Abstract
Histopathological subtyping of non-small cell lung cancer (NSCLC) into adenocarcinoma (ADC), and squamous cell carcinoma (SqCC) is of utmost relevance for treatment stratification. However, current immunohistochemistry (IHC) based typing approaches on biopsies are imperfect, therefore novel analytical methods for reliable subtyping are needed. We analyzed formalin-fixed paraffin-embedded tissue cores of NSCLC by Matrix-assisted laser desorption/ionization (MALDI) imaging on tissue microarrays to identify and validate discriminating MALDI imaging profiles for NSCLC subtyping. 110 ADC and 98 SqCC were used to train a Linear Discriminant Analysis (LDA) model. Results were validated on a separate set of 58 ADC and 60 SqCC. Selected differentially expressed proteins were identified by tandem mass spectrometry and validated by IHC. The LDA classification model incorporated 339 m/z values. In the validation cohort, in 117 cases (99.1%) MALDI classification on tissue cores was in accordance with the pathological diagnosis made on resection specimen. Overall, three cases in the combined cohorts were discordant, after reevaluation two were initially misclassified by pathology whereas one was classified incorrectly by MALDI. Identification of differentially expressed peptides detected well-known IHC discriminators (CK5, CK7), but also less well known differentially expressed proteins (CK15, HSP27). In conclusion, MALDI imaging on NSCLC tissue cores as small biopsy equivalents is capable to discriminate lung ADC and SqCC with a very high accuracy. In addition, replacing multislide IHC by an one-slide MALDI approach may also save tissue for subsequent predictive molecular testing. We therefore advocate to pursue routine diagnostic implementation strategies for MALDI imaging in solid tumor typing.
Lung cancer is the leading cause of cancer related death worldwide among both, men and woman with about 1.59 million reported deaths in 2012 (1). Two major lung cancer categories are discerned, namely small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC)1, with the latter comprising ∼85% of all cases. The two predominant histological NSCLC entities are adenocarcinoma (ADC) and squamous cell carcinoma (SqCC) accounting for ∼50 and 40% of all lung cancers, respectively (2).
Because differentiation of NSCLC subtypes is crucial for the selection of chemotherapy regimens and subsequent molecular test strategies, precise subtyping is paramount. Therapeutic targets such as ALK translocations or activating mutations of the epidermal growth factor receptor (EGFR) have been found almost exclusively in ADC and these patients benefit from the respective molecularly targeted therapy. Testing for these alterations is therefore restricted to ADC. Additionally, pemetrexed has no or little activity in SqCC, and bevacizumab, an antiangiogenic agent, has been linked to adverse side effects in these tumors (3), therefore these drugs are exclusively used in non-SqCC NSCLC. Further, it is likely that immune checkpoint blockade treatment and other biomarker test strategies such as testing for FGFR1 amplifications will also differ for NSCLC subtypes (4).
Distinction of ADC and SqCC is currently performed by histomorphological evaluation supported by immunohistochemistry (IHC). Usually a panel of markers including thyroid transcription factor 1 (TTF1), NapsinA, p63 and p40 is applied especially in carcinomas without clear cut histomorphological differentiation criteria. Although ADC usually express TTF1 and NapsinA, the majority of SqCC show a positive reaction to CK5/6, p63 and p40. However, it has been reported that on small biopsy specimens ∼20% of NSCLC cannot be classified correctly into SqCC and ADC even with extensive IHC when the tumor lacks clear morphologic differentiation (5). Despite the high importance of entity subtyping in NSCLC limited use of tissue during the diagnostic workup is mandatory to preserve as much tissue as possible for the above mentioned subsequent predictive genetic analyses (6). Therefore, current guidelines recommend using only one marker for ADC and one marker for SqCC in IHC based subtyping approaches (7). However, it is obvious, that a less extensive work-up will result in a decreased diagnostic precision (8). Such a tradeoff between diagnostic accuracy and the need for tissue preservation is clearly suboptimal; therefore, novel technologies able to resolve this dilemma are highly warranted.
Matrix-assisted laser desorption/ionization (MALDI) imaging has emerged as a powerful tool for analyzing the spatial distribution of proteins and peptides in tissue (9–11). Because hundreds of proteins and/or peptides are analyzed simultaneously on one single section without the need for target-specific reagents, MALDI emerges as a prime candidate to solve the conflict between limited tissue input and accurate subtyping of NSCLC.
However, despite recent advances in mass spectrometry (MS) imaging, this novel technology has not found its way into the routine diagnostic setting up to now. This is due to several reasons: (1) most groups applied this technology on fresh-frozen tissue and not on formalin-fixed paraffin-embedded (FFPE), the latter, being the diagnostic standard material, (2) reliable classifiers able to distinguish different tumor entities have not been firmly established, (3) validation of suggested classifiers is usually not performed. To address these topics, we conducted a large-scale study on FFPE tissue in which we evaluated the ability of MALDI imaging to reliably delineate lung ADC from lung SqCC in a routine diagnostic scenario.
