Abstract
Array comparative genomic hybridization (CGH) has been popularly used for analyzing DNA copy number variations in diseases like cancer. In this study, we investigated 82 sporadic samples from 49 breast cancer patients using 1-Mb resolution bacterial artificial chromosome CGH arrays. A number of highly frequent genomic aberrations were discovered, which may act as “drivers” of tumor progression. Meanwhile, the genomic profiles of four “normal” breast tissue samples taken at least 2 cm away from the primary tumor sites were also found to have some genomic aberrations that recurred with high frequency in the primary tumors, which may have important implications for clinical therapy. Additionally, we performed class comparison and class prediction for various clinicopathological parameters, and a list of characteristic genomic aberrations associated with different clinicopathological phenotypes was compiled. Our study provides clues for further investigations of the underlying mechanisms of breast carcinogenesis.
Key words: breast cancer, genomic aberration, array CGH, clinicopathological parameter
Introduction
Breast cancer is the most common cancer in women, comprising 23% of all female cancers, and it ranks second in overall cancer incidence when both sexes are considered. There were an estimated 1.15 million patients diagnosed with breast cancer worldwide in 2002 (1). Like other solid cancers, breast cancer presents genomic instability. The current concept is that frequently occurring regions of DNA amplification commonly harbor oncogenes, whereas regions of recurrent deletion harbor tumor suppressor genes. Classical cytogenetic methods have been used to detect such copy number changes in tumors (2), which have deepened our understanding of the genomic hallmarks of breast cancer. In recent years, array comparative genomic hybridization (CGH) has proven its value for analyzing DNA copy number variations in diseases like cancer (3). In this study, we analyzed a total of 82 sporadic samples from 49 breast cancer patients using bacterial artificial chromosome (BAC) CGH arrays with a resolution of 1 Mb on average, revealing a number of frequently recurring genomic aberrations.
Morphologically normal regions adjacent to primary tumor site might harbor genomic aberrations and these genetically altered cells, if they exist, might represent early precursors of breast cancer and/or markers of increased risk. In this study, we analyzed four “normal” tissues that were adjacent but at least 2 cm away from the primary tumor sites. Notably, some recurrent aberrations were present in these samples, which might have important clinical implications.
In breast cancer, axillary lymph node (ALN) metastases are the most common metastastic form and usually have poor prognosis (4). Comparisons of matched pairs of primary tumors and lymph node metastases show similar phenotypes in histology, proliferation activity, and gene expression 5., 6., 7., 8., 9., 10.. Our previous study based on array CGH, 2D-PAGE, and immunohistological approaches revealed that the main characteristics of the primary tumors are maintained in the ALN metastases (11). In the present study, we performed class comparison and class prediction to see whether there are some clones in the CGH profile that can distinguish the primary tumors from lymph node metastases.
At present, the classification and prognosis of breast cancer patients are based on clinicopathological parameters, such as tumor type, malignancy grade, and regional lymph node status. The selection criteria for adjuvant therapy are based on the presence or absence of the ALN metastases (12), steroid receptor status [estrogen receptor (ER) and progesterone receptor (PgR)], and whether the gene for human epidermal growth factor receptor 2 (HER2/neu) is amplified or not 13., 14.. Despite recent developments, the present clinicopathological parameters are generally considered to be not sufficient for the optimum management of patients (12). Therefore, there is a need for more accurate prognostic parameters. With the development of gene expression microarray analyses and CGH, a new molecular classification of breast cancer is being established (15). In the present study, class comparison and class prediction according to a variety of traditional parameters were performed, and the relationships between these parameters and the genomic aberration profiles obtained by array CGH were assessed. Our findings provide clues to deepen the understanding of breast cancer tumorigenesis.
Results
Genomic aberrations in primary tumors
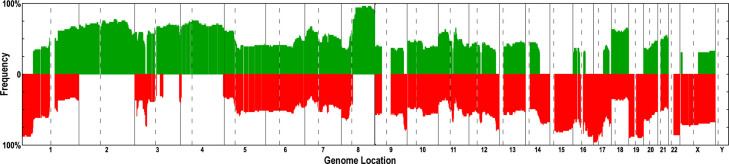
In our previous study, 29 pairs of breast cancer primary tumors and their matched ALN metastases (58 samples in total) were analyzed (11). In this study, we extended the total sample size to 82 samples from 49 patients, including 49 primary tumors, 29 ALN metastases, and 4 “normal” breast tissues. The recurring aberration regions observed in the 49 primary tumor samples are shown in Figure 1 and those frequently occurred in over 30% of the 49 samples are shown in Table S1 (see Materials and Methods for details). The frequent aberration regions were as follows: gains in 2p25.3–q37.3, 3q11.2–13.13, 3q21.1–29, 4p16.2–q35.1, and 8q11.21–q24.3, whereas losses in 1p36.31–33, 3p21.31–21.1, 9q33.3–q34.3, 14q23.2–32.33, 15q11.2–26.3, 16p11.2–q12.1, 17p13.3–q21.32, 17q25.1–25.3, 19p13.3–q13.43, 22q11.23–13.33, and Xp22.2–q21.1 (Figure 1 and Table S1). Known oncogenes and tumor suppressor genes located in the above aberration regions were listed: MYC locates in the amplified region 8q11.21–24.3 that is aberrant in 91% of the 49 primary samples. RAS family genes, such as RASA2 (3q21.1–29, aberrant in 68% samples), and other genes involved in cell proliferation, such as MAPK10, EGF, and FGF2 (4p16.2–q35.1, aberrant in 70% samples), were also found to anchor frequent regions of gain. The region of 17p13.3–q21.32, containing TP53 and BRCA1, showed a DNA copy number loss in 88% samples. Genes related to growth arrest and cell proliferation checkpoints, such as DDIT3 (12q13.11–13.3, aberrant in 67% samples), were also found to anchor frequently deleted regions.
