants related to galactosemia, selected on the basis of the presence of published references. Information about the list of gene mutations can be found in the literature (14) and in the public database of GALT mutations at genetic level (GALTdb) developed by Calderon and co-workers (3). Mutants were modeled using a Python script implemented in the MODELLER program v8.2 (13) (http://salilab.org/modeller/wiki/Mutate_model). This script implements a fully automated procedure that has been developed to model mutations in protein structures (15).
We employed protein structure analysis software DSSP (9), NACCESS (10), and HBPLUS (11) to extract information about secondary structure, relative solvent accessibility, and H-bond patterns, thus evaluating variations between each mutant and the wild type. An in-house script was also developed to predict the presence of salt bridges in the protein. Moreover, each mutant structure was submitted to two different web servers, PoPMuSiC (16) and DMUTATION (17), to predict the mutation-induced change of protein stability with respect to the wild-type enzyme. Since the two servers use different criteria to evaluate the impact of mutation on stability, we decided to consider it reliable only when the results of both predictors reached a consensus, and the mutant protein was classified into “more unstable”, “unchanged” or “more stable”, taking the wild-type protein as reference. When a consensus between the two methods is not reached, the effect is not determined.
In addition to these kinds of information, for each residue an evaluation of its conservation in the whole GALT family was performed with the AMAS server based on the algorithm by Livingstone and Barton (18).
System design and development
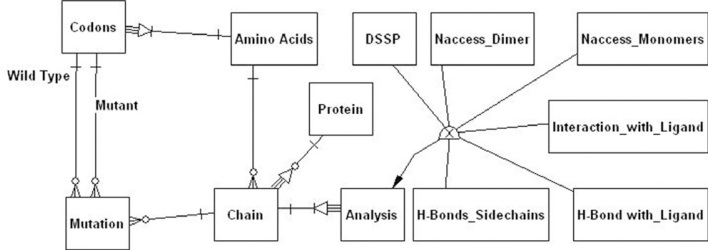
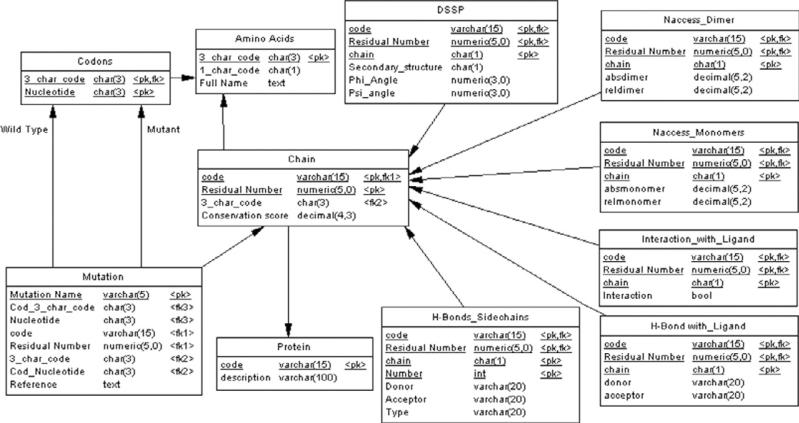
The data were firstly modeled using an entity-relationship (ER) diagram (19) where some entities are worth to be noted (Figure 3). The Protein entity, for example, is introduced to make the final database capable of storing data not just for the GALT protein. The Chain entity, a weak entity whose occurrences are identified by a code and by the corresponding protein, is used to model amino acid chains. The Analysis entity is again a weak entity identified by the element of the chain under study, and is specialized into several entities (H-bonds, DSSP, Monomer, etc). The occurrence of the Mutation entity represents a mutation to be stored. Then, we translated the ER diagram into a logical model (Figure 4). Since we are interested in using a classical relational database management system, we have to eliminate the generalization; therefore we eliminated the Analysis entity and just remained its child entities in order to avoid a lot of null values. We have also considered the performance of the whole database, introducing several indexes and some views to make queries simpler.
Figure 3.
Scheme of the entity-relationship (ER) model.
Figure 4.
Scheme of the logical model.