EXPERIMENTAL PROCEDURES
Cohort Characteristics and TMA Construction
FFPE resection specimens of ADC and SqCC resected from 2004 to 2007 were extracted from the archive of the Institute of Pathology, Heidelberg University with the support of the tissue bank of the National Center for Tumor Diseases (NCT; project: #206). Tissues were used in accordance with the ethical regulations of the NCT tissue bank established by the local ethics committee. A cohort of 326 patients was identified for tissue microarray (TMA) construction: 168 patients with primary ADC and 158 patients with primary SqCC of the lung. Diagnoses were made according to the recommendations of the World Health Organization classification for lung cancer 2015 (12) and the IASLC/ATS/ERS classification of lung ADC. Tumor histotypes were confirmed for all cases by two experienced pulmonary pathologists (A.W., W.W.). Prior to TMA construction, a hematoxylin and eosin (H&E)-stained slide of each block was analyzed in order to select representative tumor-containing regions. A TMA machine (AlphaMetrix Biotech, Rödermark, Germany) was used to extract a tandem 1.0 mm cylindrical core sample from each tissue donor block. In total 8 TMA blocks were created.
Experimental Design and Statistical Rationale
The sample cohort was randomly separated in a training set (5 TMAs: 208 patients: 110 ADC, 98 SqCC) and a validation set (3 TMAs; 118 patients; 58 ADC, 60 SqCC). Both sets had no case/patient overlap. The layout of the TMAs analyzed included duplicate tissue cores for each case. A summary of the workflow is depicted in Fig. 1.
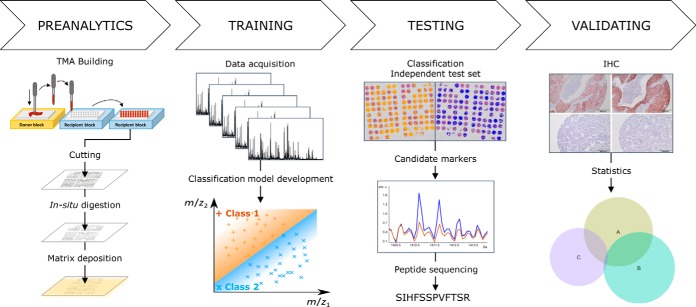
Fig. 1.
Workflow: The workflow of the study is depicted. After TMA construction and preparation of the tissue samples, a test set was analyzed by MALDI imaging in order to find subgroup specific MALDI profile differences. The MALDI algorithm was subsequently validated on a separate cohort. Differentially expressed peptide-ions were identified by MS/MS and validated by IHC.
Technical Setup
Data were acquired with a spatial resolution of 100 μm. To test for the optimal spatial resolution, prior to starting our study MALDI imaging analysis with 50 μm and 100 μm spatial resolution has been performed on one of our TMA sections. However, the higher resolution of 50 μm was not able to add relevant discriminating information to our probatory classifier design. In addition, the MALDI acquisition time at 50 μm substantially increased (about fourfold) when using our mass spectrometer. We also tried to decrease the total number of spectra per spot position (800–1000 shots) to reduce the acquisition time at a higher spatial resolution, but this resulted in a reduction of the quality of the spectra. Thus, we decided that a spatial resolution of 100 μm was optimal to perform this study with the available instrumentation.
MALDI Imaging Analysis
Preanalytics
Five μm thick FFPE sections were cut from each TMA block and mounted onto ITO-conductive slides (Delta Technologies, Loveland, CO). Sections were prepared for MALDI analysis following a published protocol of xylene and graded ethanol (Fischer Scientific, Schwerte, Germany) washes for paraffin removal, followed by heat induced antigen retrieval in Tris-HCl buffer (10 mm, pH 9.0) at 95 °C for 20 min (13). Trypsin solution (0.1 μg/μl) (Promega, Mannheim, Germany), prepared in 200 μl of 40 mm ammonium bicarbonate (Sigma-Aldrich, Taufkirchen, Germany), was dispersed onto the section using an automatic reagent sprayer (ImagePrep, Bruker Daltonik, Bremen, Germany) in 25 cycles with a fix nebulization time of 1.2 s. On-tissue trypsin digestion was carried out in a humidity chamber at 37 °C for 1.5 h.
MALDI-MS Acquisition
A solution of 7 mg/ml alpha-cyano-4-hydroxycinnamic acid (CHCA) matrix (Sigma-Aldrich) in 50/50 ACN/0.5% trifluoroacetic acid (Fischer Scientific) was applied onto digested sections by the ImagePrep device, using a Bruker default method optimized for sensor controlled nebulization of the matrix. The method consisted of five phases in total, each including individual cycles of nebulization, incubation and dehydration. Phases two to five were controlled by an optical sensor, which measured the moisture, matrix layer thickness, and dehydration parameters, by monitoring the scattered light intensity by refraction index matching. The amount of matrix was then applied in a defined range (minimum/maximum number) of cycles based on a given sensor signal or the defined range of spray cycles that are specified within each phase. Spray time was ∼2 s, and incubation time was 30 s except for the first phase which was 10 s. We modified phase 5 by increasing the number of spray cycles from the default-defined range 8/64, to the optimized range 36–40. Thus, tissue sections were coated with a higher number of matrix layers (∼70). Data were acquired in the range of m/z 500–5000, at a spatial resolution of 100 μm using an Autoflex Speed (Bruker Daltonik, Bremen, Germany) MALDI-TOF/TOF instrument equipped with a SmartBeam laser beam, operated in reflectron mode. A total of 1600 spectra in 200-shots increment were summed for each laser spot position using the Random Walk raster pattern as described previously (supplemental Fig. S1) (9, 14). A peptide calibration standard mix including angiotensin II, angiotensin I, substance P, bombesin, ACTH clip 1–17, ACTH clip 18–39, and somatostatin 28 (Bruker Daltonik) was used for external calibration.