Figure 1.
Recurrent genomic abnormalities in 49 primary breast tumor samples revealed by array CGH. Frequencies of genome copy number gains and losses are plotted as a function of genome location with chromosome 1pter to the left and chromosomes 22 and X to the right. Vertical lines indicate chromosome boundaries and vertical dashed lines indicate centromere locations. Green and red columns indicate frequencies of tumors showing copy number increases and decreases, respectively.
Genomic aberrations in “normal” breast tissues
Notably, the four “normal” tissue samples obtained more than 2 cm away from the primary site of the breast tumors were also found to contain aberrant genomic regions. These regions were largely consistent with the regions having a high frequency of aberrations found in over 30% of the 49 primary tumor samples. The shared aberrant regions, including 1p36.32–34.1, 3p22.1–21.1, 9q33.3–34.3, 11q12.2–13.1, 16p13.3, 16q11.2–12.1, 16q21–24.3, 17p13.3–q25.3, 19p13.3–q13.43, and 22q11.23–13.33, are presented in Table S2, together with the cancer-related genes harbored in these regions.
Unsupervised cluster analysis
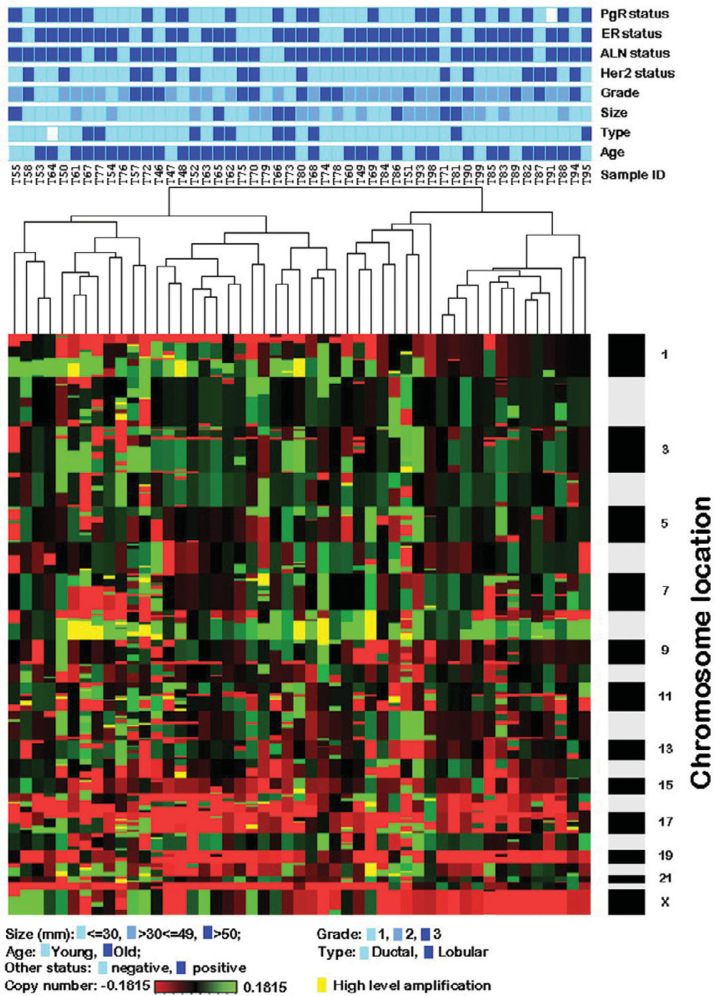
Unsupervised cluster analysis focuses on the identification of novel subtypes of samples that are biologically homogeneous and whose genomic profiles may reflect differences in tumorigenesis (16). This objective is based on the idea that important biological differences among specimens that are clinically and morphologically similar may be discernible at the molecular level (16). In this study, the overview of the distribution of various clinicopathological parameters based on the whole genomic profiles of the 49 primary tumor samples is presented in Figure 2. Furthermore, we used class comparison analysis to identify the clones that can distinguish the different clinicopathological parameters.
Figure 2.
Unsupervised hierarchical clustering of genome copy number profiles measured for 49 primary breast tumor samples. Green indicates increased genome copy number, and red indicates decreased genome copy number. The bar to the right indicates chromosome locations with chromosome 1pter at the top and 22 and X at the bottom. The locations of the odd-numbered chromosomes are indicated. The upper color bars indicate biological and clinical aspects of the tumors. Color codes are indicated at the bottom of the figure. Dark blue indicates positive status, and light blue indicates negative status for ALN, ER, PgR, and HER2. For tumor type, dark blue indicates lobular, and light blue indicates ductal. For age, dark blue indicates old (≥50 years), and light blue indicates young (<50 years). Color codes for grade are as follows: light blue, grade 1; middle blue, grade 2; dark blue, grade 3. For tumor size, light blue indicates size ≤30 mm; middle blue indicates >30 mm ≤49 mm; dark blue indicates >50 mm. Yellow color indicates the high level amplification (log based 2 ratio higher than 3×0.1815).
Class comparison
Class comparison is mainly applied for determining whether genomic profiles differ among samples selected from predefined classes and for identifying which clones are differentially presented among the classes (16). To delineate the genomic aberration patterns between primary breast carcinomas and their ALN metastases, we used the signal-to-noise method to select the 50 clones with the biggest DNA copy number changes in each group. As a result, no clone related DNA copy number changes were statistically different between primary breast carcinomas and their ALN metastases (compared with 5% level permutations), which is in agreement with our previous analysis (11).
