Abstract
The PE_PGRS family of proteins unique to mycobacteria is demonstrated to contain multiple calcium-binding and glycine-rich sequence motifs GGXGXD/NXUX. This sequence repeat constitutes a calcium-binding parallel β-roll or parallel β-helix structure and is found in RTX toxins secreted by many Gram-negative bacteria. It is predicted that the highly homologous PE_PGRS proteins containing multiple copies of the nona-peptide motif could fold into similar calcium-binding structures. The implication of the predicted calcium-binding property of PE_PGRS proteins in the light of macrophage-pathogen interaction and pathogenesis is presented.
Key words: Mycobacterium tuberculosis, virulence factors, PE_PGRS, calcium-binding motif, parallel β-roll fold
Introduction
The PE/PE_PGRS multigene family unique to mycobacteria accounts for about 5% of the Mycobacterium tuberculosis genome. The family includes 38 PE-encoding genes and 61 PE_PGRS-encoding genes scattered throughout the genome, characterized by a relatively conserved NH2-terminus of ~110 amino acids (aa) followed by a COOH-terminal glycine-rich repeat region ranging from ~100 aa to over 500 aa in length 1., 2.. Several PE_PGRS proteins have been demonstrated to be associated with the replication and survival of M. tuberculosis within macrophages of infected host tissues and are important for pathogenesis (3). One of the functions proposed for these proteins is that they are a source of antigenic variability to evade host immune responses (1). Other observations suggest the possibility that PE_PGRS proteins could either be cell surface constituents (adhesins) (4) that can influence bacterial cell structure (5) or could interfere with immune responses by inhibiting antigen processing (6). A recent study has shown that the evolution and expansion of the PE and PPE families is closely associated with the ESAT-6 (esx) gene cluster and has suggested a functional interdependence (7). As the ESAT-6 cluster encodes proteins involved in secretion and membrane pore formation, it is likely that the ESAT-6 gene cluster-encoded proteins might also be required for secretion of some of the PE family proteins.
Despite these reports, the precise function and underlying mechanism of action still remain unknown for this large family of proteins. The aim of this study was therefore to investigate all of the 61 PE_PGRS proteins in the PE subfamily to predict whether they carry any distinct function in M. tuberculosis. Attempts were made to determine their underlying mechanism of action, and an explanation was sought for answering why there are so many different PE_PGRS proteins in M. tuberculosis.
Results
Attempts to identify similar proteins in databases to provide insights into their functions have been difficult owing to the repetitive nature of these proteins. Similarity searches of PE_PGRS proteins invariably identify only other PE_PGRS family members or glycine-rich proteins such as those found in highly elastic plant cell wall proteins (8). To search specifically for non-mycobacterial microbial homologues that have so far not been identified, we analyzed the microbial genome databases (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) excluding Mycobacterium species (9). Using the widely investigated PE_PGRS protein Rv1818c as query and without masking the low complexity regions, one of the proteins revealed from the BLAST results was an RTX (repeat in toxin) toxin of Magnetococcus sp. MC-1 (COG2931), which is a related Ca2+-binding protein. The Rv1818c protein has 42% identity over 100% stretch of its PGRS domain to the glycine-rich domain of the COG2931 protein (data not shown). Although this domain corresponds to only a small region (5.8%) of the large RTX protein, the fact that some PE_PGRS proteins were reported as surface-exposed adhesin-like molecules 5., 10. prompted us to examine this similarity more carefully.