Data Processing and Linear Discriminant Analysis
Data sets were imported in flexAnalysis 3.3 (Bruker Daltonik) and preprocessed for chemical noise smoothing, and peak alignment using a statistical peptide internal calibration strategy. One-Dalton chemical noise frequency was observed in the analysis of the spectra. Thus, we used chemical noise smoothing with a width range of 5 m/z, as suggested from the manufacturer, which removed this constant frequency improving the quality of the peaks. An example of an average spectrum before and after chemical noise smoothing is shown in supplemental Fig. S2. After MALDI measurement, matrix was removed from each TMA section by two washes (4 min each) in 100% methanol (Fischer Scientific, Schwerte, Germany). Slides were subsequently stained with H&E to digitally annotate tissue regions. These annotations were superimposed with the MALDI imaging data and the regions of interest were manually marked on each core of the TMA using flexImaging software (Bruker Daltonik). All mass spectra from the marked areas were then imported into SCiLS Lab (SCiLS v. 2014b, Bremen, Germany) software for data analysis. Spectra were processed for baseline subtraction using the iterative convolution algorithm with Sigma = 20 and Iterations = 20, and normalized based on their own total ion count (TIC). The classification model was generated using the Linear Discriminant Analysis (LDA) algorithm model supported by SCiLS Lab software. The spectra from each annotated cancer region were grouped together for each biopsy in separated folders. All spectra were separated into two data sets: set 1 (n = 208) was used to train the LDA model, set 2 (n = 118) was used to validate the algorithm.
A nonparametric statistical test was applied to our MALDI imaging datasets obtained from the tissue cores, where mean intensities of the MS signals do not have a Gaussian distribution. Wilcoxon/Kruskal-Wallis test was chosen as a nonparametric statistical test to establish a significant difference in the peak intensities between ADC and SqCC sample types.
Protein Identification
Whole FFPE tissue slides from two ADC and two SqCC cases, which were also included in the TMA cohorts, were used for identification. Five μm thick whole block sections were on-slide digested and sprayed with CHCA matrix solution following the same procedure described for the TMAs. MALDI MS spectra of each selected peptide were obtained using an Autoflex Speed MALDI-TOF/TOF instrument operated in reflectron mode with spectra acquired in the range of m/z 500–5000. Each peptide peak was selected and manually fragmented in the LIFT cell of the mass spectrometer, and the obtained spectra were processed using flexAnalysis 3.3 (Bruker Daltonik). Data were submitted to Mascot Server 2.4 (Matrix Science, Boston, MA) and run against the SwissProt database (SwissProt 2014_10) to match the peptide sequences to their corresponding intact proteins. Trypsin was chosen as the protease in the search parameter. SwissProt database search included 546790 sequences, and 194613039 residues; specifically from the taxonomy “Homo sapiens (human)” 20194 entries were searched. The tandem mass spectrometric (MS/MS) spectrum search parameter included MS tolerance of 200 ppm and MS/MS tolerance of ±0.4 Da (supplemental Table S1). The search conditions also comprised variable modifications, including protein N-term acetylation, and methionine oxidation; no fixed modification were included. Up to three missed cleavages were considered.
Immunohistochemistry
IHC staining was performed with commercially available antibodies (supplemental Table S2). All standard routine antibodies used (CK7, CK5) were applied according to quality-controlled protocols that are regularly evaluated in round-robin trials (http://www.nordiqc.org). Cytokeratin-15 (Keratin-15, CK15) and heat-shock protein 27 (HSP27), also known as heat-shock protein beta-1 (HSPB1), were stained according to the manufacturers protocols. In brief, TMA slides were deparaffinized and pretreated with antigen retrieval buffer. Subsequent steps were performed on an immunostaining device (Ventana Benchmark Ultra, Tuscon, AR) according to manufacturer's protocols. Pretreatment and dilution specifications are summarized in supplemental Table S2. Evaluation of IHC was performed blinded to the resection specimen diagnoses, according to a dichotomous scoring scheme (15); 1% stained tumor cells were considered to indicate positivity. Evaluation of IHC was done by M.K. All samples of the validation set were stained with antibodies against CK5/6, CK15, HSP27 and CK7. In one patient (ADC) only CK5/6 and CK7 could be evaluated (drop-out rate: 0.2%) by IHC because there were either no vital tumor cells or lost tissue cores during the staining process.
In addition, the resection specimens of the three discordant cases (see below) were additionally (re)stained for p40, p63, TTF1, and NapsinA.
RESULTS
Classifier Generation and Application in the NSCLC Validation Set
110 ADC and 98 SqCC core biopsies were used to generate a MALDI NSCLC classification model using the LDA algorithm. The algorithm generation resulted in a total of 339 peaks that were automatically selected to create a bimodal classification model. Because we did not want to lose any discriminating information we did not try to reduce the complexity of the classifier to a smaller number of m/z values.
118 NSCLC samples (58 ADC, 60 SqCC) were used for the validation of our classifier. The generated model was applied to sort each spectrum of these tumors into one of the two different classes based on the distribution of the 339 peaks from our generated model. The MALDI classifier developed in the training set classified 58 of 58 (100%) ADC and 59 of 60 (98.3%) SqCC correctly. Overall, 117 out of 118 (99.1%) tumors in the validation set were classified correctly by measuring MALDI profiles on biopsy equivalent TMA cores, with the initial pathological diagnosis (including auxiliary IHC where necessary) made on the corresponding TMA donor resection specimens taken as the “gold standard” reference. Sensitivity and specificity for the categorization of an NSCLC as ADC was 100 and 98% and for SqCC 98 and 100%, respectively.