Similarly, we selected the 50 clones that revealed the greatest differences in DNA copy number changes between the following phenotypic classes: invasive ductal carcinoma (IDC) vs invasive lobular carcinoma (ILC), ER+ vs ER−, IDC ER− vs IDC ER+ vs ILC ER+, PgR+ vs PgR−, ER−PgR− vs ER+PgR− vs ER+PgR+, HER2/neu+ vs HER2/neu−, size (small vs moderate vs large), size (small vs large), grade 1 vs 2 vs 3, grade (1+2) vs 3, grade 1 vs (2+3), and high grade vs low grade. The numbers of selected clones are presented in Table 1. Cancer-related genes of the selected clones are also noted.
Table 1.
Summary of class comparison (marker selection) in relation to clinicopathological parameters
| Class | Marker selection* | Related genes |
|---|---|---|
| ALN metastases vs primary tumors | none in both groups | |
| ALN+ vs ALN− | none in both groups | |
| Ductal vs Lobular | 9 in ductal, 0 in lobular | NF-AT5 |
| ER+ vs ER− | 49 in ER+, 0 in ER− | GDF-2, GDF-10, JNK-46, CHN1, RCK, JNKK, FSH-R |
| IDC ER− vs IDC ER+ vs ILC ER+ | 50 in IDC ER−, none in the other two groups | GDF-2, GDF-10, JNK-46, CHN1, Sts-1, JNKK |
| PgR+ vs PgR− | 38 in PgR+, 0 in PgR− | |
| ER−PgR− vs ER+PgR− vs ER+PgR+ | 50 in ER−PgR−, 41 in ER+PgR+ | |
| HER2/neu+ vs HER2/neu− | 2 in HER2/neu+, 0 in HER2/neu− | CK-12, CK-20, CK-23 |
| Size (small vs moderate vs large) | none in the three groups | |
| Size (small vs large) | none in both groups | |
| Grade 1 vs 2 vs 3 | 50 in Grade 3, none in the other two groups | |
| Grade (1+2) vs 3 | 50 in Grade 3, none in Grade (1+2) | |
| Grade 1 vs (2+3) | 24 in Grade (2+3), none in Grade 1 | |
| High grade vs Low grade | 50 in high grade, none in low grade |
T-testing was used for marker selection (class comparison). The number of markers with scores higher than the 5% level in 500 permutations (of the selected 50 markers in each group) was counted.
Class prediction
Class prediction is similar to class comparison except that the emphasis is on developing a statistical model that can predict to which class a new sample belongs based on its genomic profile (16). In this study, we constructed classifiers that should distinguish between different tumor genotypes in relation to clinicopathological phenotypes and thereby reveal the underlying relationships between the genotypes and phenotypes. We used the leave-one-out cross validation (LOOCV) method to evaluate the overall performance of classifiers, and classifiers related to the different phenotypes were built and estimated. Classifiers were chosen if the clones give contributions for prediction in at least 70% samples. The clone lists of the final classifiers as well as the overall performance of the final classifiers and the significance of statistical tests (accuracy, sensitivity, and specificity) are summarized in Table S3. Poor classification performance was seen for ALN metastases vs primary tumors, ALN status, tumor size, IDC ER− vs IDC ER+ vs ILC ER+, and PgR status, indicating that these phenotypes present a large variety of genomic aberration profiles not directly correlated to some specific genotypes. Moderate classification performance (73.9% accuracy) was observed for high-grade vs low-grade tumors. Notably, relatively good classification performance was achieved for tumor type (IDC vs ILC) (94.4% accuracy), HER2/neu status (90% accuracy), and ER status (85.7% accuracy).
Discussion
In our study, gains in 3q21.1–29, 4p16.2–q35.1, and 8q11.22–24.3 while losses in 1p36.31–33, 16q12.2–24.3, 17p13.3–q21.32, 19p13.3–q13.43, and 22q11.23–13.33 were the most frequent alterations; this result is consistent with previous findings 17., 18.. Some tumor suppressor genes and oncogenes are located in the loss and gain regions, respectively. For example, the TP53 gene located in 17p13.3–q21.32 deletion region and the MYC proto-oncogene (c-MYC) (transcription factor p64) anchored in 8q11.21–24.3 amplification region are obviously important for sporadic breast cancer carcinogenesis. The frequent genomic aberration regions are expected to have more important biological meanings than the randomly occurring aberrations do, because the recurring abnormalities indicate the presence of “drivers” of the tumor progression, rather than the random “passengers” elsewhere.
Notably, four “normal” tissues far from the primary site shared recurrent aberration regions with the primary tumors. Many genes in these regions are related to tumor development, indicating that these genes are important at the beginning of tumorigenesis. These genes include tumor necrosis factor receptor superfamily members TNFRSF1B, TNFRSF8, TNFRSF9, TNFRSF12A, TNFRSF17, TNFRSF25, ligand members 7, 9, 13, 14, tumor suppressor candidates TUSC2, TUSC4, TUSC5, natural killer-tumor recognition sequence NKTR, and breast cancer metastasis-suppressor 1. Genes related to cell proliferation, DNA repair, cell cycle, and apoptosis regulation, such as TP53 and the genes for cell division protein kinase 9, programmed cell death protein 5 (TFAR19), DNA-repair protein (XRCC1), and apoptosis regulator BAX, also seem to be involved. The genomic aberrations involved with these genes are deletions, suggesting that losses of the genes related to proliferation and apoptosis may play an important role in tumor initiation, giving a selective advantage to the aberrant cells. Notably, Beckmann et al. suggested that the second event in multistep carcinogenesis is usually chromosome loss, mitotic recombination, or partial chromosome deletion after oncogene amplifications and tumor suppressor gene mutations (19). They also mentioned that chromosome loci 16q and 17p seem to be pathognomonic for the development or progression of a specific histological subtype (19). The losses in 16q and 17p were highly consistent with aberrations in “normal” tissues, primary breast carcinomas, and their matched ALN metastases in the present study. Our finding that “normal” cells harbor such alterations suggests that the aberrations in question might be important in relation to tumor initiation and development and also be responsible for breast cancer relapse after surgery.