The family of RTX toxins includes haemolysin, cyclolysin, leukotoxin, and metallopeptidase, and these proteins seem to have two properties in common. First they bind calcium, and second they contain a multiple tandem repeat of nona-peptides, GGXGXD/NXUX, where X = any amino acid and U = unpolar/large hydrophobic residue at the C-terminal end of each protein 11., 12.. It was subsequently found that these sequence repeats constitute a Ca2+-binding structure called a parallel β-helix or parallel β-roll and might participate in host cell binding (13). To examine whether Rv1818c also contains this nona-peptide repeat corresponding to a Ca2+-binding motif, we searched for GGXGXD/NXUX as the calcium-binding motif and detected numerous such motifs in Rv1818c (Figure 1). Subsequently the presence of this motif was examined in all the 61 PE_PGRS proteins. Among the different PE_PGRS proteins grouped according to domains (not shown), 56 PE_PGRS proteins were found to contain this motif. The other five PE_PGRS proteins that lacked this motif were Rv0742, Rv0832, Rv0978c, Rv3652, and Rv3812. However, among the five proteins, Rv0832 (137 aa) may be frame-shifted in M. tuberculosis H37Rv to be fused with a protein of 749 aa encoded by Rv0833; Rv0978c is a protein of 331 aa that contains an unusually short (78 aa) PRGS motif; while Rv3652 (104 aa) may also be a frame-shifted PE_PGRS protein. All these proteins belong to both classical and non-classical types of PE_PGRS proteins. The maximum number of repeats was found in Rv3345c where 77 copies of this calcium-binding parallel β-helix or β-roll motif GGXGXD/NXUX were present (Table S1). A total of 911 such calcium-binding motifs, among which 403 are GGXGXDXUX type and the remaining 508 are GGXGXNXUX type, were detected among all the PE_PGRS family members. The number of repetitive motifs varied according to the length of the open reading frames (ORFs), and the inter-motif distance was not fixed. For example, Rv1818c (498 aa) contains 9 motifs and the inter-motif distance varies from a minimum of 3 aa to as high as 67 aa (Figure 1). This is unlike RTX toxins where the calcium-binding motifs are not distributed so randomly. In this context, it is worth mentioning that the U residues of GGXGXD/NXUX motifs in PE_PGRS proteins are not always unpolar/large hydrophobic residues in nature (Table S1). In more than 70% of these cases, U = G. In the remaining cases, U could be any arbitrary residue except for cystein (data not shown). We also determined the secondary structure of all the 61 PE_PGRS proteins including Rv1818c to reaffirm the presence of β-strands intercepted by coils in the C-terminal PGRS domain, which is a characteristic of parallel β-roll structure (data not shown).
Fig. 1.
The GGXGXD/NXUX calcium-binding motifs (bold) identified in Rv1818c and their alignments.
To identify structurally important folds of PE_PGRS proteins, we attempted to associate them with structurally similar bacterial protein(s). Employment of the 3D-Jury system (14) to predict a possible protein fold of known function, using the C-terminal PGRS domain (382 aa) of Rv1818c as query sequence, revealed a Ca2+-binding fold with a statistically moderate score (Table S2). The highest fold recognition scores were compiled next for the PGRS domains of all 61 proteins (Table S2). Interestingly, ~70% of the total PE_PGRS proteins exhibited a common fold to the C-terminal β-roll Ca2+-binding domain of Serratia marcescens metalloprotease [PDB ID: 1SRP (15) and 1SAT (16)] and the alkaline protease of Pseudomonas aeruginosa IFO3080 [PDB ID: 1AKL (17) and 1KAP (18)], all of them belong to the RTX family.
The Rv3344c protein with fold prediction data greater than the confidence threshold limit set by 3D-Jury system (Table S2) was used subsequently to generate an optimized 3D molecular model using the alkaline protease of P. aeruginosa (PDB ID: 1AKL)(17) as the template. It is important to mention here that although according to the GenBank report, the Rv3344c gene (Accession No. YP_177961) might be a gene fragment that should be in-frame with a following ORF (MTV016.45c), no frame-shift was found when checked in BAC and cosmid clones (as of GenBank latest update: 24-May-2007). The MODELER program was used to create the model 18., 19.. The predicted model (Figure 2) was found to adopt Ca2+-binding parallel β-helix or parallel (β-roll structures (13) located at the turn of coil regions. The models 18., 19. created for the PE_PGRS proteins with 1AKL/1KAP/1SAT/1SRP (PDB IDs) fold scores greater than 25.0 were also predicted to hold calcium ions in the individual glycine-rich nona-sequence motifs (data not shown).
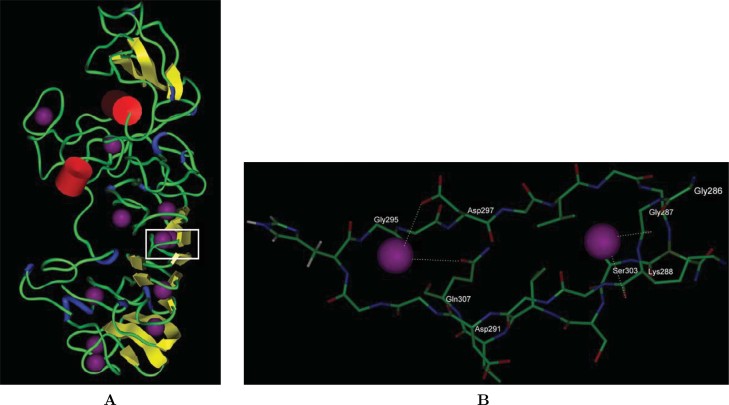
Fig. 2.