Protein Identification
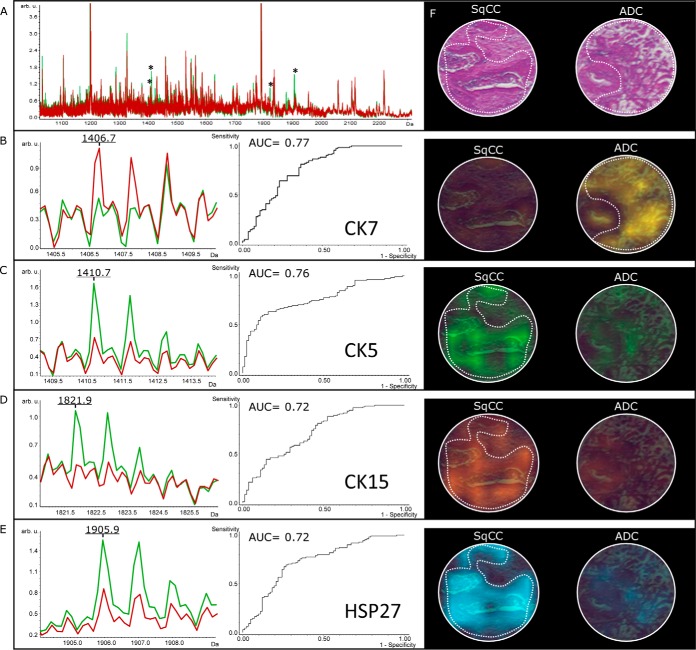
Comparison of the average relative intensities at each m/z value between the two sample types of our validation cohort revealed five differential m/z peaks from four proteins exhibiting a weight statistical significance p < 0.000001, and an area under the Receiver Operating Characteristic (ROC) curve (AUC) >0.7. The overlay of average spectra from both classes is shown in Fig. 2. The discriminatory peaks were partly correlated (for detailed correlation analyses see supplemental Fig. S3).
Fig. 2.
MALDI imaging: a, A full average spectrum of SqCC (green) and ADC (red) with a significantly different peptide pattern (asterisk) is shown. b, A peptide average profile of CK7 (m/z 1406.7) with high expression in ADC, but low expression in SqCC is depicted. c–e, Likewise average peptide profiles of CK5, CK15 and HSP27 with high expression in SqCC, but low expression in ADC are demonstrated. f, H&E and corresponding color encoded MALDI images of the mentioned peptide-ions show the distribution in tumor areas from SqCC and ADC (dashed line).
Identification of the respective differentially expressed peptide-ions was done directly by MS/MS from the tissue digest. Four peptides, namely fragments from CK5, CK7, CK15, and HSP27 could be identified and associated to their respective parent proteins through database matching. The mass resolutions from the parent peptides were as follows: m/z 1406.6 (4145 m/dm), m/z 1410.7 (8410 m/dm), 1821.8 (11233 m/dm), 1877.8 (9129 m/dm), and 1905.9 (12449 m/dm). Details of the identification analyses are given in supplemental Table S1. The zoomed average spectra for these four peptide-ions significantly discriminant between ADC and SqCC are illustrated in Fig. 2.
Immunohistochemistry for the Identified Proteins in the Validation Cohort
Expression of the Identified Discriminator Proteins in SqCC
CK5/6 (93.3%), HSP27 (83.3%), and CK15 (58.3%) were commonly expressed in SqCC and less often in ADC (1.7%, 40.0%, 0%). All three markers were positive in 29 (49.2%) SqCC cases and all were negative in one (1.7%) case. CK5/6 often showed coexpression with HSP27 (85.7%), and CK15 (62.5%). The same was true for HSP27 (CK5/6 (96.0%) and CK15 (62.0%)) and CK15 (CK5/6 (100%) and HSP27 (88.6%)). Expression of the ADC marker CK7 was found in 10.0% of SqCC samples. The marker overlap is depicted in supplemental Fig. S4.
Expression of the Identified Discriminator Proteins in ADC
As could be expected, opposing results for the expression of the respective proteins were seen in ADC, where 96.6% of tumors were CK7 positive. CK7 expression was not associated with CK15 positivity (0%) but showed a coexpression with HSP27 in a number of cases (42.9%). Not surprisingly, expression of the typical SqCC markers was less often found in ADC (CK5/6 (1.7%), CK15 (0%), and HSP27 (42.9%)).
Correlation Between MALDI Imaging Data, Histology, and Immunohistochemistry
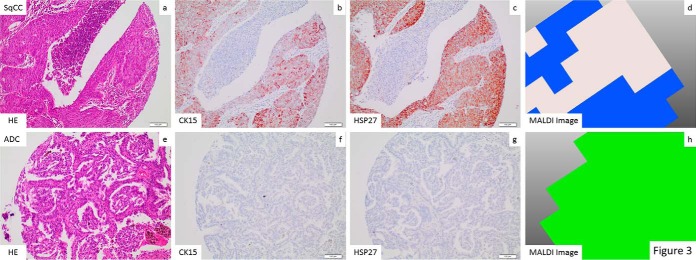
Molecular images of the peptides identified were mapped over tissue cores from ADC and SqCC. Ion-density maps of the detected discriminator peptide species were in excellent concordance with the histological annotations demonstrating epithelial tumor regions (Fig. 3). As expected, the MALDI image distribution of our classifier was also matching IHC expression profiles in the respective tissue cores. An excellent correlation of the SqCC and the ADC classifier (MALDI imaging) with the IHC staining for CK15 and HSP27 was observed (Fig. 3).