Decisions regarding postoperative treatment of primary breast cancer are based on clinical (age), histopathological (lymph node status, tumor size, and malignancy grade), and cell biological (ER and PgR) parameters (20). Markers from gene expression micorarray analyses have also shown some promise as a prognostic tool in breast cancer 21., 22.. Thus, novel molecular profiling and classification of breast cancers should eventually give stronger correlation with clinical outcome and patient survival.
The most widely used prognostic factor in breast cancer is ALN status. In one study, node-negative patients had a longer 5-year survival than node-positive patients (23), but no mention was made in that study of either selected markers or a classifier showing statistically significant differences between ALN+ and ALN−. No classifier was constructed that could make a correct prediction of ALN status through analysis of expression profiles of 151 ALN− and 144 ALN+ patients using cDNA expression microarray analysis (10). Similarly, results from array CGH analysis of prostate cancer show genomic profile similarity between the primary prostate cancers and their matched lymph node metastases (24). In this study, we tried to build a CGH-based classifier for primary tumors vs ALN metastases, but no classifier was able to distinguish ALN metastases from the primary tumors, since in general the ALN metastases shared the genomic profiles with their primary carcinomas. The strong similarity of ALN metastases and their primary tumors is obvious at both genomic and proteomic levels as documented and discussed in our previous study (11). This implies that important biological characteristics are already present in the primary breast tumors and maintained in their lymph node metastases. Detailed analyses of the primary tumors should thus be prognostically informative.
Tumor size is one of the common prognostic factors for breast cancer in the clinic (25). Quiet et al. reported that patients with negative ALNs and tumors less than 2 cm, not receiving adjuvant therapy, had a higher disease-free survival rate and longer median survival time than patients with a tumor larger than 2 cm in a 20-year follow-up study (26). In our study, we compared three groups based on tumor size. However, the performance of our classifier was poor, suggesting that there was no significant difference among the three groups related to gross genomic aberrations. Thus, there was no significant correlation between our genomic profiles and tumor size.
For breast cancers, amplification and/or overexpression of HER2/neu occurs in up to 30% of the cases and is associated with aggressive biological behavior that reduces relapse-free and survival time (27). HER2/neu is therefore used for the selection of patients for adjuvant therapy in breast cancer (12). The differential gene expression patterns in HER2/neu amplified and non-amplified breast cancer cell lines and tumors have been investigated previously (28). In our present study, a set of 19 clones were included as a final classifier for HER2/neu (Table S3). Genes located in the classifier clones code for proteins such as tumor endothelial marker 7 (PLXDC1), metastatic lymph node protein 64 (STARD3), epidermal growth factor receptor GRB-7 (GRB7), suppression of tumorigenicity protein 13 (FAM10A6), proliferation-associated nuclear element protein 1 (CENPM), cytokeratin-12 (CK12), cytokeratin-20 (CK20), and cytokeratin-23 (CK23). More importantly, the HER2/neu gene is also located in these clones, the amplification and expression level of which are regarded as criteria for determining HER2/neu status. In the present study, the classifier of HER2/neu showed good performance on cross validation (90% accuracy), suggesting that the difference in HER2/neu status is associated with different genomic aberrations in carcinogenesis.
ER status was reported to be a fundamental differentiating characteristic of breast cancer in gene expression micorarray (9) and CGH studies (29). ER-negative tumors are more aggressive than ER-positive tumors, and the loss of ERs in tumor cells is associated with poor prognosis and poor response to hormonal therapy (30). In previous studies, ER-negative tumors predominantly had amplifications in 17q12, whereas ER-positive tumors had amplifications in 8q, gains in 1q, and losses in 1p and 16q (31). In this study, 25 clones were chosen as the final classifier to predict the ER status (Table S3). Genes related to cell growth and differentiation regulation, including GDF-2 and GDF-10 (coding for growth/differentiation factor 2 and 10 precursors), and genes related to cell proliferation, such as MAPK8 (coding for mitogen-activated protein kinase 8), are located in these clones of this final classifier.
PgR is reported to be another important clinical prognostic parameter in breast cancers. Genes related to tumorigenesis, such as RASAL2 (coding Ras GTPase-activating protein nGAP), PIK3C2B, MDM4 (p53-binding protein Mdm4), RASSF5 (coding Ras association domain-containing family protein 5), IL24 (coding suppression of tumorigenicity 16 protein), FAIM3 (Fas apoptotic inhibitory molecule 3), and PIGR (coding hepatocellular carcinoma associated protein TB6), are harbored in the 24 clones of the final classifier (Table S3). However, the performance of the classifier for PgR is poor (45.8% accuracy).
ER and PgR are mainly used to select patients for endocrine therapy (32). Ma et al. reported in their epidemiological study that ER+PgR+ and ER−PgR− tumors show different association with risk factors, suggesting that these two types of breast cancers have etiologically different hormonal mechanisms (33). In the present study, there were two shared clones (bA534N5 and bA432I13) between ER status and the combination of ER and PgR classifiers, and the performance level of the classifier for ER+PgR+, ER+PgR−, and ER−PgR− (66.7% accuracy) was in the middle of the performance levels of the ER classifier (85.7% accuracy) and the PgR classifier (45.8% accuracy), suggesting that the difference of the three classes of ER+PgR+, ER+PgR−, and ER−PgR− is actually derived from the difference of ER+ and ER−, instead of PgR status. Only one potentially important oncogene RHOT1 (Ras homolog gene family member T1) was located in the six clones of the combined classifier (Table S3).