The model of parallel β-roll structure of PGRS domain of Rv3344c with potential calciums interacting with glycine-rich nona-peptide motifs. A. Secondary structure rendering by InsightII software (Purple: calciums; Red: helices; Yellow: β-strands; Blue: turns; Green: random coils). The calcium-complexed structure was stabilized by energy minimization using InsightII. B. Magnified top view (of the box marked in Panel A) of one complete ring (aa 286–307) containing overlapping motifs (aa 286–294 and aa 292–300) interacting with calcium. In one motif, it is interacted via the side chain of Asp297, Gln307, and Gly295. In another motif, the side chain of Gly287, Lys288, and Ser303 (from below) holds onto the second calcium in the ring.
Discussion
Our results presented here suggest that the highly homologous PGRS domain of the majority of the PE subfamily proteins have calcium-binding motifs and are therefore likely to be calcium-binding proteins. Calcium-dependent adhesins are known to exist in the soil bacterium Rhizobium species where their attachment to the developing root hairs of leguminous plants is considered to be the first step in the host-specific infection process that leads to a nitrogen-fixing symbiosis (20). Analogous to the Rhizobium nodulation gene nodO that encodes a Ca2+-binding protein involved in interactions with plant root cells in a Ca2+-dependent way, it is possible that a similar Ca2+-dependent PE_PGRS-mediated interaction may exist in M. tuberculosis with host cells 4., 10..
The earliest interactions of M. tuberculosis with macrophages are known to result in a number of alterations in Ca2+ signaling events critical for phagosome maturation 21., 22., 23.. In macrophages, activation of cytosolic Ca2+-regulated enzyme Ca2+/calmodulin-dependent protein kinase II (CaMKII) is essential for the phagosome-lysosome fusion (24). Other studies have demonstrated that M. tuberculosis blocks this pathway via inhibition of sphingosine kinase, a macrophage enzyme that increases cytosolic Ca2+ levels 22., 25.. Furthermore, the maturation and the acidification of myctobacterial phagosome could be restored by artificially raising the cytosolic Ca2+ levels using Ca2+ ionophores (21) or receptor stimulation with ATP (26). This has suggested that M. tuberculosis depletes internal Ca2+ stores in infected human macrophages 21., 26..
In the light of these observations, a role for Ca2+-binding PE_PGRS proteins could be envisaged as follows. The initial non-specific attachment of M. tuberculosis to the host alveolar macrophages via Ca2+-dependent PE_PGRS proteins will cause a sudden dip in calcium concentration at the focal point of host pathogen interaction 27., 28.. Such an event would lead to a fall in the cytosolic Ca2+ levels in macrophages, ultimately preventing phagolysosome fusion. We thus postulate that the initial host-pathogen interactions could play a very crucial role to the sensing and establishment of the bacilli’s intracellular pathogenesis.
The generalized calcium-dependent adhesion ability of PE_PGRS proteins does not rule out the possibility that these proteins have additional functions. Existence of “non-classical” type of PE_PGRS proteins (data not shown) with protein folds other than “conserved parallel β-roll calcium-binding folds” (Table S2) indicates that some PE_PGRS proteins might have yet undiscovered additional virulence-related functions that help the bacilli to survive in infected host (6).
Additionally, it will also be of worth to see if these initial PE subfamily protein(s)-host cell interactions can promote membrane cholesterol accumulation at the site of mycobacterial entry (29) that might eventually modulate the membrane depolarization event needed for the entry of external Ca2+.
Materials and Methods
The genome information of M. tuberculosis strain H37Rv was retrieved from NCBI genome database as Accession NC_000962 (1). Subsequently, the re-annotated M. tuberculosis genome sequence was consulted from Camus et al. (30). An indigenously developed algorithm was written in C++ language to extract all the 61 PE_PGRS ORF sequences.
To search specifically for non-mycobacterial microbial homologues of PE_PGRS proteins, we analyzed the microbial genome databases (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) excluding Mycobacterium species (9). Classification of all the 61 PE_PGRS proteins from M. tuberculosis H37Rv based on domain patterns was performed using the PROSITE database (31) at http://au.expasy.org/prosite/. Fold recognition data for these proteins were compiled using the 3D-Jury system (14) available via the Structure Prediction Meta Server (http://meta.bioinfo.pl/) to predict a possible protein fold of known function in each of the PE_PGRS proteins. No predictions were made for Rv0832 and Rv3652. The model of parallel β-roll structure of PGRS domain of Rv3344c with potential calciums interacting with glycine-rich nona-peptide motifs was generated by InsightII software (Accelrys Inc., San Diego, USA). The calcium-complex structure was stabilized by energy minimization using the same software.
Authors’ contributions
NB conceived and supervised the study, collected and analyzed the data, and prepared the manuscript. BS collected the data relating to the fold, created the models, and assisted in manuscript preparation. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
NB thanks Prof. Samir K. Brahmachari for his encouragement and support during the course of this work, and also thanks Davinder Kohli for all his help. NB is a recipient of the IGIB Knowledge Center(CSIR) Fellowship.