Fig. 3.
Correlation between H&E, IHC, and MALDI imaging: An excellent correlation between the identification of tumor areas by morphology H&E stained tissue slides, (a, e), CK15 IHC (b, f), HSP27 IHC (c, g) and imaging classification (d, h) is seen in an exemplary SqCC (a–d) and ADC (e–h). Histological assignment from MALDI is shown in a color-encoded visualization: Blue was assigned to SqCC and green to ADC.
Discordances Between MALDI Classification and Initial Pathological Subtyping
When the training cohort (2/208) and the validation cohort (1/118) were taken together, a total of three cases were discordant between MALDI classification done on tissue cores and the initial “gold standard” tumor typing done by morphology and an extended auxiliary IHC panel (where necessary) on the many times larger corresponding whole “donor” resection specimen. The respective discordant cases were one solid predominant ADC and two SqCC, according to initial pathological typing.
The discordant putative “ADC case,” however, upon reevaluation of the whole slide set from routine diagnostics consisted of pleomorphic, medium-sized tumor cells with pure solid growth. A post hoc extended IHC panel showed negativity for TTF1, NapsinA, CK7, p40, and CK15 with focal expression of CK5/6 and p63. We therefore concluded that the tumor was initially misclassified by us by histological means and reclassified the tumor as SqCC.
The first discordant “SqCC case” was strongly positive for CK5/6, p63, p40 and weakly positive for CK15 and HSP27. NapsinA, CK7 and TTF1 remained negative. When evaluating the respective resection specimen we saw a tumor composed of medium to large sized tumor cells with intercellular bridges and focal keratinization. We concluded that this case had been truly misclassified by our MALDI algorithm as ADC despite classical morphological and IHC properties in favor of SqCC.
Interestingly, the second discrepant “SqCC case” had a discordant IHC staining pattern in both tissue cores. One core was positive for CK7, TTF1 and NapsinA and negative for SqCC markers. The other tissue core was positive for CK5/6 and p63 but negative for ADC markers. MALDI imaging when viewed for both cores separately revealed discordant ADC and SqCC profiles similar to the IHC data. The overall classification of integrated spectra, however, had obviously favored ADC. When we got back to the resection specimen, we found a solid tumor both with intercellular bridges and focal keratinization but also with focal gland forming areas. We therefore concluded that this tumor would be better classified as adenosquamous carcinoma. When both morphologically and immunohistochemically divergent cores would have been classified separately they would have been classified correctly by the MALDI algorithm. Taken together, our MALDI approach, after reanalyzing our three putatively misclassified cases, resulted in one clear misclassification out of 326 cases (0.3%).
DISCUSSION
Stratification according to the histological subtype is important for NSCLC therapy (16). Because genetic analyses of therapeutic targets require increasing amounts of tissue material, especially on small biopsies the initial diagnosis must be made in a reliable and tissue saving manner. To achieve this goal, the implementation of novel technologies into routine pathological diagnostics is mandatory. By investigating an overall of more than 300 lung cancer cases, we demonstrate that MALDI imaging can be used for this purpose on formalin-fixed paraffin-embedded biopsy equivalents with almost 100% diagnostic accuracy. After detailed reanalysis of the very few divergently classified cases (n = 3) when compared with “gold standard” IHC aided morphological classification on corresponding resection specimen, only one case was finally found to be classified wrong by MALDI imaging.
In comparison, others reported that when a conventional stratifying approach based on serial IHC using three immune markers is used on biopsies, accurate subtyping can only be achieved in about 80% of cases (17). The accuracy can be increased up to 90% by employing a panel of six markers (18). Our data suggests that MALDI stratification in the biopsy situation may compete favorably even to the latter approach. However, this certainly has to be proven in a prospective head-to-head comparison of both methodologies in the same cohorts, which is something we are planning to do in the future.
Lung cancer has been studied by both, liquid chromatography based MALDI (LC-MS/MS) (19, 20) and MALDI imaging (21). In a previous exploratory study Yanagisawa and coworkers using fresh frozen tissue material already demonstrated that in principle the differentiation between lung SqCC and ADC is possible by MALDI and suggested that the risk for nodal involvement as well as survival may be predicted by MALDI profiles, as well (20). In this study 13 of 13 ADC and 16 of 16 SqCC were classified correctly. Although Yanagisawa and colleagues showed that MALDI is generally applicable for stratifying purposes in lung cancer, the sample size was extremely small, therefore it is difficult to draw any diagnostically relevant conclusions from their data set. In addition, fresh frozen material was used but in routine diagnostics only FFPE tissue is at hand as the analysis of fresh frozen material requires special logistics, sample handling and storage. In a second study Groseclose et al. (21) investigated lung ADC (n = 12) and SqCC (n = 14) FFPE samples in a TMA setting and demonstrated the feasibility of MALDI imaging to discriminate both tumor families on FFPE material. However, also from this study definite conclusions could not be drawn because of the limited sample size and the lack of validation. Our study avoided these shortcomings using FFPE tissue under diagnostically relevant conditions.
We limited our analysis to the by far most common NSCLC subtypes - ADC and SqCC - for two reasons: (1) to avoid statistical shortcomings associated with a low number of patients in entity subgroups that have a low prevalence and (2) to avoid background noise resulting from rarer subgroups that most likely belong to either ADC, SqCC, or a mixed lineage such as adenosquamous carcinoma, sarcomatoid carcinoma, or large cell carcinoma. However, applying (and extending) our classifier to these entities is clearly one of the next steps.