IDC and ILC are the major histological types of breast cancer. IDC is more predominant, ranging from 47% to 79%, whereas ILC accounts for 2% to 15% (34). Although histologically disparate, these tumor types show similarities in the clinic. In fact, IDC and ILC patients receive similar treatment. However, women with ILC have a risk of mortality that is 11% lower than women with IDC, and the magnitude of this difference has increased over the past 10 years (35). In addition, in previous chromosomal CGH studies, gains of 8q and 20q were often seen in IDC, whereas losses of 16q and 22q were found in ILC 35., 36., 37.. In array CGH studies, Loo et al. reported that 1q and 11q aberrations showed different frequencies between these two types; however, this difference was not statistically significant (18). Stange et al. identified 1q and 16p aberrations as significant classifiers of the two subtypes (38). In this study, we used a marker selection method and found frequent gains in 12q23.3–24.21 and 16q12.1–23.2 for IDC and even more frequent gains in 2q36.3–37.1, 9q13–22.32, 11p13–12, and 11q13.1 for ILC. Additionally, we built a classifier achieving 94.4% accuracy, 100% sensitivity, and 30% specificity (Table S3). The classifier presented more frequent gains in 16q12.1–24.1 in IDC than in ILC, which is consistent with previous studies 36., 37.. The gene CDH1, coding for the epithelial-cadherin precursor (E-cadherin), is included in the final classifier for the discrimination of IDC and ILC. Notably, the difference of E-cadherin copy number between IDC and ILC in our study was consistent with a previous expression profile study of Korkola et al. who reported that lobular tumors showed low expression levels of E-cadherin 39., 40..
In addition to the genomic aberration difference, IDC and ILC differ in hormone receptor statuses: 55%–72% of IDCs present ER+ compared with 70%–92% of ILCs, and 33%–70% of IDCs are PgR+, in contrast to 63%–67% of ILCs (41). Zhao et al. reported some genes whose expressions distinguished between IDC ER+, IDC ER−, and ILC ER+ (41). In our study, we tried to find classifiers that were able to classify the three groups, but the performance of the chosen classifier was not good (data not shown), and the classifier of the three groups has no clones in common with either the classifier of ER or the classifier of IDC vs ILC. Our results imply that there was no consistent difference in DNA copy number changes among IDC ER+, IDC ER−, and ILC ER+.
The malignancy grade has also been considered to have an independent prognostic value (42). Patients classified as grade 1 were reported to have a 95% 9-year survival (43). In the present study, we built classifiers based on grade 1 vs 2 vs 3, grade (1+2) vs 3, and grade 1 vs (2+3). The latter two classifiers did not perform well, whereas the performance of the classifier of grade 1 vs 2 vs 3 was better, but still unsatisfactory (Table S3).
Recently, Simpson et al. mentioned that low- and high-grade invasive breast cancers might represent distinct major pathways of tumor evolution (15), whereas the boundaries between the evolutionary pathways of well-differentiated/low-grade ductal and lobular carcinomas have been blurred (15). In this study, we compared the two groups (high- and low-grade) and performed class prediction. Our result showed that 16 clones (mainly located on chromosomes 1 and 22) composed a classifier achieving a relatively good prediction for high- and low-grade invasive breast cancers (Table S3). Indeed, we observed that 6/8 grade 1 samples present 16q loss in contrast to 3/15 grade 3 samples, which is consistent with Simpson’s report (15).
Breast cancer is a heterogeneous disease encompassing a wide variety of cell subpopulations. Thus a comprehensive and clear delineation of the relationship between clinicopathological parameters and DNA copy number aberrations will depend on new knowledge of tumor heterogeneity. In a continuing study, we are building tumor heterogeneity models based on array CGH data. Integration of many different types of data should deepen our understanding of breast cancer tumorigenesis.
Conclusion
In the present breast cancer study, frequently recurring genomic aberration regions were revealed, and oncogenes and tumor suppressor genes located in the corresponding regions were listed. Importantly, similar recurrent aberrations were found between primary breast carcinomas, their matched ALN metastases, and “normal” tissue samples. We screened the common clinicopathological prognostic parameters, such as ALN, HER2/neu, malignancy grade, tumor size, histological type, ER, PgR and their corresponding combinations, and built up a series of classifiers for the above parameters. The genomic aberration patterns relative to different clinicopathological parameters are presented, providing genetic clues for the study of the underlying mechanisms of tumor development.
Materials and Methods
Sample collection and handling
We collected 82 samples from 49 breast cancer patients, including 49 primary tumors, 29 ALN metastases, and 4 “normal” breast tissues. A summary of the clinicopathological information for the 49 cases is shown in Table 2. All samples were provided by Rigshospitalet (Copenhagen)/Danish Center for Translational Breast Cancer Research and were evaluated using consistent pathological criteria by an experienced pathologist (44). None of the patients received any treatment prior to the sample collection. The project was approved by the Scientific and Ethical Committee of the Copenhagen and Frederiksberg Municipalities (KF 01–069/03).
Table 2.