Supporting Online Material
http://www.imtech.res.in/bvs/PE-PGRS-Mtb/
Tables S1 and S2
References
- 1.Cole S.T. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Poulet S., Cole S.T. Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch. Microbiol. 1995;163:87–95. doi: 10.1007/BF00381781. [DOI] [PubMed] [Google Scholar]
- 3.Ramakrishnan L. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 4.Brennan M.J. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 2001;69:7326–7333. doi: 10.1128/IAI.69.12.7326-7333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delogu G. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 2004;52:725–733. doi: 10.1111/j.1365-2958.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 6.Brennan M.J., Delogu G. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 2002;10:246–249. doi: 10.1016/s0966-842x(02)02335-1. [DOI] [PubMed] [Google Scholar]
- 7.Gey van Pittius N.C. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsui M. Novel N-terminal sequence of a glycine-rich protein in the aleurone layer of soybean seeds. Biosci. Biotechnol. Biochem. 1994;58:1920–1922. doi: 10.1271/bbb.58.1920. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachdeva G. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005;21:483–491. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coote J.G. Structural and functional relationships among the RTX toxin determinants of Gram-negative bacteria. FEMS Microbiol. Rev. 1992;8:137–161. doi: 10.1111/j.1574-6968.1992.tb04961.x. [DOI] [PubMed] [Google Scholar]
- 12.Baumann U. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilie H. Folding of a synthetic parallel beta-roll protein. FEBS Lett. 2000;470:173–177. doi: 10.1016/s0014-5793(00)01308-9. [DOI] [PubMed] [Google Scholar]
- 14.Ginalski K. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 15.Hamada K. Crystal structure of Serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a beta-sheet coil motif at 2.0 Å resolution. J. Biochem. 1996;119:844–851. doi: 10.1093/oxfordjournals.jbchem.a021320. [DOI] [PubMed] [Google Scholar]
- 16.Baumann U. Crystal structure of the 50 kDa metallo protease from Serratia marcescens. J. Mol. Biol. 1994;242:244–251. doi: 10.1006/jmbi.1994.1576. [DOI] [PubMed] [Google Scholar]
- 17.Miyatake H. Crystal structure of the unliganded alkaline protease from Pseudomonas aeruginosa IFO3080 and its conformational changes on ligand binding. J. Biochem. 1995;118:474–479. doi: 10.1093/oxfordjournals.jbchem.a124932. [DOI] [PubMed] [Google Scholar]
- 18.Marti-Renom M.A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 19.Sali A. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 20.Smit G. Involvement of both cellulose fibrils and a Ca2+-dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J. Bacteriol. 1987;169:4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik Z.A. Inhibition of Ca2+ signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. J. Exp. Med. 2000;191:287–302. doi: 10.1084/jem.191.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusner D.J. Mechanisms of mycobacterial persistence in tuberculosis. Clin. Immunol. 2005;114:239–247. doi: 10.1016/j.clim.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Vergne I. Cell biology of Mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 2004;20:367–394. doi: 10.1146/annurev.cellbio.20.010403.114015. [DOI] [PubMed] [Google Scholar]
- 24.Malik Z.A. Mycobacterium tuberculosis phagosomes exhibit altered calmodulin-dependent signal transduction: contribution to inhibition of phagosome-lysosome fusion and intracellular survival in human macrophages. J. Immunol. 2001;166:3392–3401. doi: 10.4049/jimmunol.166.5.3392. [DOI] [PubMed] [Google Scholar]
- 25.Malik Z.A. Cutting edge: Mycobacterium tuberculosis blocks Ca2+ signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J. Immunol. 2003;170:2811–2815. doi: 10.4049/jimmunol.170.6.2811. [DOI] [PubMed] [Google Scholar]
- 26.Stober C.B. ATP-mediated killing of Mycobacterium bovis bacille Calmette-Guérin within human macrophages is calcium dependent and associated with the acidification of mycobacteria-containing phagosomes. J. Immunol. 2001;166:6276–6286. doi: 10.4049/jimmunol.166.10.6276. [DOI] [PubMed] [Google Scholar]
- 27.Wagner D. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 28.Wagner D. Changes of the phagosomal elemental concentrations by Mycobacterium tuberculosis Mramp. Microbiology. 2005;151:323–332. doi: 10.1099/mic.0.27213-0. [DOI] [PubMed] [Google Scholar]
- 29.Gartfield J., Pieters J. Essential role for cholesterol in entry of mycobacteria into macrophage. Science. 2000;288:1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 30.Camus J.C. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 31.Hulo N. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2