In our study, tissue cores with a diameter of one mm were analyzed. Although differentiation of both subtypes by MALDI profiling (acquisition of single spectra per core) would possibly be feasible, we used standard imaging of the whole tissue core (multiple spectra per core) and sum spectra to increase the robustness and overall accuracy. Whether single spectra are specific enough to predict the subtype by the application of our MALDI classifier has to be shown in further technically focused studies.
Despite the fact that using FFPE tissue material has its downsides such as limited resolution, limited sensitivity with respect to the detection of low abundant peptides and the need of special knowledge for tissue preparation recent developments show that the analysis of FFPE tissues by MALDI imaging becomes more and more established (13). Promising results using this technology could be obtained in several diseases and different types of cancer (9, 22–24).
For our MALDI imaging-based differential diagnosis only one tissue slide was required in the biopsy “like” TMA setting that may even be stained by H&E after the MALDI analysis. Thus, only minimal amounts of tissue are needed for diagnostic MALDI evaluation. Sparing tissue during entity subtyping is of utmost importance in the lung cancer setting because usually only biopsies but not resection specimen are available and a variety of subsequent prognostic and predictive analyses have to be performed on this material in a subentity stratified manner (e.g. sequencing and translocation analyses or IHC for new predictive markers such as PD-L1).
A MALDI imaging analysis required 24h with the MS instrument used in this study, a time frame, which is comparable to IHC. MS technological development, however, is rapid and MS analyses are to become faster in the very near future, with first MALDI prototypes reducing the run time to less than one hour.
The dropout rate in our study was zero because all TMA cores on several different slides could be analyzed by MALDI; therefore, the analysis appears fairly robust with respect to varying preanalytic conditions. However, we have to acknowledge that the well-controlled TMA situation might substantially differ from sequential MALDI analyses of “true” biopsies. This has to be addresses, ideally by a prospective head-to-head comparison of different stratification approaches, something we are currently planning to do.
Operational costs for MALD imaging are modest. However, to truly answer the question whether MALDI analysis in the lung cancer setting may be cost-effective, one also has to factor in the high price for MALDI equipment and some additional cost factors. A comprehensive economical analysis comparing MALDI to multi marker IHC–which is also fairly expensive–would certainly be of interest but is beyond the scope of this article (25).
Overall, to us it seems attractive to pursue strategies aiming on a broader implementation of MALDI technology into routine pathology diagnostics. In microbiology and for the subtyping of amyloidosis, MS has already been successfully implemented into routine diagnostics on patient material (26–31).
We set out to identify the proteins corresponding to the strongest differentially expressed peaks of our algorithm classifier. We did so because first and foremost we wanted to know whether our stratifier included any already known proteins used for subtyping of ADC and SqCC by IHC, which would additionally underline the validity of the experimental approach.
Four peptides could be identified as very strongly differentially expressed, one of the peptides was highly expressed in ADC (CK7), the other three peptides were highly expressed in SqCC (CK5, CK15, and HSP27). All four candidate protein biomarkers were subjected to IHC validation (32). Of these four markers, two (CK5, CK7) are already well-known discriminative IHC markers in the routine diagnostic NSCLC setting (18). This strongly supported the validity of our algorithm for the identification of differentially expressed proteins. CK7 was previously reported to have a sensitivity of >90% and a specificity of 57–94% for ADC (15, 18). CK5, a sensitive (75–100%) and specific (79–92%) immunomarker for a squamous lineage, was also previously identified by MALDI imaging and LC-MS/MS (15, 33).
Of note, two new potential markers for a lung SqCC lineage (when compared with pulmonary ADC) were also detected by our MALDI imaging approach: CK15 and HSP27. IHC data in our validation cohort showed 58.3% sensitivity and 100% specificity of CK15 positivity as well as 83.3% sensitivity and 57.9% specificity of HSP27 positivity for the diagnosis of SqCC.
CK15 is a cytoskeletal protein that has been found in basal keratinocytes of all stratified squamous epithelia, and has been suggested as a stem cell marker in the bulge of hair follicles (34). Studies in mice demonstrate that CK15 positive cells can give rise to progeny that express CK5 (35). Besides its presence in cutaneous neoplasms (36, 37), CK15 has also been found up-regulated in gene expression studies in SqCC of the lung (38, 39) and in one MALDI imaging study on lung cancer samples (21). In our study, CK15 was a valid classifier of SqCC. Although none of the ADC showed expression of CK15 by IHC, almost 60% of the SqCC showed at least focal staining.
HSP27 is a member of the heat-shock protein family and commonly overexpressed in cancers. It is activated through processes that induce cell stress such as heat, hypoxia, radicals, or carcinogens (40). HSP27 has a role in the sequestration of damaged proteins thereby protecting cells against protein aggregates (41). Moreover, interactions with components of apoptotic pathways and oncogenic signaling pathways have been described (40). In addition, HSP27 functions as a pivotal protein in cell organization and cell migration and it has been linked to cancer invasiveness, metastasis, and poor prognosis (40, 42). Therefore, HSP27 has been suggested as a therapeutic target. Down-regulation or inhibition of HSP27 resulted in increased sensitivity of different cancer subtypes to chemotherapy (colorectal, bladder, prostate, ovarian, and uterine cancer) (40) or radiotherapy (head and neck squamous cell carcinoma) (43).