Summary of the clinicopathological information for the 49 primary tumor samples
| No. of cases |
Percentage |
|
|---|---|---|
| Total patients | 49 | |
| Mean age (range) | 61 (27–99) years | |
| Histological Type*1 | ||
| Ductal, grade 1 | 4 | 8.2% |
| Ductal, grade 2/3 | 34 | 69.4% |
| Lobular, classic type, grade 1 | 3 | 6.1% |
| Lobular, grade 2/3 | 7 | 14.3% |
| Size (mm)*2 | ||
| <30 mm | 19 | 38.8% |
| 30–49 mm | 23 | 46.9% |
| >50 mm | 7 | 14.3% |
| Grade | ||
| 1 | 8 | 16.3% |
| 2 | 26 | 53.1% |
| 3 | 15 | 30.6% |
| HER2/neu*3 | ||
| 0 | 6 | 12.2% |
| 1+ | 15 | 30.6% |
| 2+, not amplified | 13 | 26.5% |
| 2+, amplified | 3 | 6.1% |
| 3+ | 12 | 24.5% |
| Positive | 15 | 30.6% |
| Negative | 34 | 69.4% |
| Axillary lymph node | ||
| Positive | 40 | 81.6% |
| Negative | 9 | 18.4% |
| Estrogen receptor | ||
| Positive (10% or more) | 13 | 26.5% |
| Negative | 36 | 73.5% |
| Progesterone receptor*4 | ||
| Positive (10% or more) | 22 | 44.9% |
| Negative | 26 | 53.1% |
One sample’s histological type is Tu/Kr (tubular/cribriform), a sort of well differentiated ductal variant. The rest are ductal or lobular.
In the clinic, 20 mm is the most common criterion for tumor size. However, in our study, we use 30 and 50 mm thresholds as class criteria, because 20 mm will assign 6 patients in one group and 43 patients in the other group, which will lead to sampling problem for statistic analysis.
HER2/neu positive and negative statuses were determined by both immunohistochemical tests and fluorescence in situ hybridization (FISH) following DAKO criteria [that is, 0, 1+, and 2+ (not amplified) are considered negative, whereas 2+ (amplified) and 3+ are considered positive].
One sample presents a PgR positive phenotype in some staining sections, whereas it is negative in the others.
Immunohistochemistry
Following surgery, fresh tissue blocks were immediately placed in formalin fixative and paraffin embedded for archival use. Antigen was detected with a relevant primary antibody followed by a suitable secondary antibody conjugated to a peroxidase complex (HRP conjugated goat anti-rabbit or antimouse antibody; DakoCytomation, Glostrup, Denmark). Finally, color development was done with 3,3′-diaminobenzidine (Pierce Biotechnology, Inc., Rockford, USA) as a chromogen to detect bound antibody complex. Slides were counterstained with hematoxylin. Standardization of the dilution, incubation, and development times appropriate for each antibody allowed an accurate comparison of expression levels in all cases. At least three independent stainings of the samples were performed for each antibody. Sections were imaged using either a standard bright field microscope (Leica DMRB) equipped with a high-resolution digital camera (Leica DC500), or a motorized digital microscope (Leica DM6000B) controlled by Objective Imaging’s Surveyor Software (Objective Imaging Ltd., UK) for automated scanning and imaging, which enables tiled mosaic image creation. Original magnification for all images was 200x.
Array CGH
The process of array CGH was followed as described previously 11., 45.. Briefly, all the tumor and ALN metastasis samples collected for the study were histologically analyzed and found to contain less than 40% of non-tumor cells. Total genomic DNA was isolated with a DNA isolation kit (NucleoSpin® Tissue, MACHEREY-NAGEL, France) following the manufacturer’s instructions. Reference DNA was derived from a healthy male’s peripheral blood. Arrays for 1-Mb resolution coverage of the whole genome contained elements produced from BAC clones that were obtained from Wellcome Trust Sanger Institute (45). Tumor and reference DNA was labeled by a random priming method with the BioPrime® DNA labeling system (Invitrogen). Cy3-dCTP and Cy5-dCTP (Amersham Biosciences) were used for labeling of the tumor samples and reference DNA, respectively. An amount of 40 uL (1 mg/mL) of Cot-1 DNA (Invitrogen) was used for the suppression of hybridization to repetitive sequences. Tumor and reference DNA was mixed and co-hybridized to the denatured target DNA of the array. The array was placed on a slowly rocking table at 37°C for 48–60 h (45). Some arrays were also hybridized in a TECAN HS 4800 Pro hybridization station. After hybridization, arrays were washed with a series of washing solutions and dried out with nitrogen. The arrays were imaged in a charge-coupled device arrayWorxe scanner (Applied Precision Company) with the Cy3 and Cy5 channels. Two singlechannel 16-bit images were combined for analysis using the image analysis software Tracker (Applied Precision Company). After filtering, clones representing the same DNA sequence were averaged and subjected to base 2 log transformation. Data were then sent to the “DNAcopy” R/Bioconductor package, which uses the circular binary segmentation (CBS) method (46). The output clone segments from “DNAcopy” were merged using a MergeLevel procedure (47). In this process, segmental values across the genome were merged to create a common set of copy number levels for each individual tumor sample. The segments corresponding to the copy number level with the smallest absolute median value were declared unchanged. All the segments for each sample were then normalized by subtracting their corresponding normal level values. In this way, the normal level value would be 0 (log transformation base 2 scale).
Unsupervised clustering
An unsupervised hierarchical clustering method was applied to analyze genomic aberration similarities across the 49 primary tumor samples by using Cluster software v3.0 (48). The correlation algorithm was employed for similarity metric calculation. Complete linkage clustering was chosen to organize samples in a tree structure. TreeView software was utilized for visualization of the results of the cluster analysis (48).
Marker selection and permutation test
The Whitehead’s GeneCluster software package v2.0 (49) was used for the supervised selection of markers and permutation testing. We used t-test statistics to select a certain number of significant marker clones in each group. Permutation of the sample profiles for 500 times was used to test whether the clones in different groups were significantly different or more likely found by chance. All of the t-test scores of markers were compared with the corresponding scores at the 5% level from the 500 random datasets. The markers that were significant at a 5% confidence level (higher than 5% level from the permutation of the class labels) were selected.