To realize the web application, we used Java as the coding language and employed Struts (20), a framework that implements the Model 2 approach (a widely adopted variant of the Model-View-Controller design paradigm). Here a Controller servlet acts as a controller for the whole application while the business logic resides into java beans and other helper classes (the Model). The presentation layer (the View) is realized using JSP pages and tag libraries. Eclipse (21) and Exadel (http://www.exadel.com) are used as development tools.
Authors’ contributions
AdA designed and developed the database and the web application. AF created the web pages related to the mutants and the scripts to perform the analyses. Both authors participated in drafting the manuscript. AM performed the analyses on the wild-type and mutant proteins, prepared the manuscript and supervised the project. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank Dr. Ing. Michele Festa for his involvement in the first phases of this project, and Dr. Andrew C.R. Martin for fruitful discussions during the first planning of the database. This work has been developed in the frame of the CNR-Bioinformatics Project.
References
- 1.Beaudet A.L. Genetics, biochemistry and molecular bases of variant human phenotypes. In: Scriver C.R., editor. The Metabolic and Molecular Bases of Inherited Disease. eighth edition. McGraw-Hill; Columbus, USA: 2001. pp. 3–45. [Google Scholar]
- 2.Wishart D.S. Metabolism and metabolic disease resources on the web. In: Valle D., editor. The Online Metabolic and Molecular Basis of Inherited Disease (OMMBID) McGraw-Hill; Columbus, USA: 2008. [Google Scholar]
- 3.Calderon F.R. Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum. Mutat. 2007;28:939–943. doi: 10.1002/humu.20544. [DOI] [PubMed] [Google Scholar]
- 4.Holton J.B. Galactosemia. In: Scriver C.R., editor. The Metabolic and Molecular Bases of Inherited Disease. eighth edition. McGraw-Hill; Columbus, USA: 2001. pp. 1553–1587. [Google Scholar]
- 5.Segal S. Galactosaemia today: the enigma and the challenge. J. Inher. Metab. Dis. 1998;21:455–471. doi: 10.1023/a:1005402618384. [DOI] [PubMed] [Google Scholar]
- 6.Tyfield L. Classical galactosemia and mutations at the galactose-1-phosphate uridyl transferase (GALT) gene. Hum. Mutat. 1999;13:417–430. doi: 10.1002/(SICI)1098-1004(1999)13:6<417::AID-HUMU1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Tyfield L. Galactosaemia and allelic variation at the galactose-1-phosphate uridyltransferase gene: a complex relationship between genotype and phenotype. Eur. J. Pediatr. 2000;159:S204–S207. doi: 10.1007/pl00014404. [DOI] [PubMed] [Google Scholar]
- 8.Marabotti A., Facchiano A.M. Homology modeling studies on human galactose-1-phosphate uridylyltransferase and on its galactosemia-related mutant Q188R provide an explanation of molecular effects of the mutation on homo- and heterodimers. J. Med. Chem. 2005;48:773–779. doi: 10.1021/jm049731q. [DOI] [PubMed] [Google Scholar]
- 9.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard S.J. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 11.McDonald I.K., Thornton J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 12.UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 14.Elsas L.J., 2nd, Lai K. The molecular biology of galactosemia. Genet. Med. 1998;1:40–48. doi: 10.1097/00125817-199811000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Feyfant E. Modeling mutations in protein structures. Protein Sci. 2007;16:2030–2041. doi: 10.1110/ps.072855507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilis D., Rooman M. PoPMuSiC, an algorithm for predicting protein mutant stability changes: application to prion proteins. Protein Eng. 2000;13:849–856. doi: 10.1093/protein/13.12.849. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H., Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livingstone C.D., Barton G.J. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 19.Chen P.P.S. The entity-relationship model—toward a unified view of data. ACM Trans. Database Syst. 1976;1:9–36. [Google Scholar]
- 20.Holmes J. Struts: The Complete Reference. second edition. McGraw-Hill; Columbus, USA: 2006. [Google Scholar]
- 21.Gallardo D. Eclipse in Action: A Guide for the Java Developer. seventh edition. Manning Publications; Greenwich, USA: 2003. [Google Scholar]