HSP27 and functional promoter variants have also been described as favorable prognostic markers in SqCC and large cell carcinoma of the lung (44, 45). Furthermore, a certain genetic variant has been reported to be associated with radiation pneumonitis and esophageal toxicity after radiotherapy (46, 47). One group investigated serum levels of HSP27 by ELISA and found significantly higher levels in NSCLC compared with healthy controls (48). Interestingly, HSP27 expression levels have been described as a discriminator between SqCC and normal bronchial epithelium, squamous metaplasia, atypical hyperplasia, and carcinoma in situ (49). However, HSP27 has also been detected in ADC by IHC (50). We found HSP27 expression in both, SqCC and ADC, but significantly more often in SqCC specimens (p < 0.001). However, because of the overlap in expression patterns, this marker is unlikely to enter the routine diagnostic setting.
In summary, we provide evidence that differentiation between SqCC and ADC in biopsy-like FFPE lung cancer samples by MALDI imaging is feasible with extremely high sensitivity and specificity in a fast and reliable way using a single tissue section. Based on our data, it seems attractive to pursue strategies for an implementation of MALDI technologies into routine pathology diagnostics.
Supplementary Material
Acknowledgments
Tissue samples were provided by the tissue bank of the National Center for Tumor Diseases (NCT, Heidelberg, Germany) in accordance with the regulations of the tissue bank and the approval of the ethics committee of Heidelberg University. Bruker Daltonics provided the Autoflex MS instrument.
Footnotes
Author contributions: M.K. carried out experiments and analyzed the data. R.C. carried out experiments and analyzed data. J.K., A.W., and W.W. conceived experiments and participated in data interpretation. M.K. and W.W. wrote the manuscript. H.D., P.S., J.K., K.S., and A.S. participated in data interpretation and revised the manuscript critically. All authors were involved in correcting the paper and had final approval of the submitted version.
* MK is supported by the Post-doc program of the medical faculty of the University of Heidelberg.
 This article contains supplemental material.
This article contains supplemental material.
Emails: mark.kriegsmann@med.uni-heidelberg.de, rita.casadonte@molekularpatho-trier.de, kriegsmann@patho-trier.de, kobarg@scils.de, kschwamborn@tum.de, hendrik.dienemann@med.uni-heidelberg.de, peter.schirmacher@med.uni-heidelberg.de, albrecht.stenzinger@med.uni-heidelberg.de, arne.warth@med.uni-heidelberg.de, wilko.weichert@tum.de.
1 The abbreviations used are:
- NSCLC
- non-small cell lung cancer
- ADC
- adenocarcinoma
- SqCC
- squamous cell carcinoma
- IHC
- immunohistochemistry
- MALDI
- matrix-assisted laser desorption/ionization
- FFPE
- formalin-fixed paraffin-embedded.
REFERENCES
- 1. Stewart B., and Wild C. (2014) World Cancer Report. [Google Scholar]
- 2. Chen Z., Fillmore C. M., Hammerman P. S., Kim C. F., and Wong K. K. (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. 14, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reck M., Heigener D. F., Mok T., Soria J. C., and Rabe K. F. (2013) Management of non-small-cell lung cancer: recent developments. Lancet 382, 709–719 [DOI] [PubMed] [Google Scholar]
- 4. Garon E. B., Rizvi N. A., Hui R., Leighl N., Balmanoukian A. S., Eder J. P., Patnaik A., Aggarwal C., Gubens M., Horn L., Carcereny E., Ahn M. J., Felip E., Lee J. S., Hellmann M. D., Hamid O., Goldman J. W., Soria J. C., Dolled-Filhart M., Rutledge R. Z., Zhang J., Lunceford J. K., Rangwala R., Lubiniecki G. M., Roach C., Emancipator K., Gandhi L., Investigators K-.(2015) Pembrolizumab for the treatment of non-small-cell lung cancer. New Engl. J. Med. 372, 2018–2028 [DOI] [PubMed] [Google Scholar]
- 5. Mukhopadhyay S., and Katzenstein A. L. (2011) Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: Utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. Am. J. Surgical Pathol. 35, 15–25 [DOI] [PubMed] [Google Scholar]
- 6. Travis W. D., Brambilla E., Noguchi M., Nicholson A. G., Geisinger K., Yatabe Y., Ishikawa Y., Wistuba I., Flieder D. B., Franklin W., Gazdar A., Hasleton P. S., Henderson D. W., Kerr K. M., Petersen I., Roggli V., Thunnissen E., and Tsao M. (2013) Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch. Pathol. Lab. Med. 137, 668–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kerr K. M., Bubendorf L., Edelman M. J., Marchetti A., Mok T., Novello S., O'Byrne K., Stahel R., Peters S., Felip E., Panel M., and Panel M. (2014) Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol. 25, 1681–1690 [DOI] [PubMed] [Google Scholar]
- 8. Vincenten J. P., Smit E. F., Grunberg K., Postmus P. E., Snijders P. J., Witte B. I., Heideman D. A., and Thunnissen E. (2015) Is the current diagnostic algorithm reliable for selecting cases for EGFR- and KRAS-mutation analysis in lung cancer? Lung Cancer 89, 19–26 [DOI] [PubMed] [Google Scholar]