KNN and LOOCV
The “build predictor” analysis in the Whitehead’s GeneCluster software (49) was applied to build a classifier for each clinicopathological parameter. Class prediction usually requires identifying which genes are informative for distinguishing the predefined classes, using these clones to develop a statistical prediction model, and estimating the accuracy of the predictor (16). In the present study, a number of features (clones) that were informative for distinguishing the predefined classes were identified using the signal-to-noise method. The setting of the number of features was tested from 1 to 40. Then the k-nearest neighbor (KNN) algorithm (neighbor number set to 3 by default) was applied to develop a statistical prediction model according to the clinicopathological parameters, and the LOOCV method was used to estimate the performance of the prediction. The optimal number of features that had the lowest prediction error rate (the lowest absolute errors) was chosen. The classifiers (a set of clones) with the best performance for individual clinicopathological parameters were determined (49). In order to choose the most important clones in the classifiers, we picked the clones that gave contributions for prediction in at least 70% of the samples. The chosen clones were used to predict all samples again, and the performance of these final classifiers was estimated by absolute error rate using LOOCV. The statistical significance of the prediction results was analyzed by Fisher exact (for two classes) and Chi squire (for three classes) tests.
Information collection of relative clones and genes
All information on the clones and the genes located in the corresponding genomic regions of this study is based on the Snap database established by our group (50). The main site in Denmark is http://snap.humgen.au.dk.
Authors’ contributions
JL participated in the production of the array CGH platform, performed the array CGH study as well as the statistical analysis, and drafted the manuscript. KW carried out CBS and bioinformatic analysis to facilitate data analysis. SL carried out bioinformatic analysis. VTW and FR provided the breast and lymph node samples and performed the pathology analysis. VTW also participated in the modification of the manuscript. CW helped with the statistics and the modification of the manuscript. XZ participated in the development of the array CGH platform. HY gave suggestions for study design and coordination. LB initiated the study and helped drafting the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We would like to thank Prof. Julio E. Celis for kind support. We also thank the Wellcome Trust Sanger Institute for providing 1-Mb resolution BAC clone. This work was supported by the Danish Cancer Society through the budget of the Institute of Cancer Biology and by grants from the Danish Medical Research Council, the Natural and Medical Sciences Committee of the Danish Cancer Society, Novo Nordisk, the John and Birthe Meyer Foundation, the Solar Fonden, the Stensbygaard Fonden, the Kai Lange og Gundhild Kai Lange Fond, the will of Edith Stern, and the “Race Against Breast Cancer” Project. The support of the Marketing Department at the Danish Cancer Society is greatly appreciated. This study was also supported by a project grant from the Hi-Tech Research and Development Program of China (2006AA02A301).
Supporting Online Material
Tables S1–S3
References
- 1.Yang L. Statistics on cancer in China: cancer registration in 2002. Eur. J. Cancer Prev. 2005;14:329–335. doi: 10.1097/00008469-200508000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Struski S. Compilation of published comparative genomic hybridization studies. Cancer Genet. Cytogenet. 2002;135:63–90. doi: 10.1016/s0165-4608(01)00624-0. [DOI] [PubMed] [Google Scholar]
- 3.Pinkel D., Albertson D.G. Array comparative genomics hybridization and its application in cancer. Nat. Genet. 2005;37:S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 4.Dent D.M. Axillary lymphadenectomy for breast cancer. Paradigm shifts and pragmatic surgeons. Arch. Surg. 1996;131:1125–1127. doi: 10.1001/archsurg.1996.01430230007001. [DOI] [PubMed] [Google Scholar]
- 5.Daidone M.G. Proliferative activity of primary breast cancer and of synchronous lymph node metastases evaluated by [3H]-thymidine labelling index. Cell Tissue Kinet. 1990;23:401–408. doi: 10.1111/j.1365-2184.1990.tb01133.x. [DOI] [PubMed] [Google Scholar]
- 6.Feichter G.E. DNA index and cell cycle analysis of primary breast cancer and synchronous axillary lymph node metastases. Breast Cancer Res. Treat. 1989;13:17–22. doi: 10.1007/BF01806546. [DOI] [PubMed] [Google Scholar]
- 7.Goodson W.H., 3rd Tumor labeling indices of primary breast cancers and their regional lymph node metastases. Cancer. 1993;71:3914–3919. doi: 10.1002/1097-0142(19930615)71:12<3914::aid-cncr2820711219>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 8.Lahdesmaki H. Distinguishing key biological pathways between primary breast cancers and their lymph node metastases by gene function-based clustering analysis. Int. J. Oncol. 2004;24:1589–1596. [PubMed] [Google Scholar]
- 9.Perou C.M. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Weigelt B. No common denominator for breast cancer lymph node metastasis. Br. J. Cancer. 2005;93:924–932. doi: 10.1038/sj.bjc.6602794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J. Omics-based profiling of carcinoma of the breast and matched regional lymph node metastasis. Proteomics. 2008;8:5038–5052. doi: 10.1002/pmic.200800303. [DOI] [PubMed] [Google Scholar]
- 12.Hermsen M.A. Genetic analysis of 53 lymph node-negative breast carcinomas by CGH and relation to clinical, pathological, morphometric, and DNA cytometric prognostic factors. J. Pathol. 1998;186:356–362. doi: 10.1002/(SICI)1096-9896(199812)186:4<356::AID-PATH196>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Walker R.A. Molecular pathology of breast cancer and its application to clinical management. Cancer Metastasis Rev. 1997;16:5–27. doi: 10.1023/a:1005740222307. [DOI] [PubMed] [Google Scholar]
- 14.Gregory R.K. Prognostic relevance of cerbB2 expression following neoadjuvant chemotherapy in patients in a randomised trial of neoadjuvant versus adjuvant chemoendocrine therapy. Breast Cancer Res. Treat. 2000;59:171–175. doi: 10.1023/a:1006394317282. [DOI] [PubMed] [Google Scholar]
- 15.Simpson P.T. Molecular evolution of breast cancer. J. Pathol. 2005;205:248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 16.Simon R.M. Springer; New York, USA: 2004. Design and Analysis of DNA Microarray Investigations. [Google Scholar]
- 17.Albertson D.G. Profiling breast cancer by array CGH. Breast Cancer Res. Treat. 2003;78:289–298. doi: 10.1023/a:1023025506386. [DOI] [PubMed] [Google Scholar]
- 18.Loo L.W. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 19.Beckmann M.W. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J. Mol. Med. 1997;75:429–439. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- 20.Goldhirsch A. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. J. Natl. Cancer Inst. 1998;90:1601–1608. doi: 10.1093/jnci/90.21.1601. [DOI] [PubMed] [Google Scholar]
- 21.van’t Veer L.J. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 22.van de Vijver M.J. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 23.Nemoto T. Breast cancer in the medial half. Results of 1978 National Survey of the American College of Surgeons. Cancer. 1983;51:1333–1338. doi: 10.1002/1097-0142(19830415)51:8<1333::aid-cncr2820510802>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 24.Paris P.L. Genomic profiling of hormonenaive lymph node metastases in patients with prostate cancer. Neoplasia. 2006;8:1083–1089. doi: 10.1593/neo.06421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Eredita G. Prognostic factors in breast cancer: the predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur. J. Cancer. 2001;37:591–596. doi: 10.1016/s0959-8049(00)00435-4. [DOI] [PubMed] [Google Scholar]
- 26.Quiet C.A. Natural history of node-negative breast cancer: a study of 826 patients with long-term follow-up. J. Clin. Oncol. 1995;13:1144–1151. doi: 10.1200/JCO.1995.13.5.1144. [DOI] [PubMed] [Google Scholar]
- 27.Revillion F. ERBB2 oncogene in human breast cancer and its clinical significance. Eur. J. Cancer. 1998;34:791–808. doi: 10.1016/s0959-8049(97)10157-5. [DOI] [PubMed] [Google Scholar]
- 28.Wilson K.S. Differential gene expression patterns in HER2/neu-positive and -negative breast cancer cell lines and tissues. Am. J. Pathol. 2002;161:1171–1185. doi: 10.1016/S0002-9440(10)64394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard F. Patterns of chromosomal imbalances in invasive breast cancer. Int. J. Cancer. 2000;89:305–310. [PubMed] [Google Scholar]
- 30.Clark G. Prognostic and predictive factors. In: Harris J.R., editor. Diseases of the Breast. second edition. Lippincott Williams & Wilkins; Philadelphia, USA: 2000. pp. 489–514. [Google Scholar]
- 31.Chin S.F. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 32.Hayes D.F. Prognostic and predictive factors revisited. Breast. 2005;14:493–499. doi: 10.1016/j.breast.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Ma H. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast Cancer Res. 2006;8:R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C.I. Risk of mortality by histologic type of breast cancer among women aged 50 to 79 years. Arch. Intern. Med. 2003;163:2149–2153. doi: 10.1001/archinte.163.18.2149. [DOI] [PubMed] [Google Scholar]
- 35.Loveday R.L. Genetic changes in breast cancer detected by comparative genomic hybridisation. Int. J. Cancer. 2000;86:494–500. doi: 10.1002/(sici)1097-0215(20000515)86:4<494::aid-ijc8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 36.Nishizaki T. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int. J. Cancer. 1997;74:513–517. doi: 10.1002/(sici)1097-0215(19971021)74:5<513::aid-ijc6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Gunther K. Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J. Pathol. 2001;193:40–47. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH745>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 38.Stange D.E. High-resolution genomic profiling reveals association of chromosomal aberrations on 1q and 16p with histologic and genetic subgroups of invasive breast cancer. Clin. Cancer Res. 2006;12:345–352. doi: 10.1158/1078-0432.CCR-05-1633. [DOI] [PubMed] [Google Scholar]
- 39.Acs G. Differential expression of E-cadherin in lobular and ductal neoplasms of the breast and its biologic and diagnostic implications. Am. J. Clin. Pathol. 2001;115:85–98. doi: 10.1309/FDHX-L92R-BATQ-2GE0. [DOI] [PubMed] [Google Scholar]
- 40.Korkola J.E. Differentiation of lobular versus ductal breast carcinomas by expression microarray analysis. Cancer Res. 2003;63:7167–7175. [PubMed] [Google Scholar]
- 41.Zhao H. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elston C.W., Ellis I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 43.Joensuu H. Amplification of erbB2 and erbB2 expression are superior to estrogen receptor status as risk factors for distant recurrence in pT1N0M0 breast cancer: a nationwide population-based study. Clin. Cancer Res. 2003;9:923–930. [PubMed] [Google Scholar]
- 44.Celis J.E. Integrating proteomic and functional genomic technologies in discovery-driven translational breast cancer research. Mol. Cell. Proteomics. 2003;2:369–377. doi: 10.1074/mcp.R300007-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X. High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am. J. Hum. Genet. 2005;76:312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olshen A.B. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 47.Willenbrock H. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 48.de Hoon M.J. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 49.Golub T.R. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 50.Li S. Snap: an integrated SNP annotation platform. Nucleic Acids Res. 2007;35:D707–D710. doi: 10.1093/nar/gkl969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S3