- 9. Kriegsmann M., Seeley E. H., Schwarting A., Kriegsmann J., Otto M., Thabe H., Dierkes B., Biehl C., Sack U., Wellmann A., Kahaly G. J., Schwamborn K., and Caprioli R. M. (2012) MALDI MS imaging as a powerful tool for investigating synovial tissue. Scand. J. Rheumatol. 41, 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zimmerman T. A., Monroe E. B., Tucker K. R., Rubakhin S. S., and Sweedler J. V. (2008) Chapter 13: Imaging of cells and tissues with mass spectrometry: adding chemical information to imaging. Methods Cell Biol. 89, 361–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caprioli R. M., Farmer T. B., and Gile J. (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 12. Travis W. D., Burke B. E. A. P., Marx A., and Nicholson A. G. (2015) WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th Ed., IARC Publication; [DOI] [PubMed] [Google Scholar]
- 13. Casadonte R., and Caprioli R. M. (2011) Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protocols 6, 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kriegsmann M., Casadonte R., Randau T., Gravius S., Pennekamp P., Strauss A., Oldenburg J., Wieczorek K., Deininger S. O., Otto M., and Kriegsmann J. (2014) MALDI imaging of predictive ferritin, fibrinogen and proteases in haemophilic arthropathy. Haemophilia 20, 446–453 [DOI] [PubMed] [Google Scholar]
- 15. Terry J., Leung S., Laskin J., Leslie K. O., Gown A. M., and Ionescu D. N. (2010) Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am. J. Surgical Pathol. 34, 1805–1811 [DOI] [PubMed] [Google Scholar]
- 16. Ettinger D. S., Wood D. E., Akerley W., Bazhenova L. A., Borghaei H., Camidge D. R., Cheney R. T., Chirieac L. R., D'Amico T. A., Demmy T. L., Dilling T. J., Dobelbower M. C., Govindan R., Grannis F. W. Jr, Horn L., Jahan T. M., Komaki R., Krug L. M., Lackner R. P., Lanuti M., Lilenbaum R., Lin J., Loo B. W. Jr, Martins R., Otterson G. A., Patel J. D., Pisters K. M., Reckamp K., Riely G. J., Rohren E., Schild S. E., Shapiro T. A., Swanson S. J., Tauer K., Yang S. C., Gregory K., and Hughes M. (2015) Non-small cell lung cancer, version 6.2015. J. Natl. Comprehensive Cancer Network 13, 515–524 [DOI] [PubMed] [Google Scholar]
- 17. Koh J., Go H., Kim M. Y., Jeon Y. K., Chung J. H., and Chung D. H. (2014) A comprehensive immunohistochemistry algorithm for the histological subtyping of small biopsies obtained from non-small cell lung cancers. Histopathology 65, 868–878 [DOI] [PubMed] [Google Scholar]
- 18. Warth A., Muley T., Herpel E., Meister M., Herth F. J., Schirmacher P., Weichert W., Hoffmann H., and Schnabel P. A. (2012) Large-scale comparative analyses of immunomarkers for diagnostic subtyping of non-small-cell lung cancer biopsies. Histopathology 61, 1017–1025 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W., Wei Y., Ignatchenko V., Li L., Sakashita S., Pham N. A., Taylor P., Tsao M. S., Kislinger T., and Moran M. F. (2014) Proteomic profiles of human lung adeno and squamous cell carcinoma using super-SILAC and label-free quantification approaches. Proteomics 14, 795–803 [DOI] [PubMed] [Google Scholar]
- 20. Yanagisawa K., Shyr Y., Xu B. J., Massion P. P., Larsen P. H., White B. C., Roberts J. R., Edgerton M., Gonzalez A., Nadaf S., Moore J. H., Caprioli R. M., and Carbone D. P. (2003) Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 [DOI] [PubMed] [Google Scholar]
- 21. Groseclose M. R., Massion P. P., Chaurand P., and Caprioli R. M. (2008) High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 8, 3715–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Casadonte R., Kriegsmann M., Zweynert F., Friedrich K., Baretton G., Otto M., Deininger S. O., Paape R., Belau E., Suckau D., Aust D., Pilarsky C., and Kriegsmann J. (2014) Imaging mass spectrometry to discriminate breast from pancreatic cancer metastasis in formalin-fixed paraffin-embedded tissues. Proteomics 14, 956–964 [DOI] [PubMed] [Google Scholar]
- 23. Caprioli R. M. (2014) Imaging mass spectrometry: molecular microscopy for enabling a new age of discovery. Proteomics 14, 807–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kriegsmann J., Kriegsmann M., and Casadonte R. (2015) MALDI TOF imaging mass spectrometry in clinical pathology: A valuable tool for cancer diagnostics (Review). Int. J. Oncol. 46, 893–906 [DOI] [PubMed] [Google Scholar]
- 25. Kriegsmann J., Casadonte R., Zweynert F., Kriegsmann M., Otto M., and Deininger S. (2013) [MALDI-TOF imaging]. Zeitschrift fur Rheumatologie 72, 724–728 [DOI] [PubMed] [Google Scholar]
- 26. Casadonte R., Kriegsmann M., Deininger S. O., Amann K., Paape R., Belau E., Suckau D., Fuchser J., Beckmann J., Becker M., and Kriegsmann J. (2015) Imaging mass spectrometry analysis of renal amyloidosis biopsies reveals protein co-localization with amyloid deposits. Anal. Bioanal. Chem. 407, 5323–5331 [DOI] [PubMed] [Google Scholar]
- 27. Sethi S., Vrana J. A., Theis J. D., Leung N., Sethi A., Nasr S. H., Fervenza F. C., Cornell L. D., Fidler M. E., and Dogan A. (2012) Laser microdissection and mass spectrometry-based proteomics aids the diagnosis and typing of renal amyloidosis. Kidney Int. 82, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vrana J. A., Gamez J. D., Madden B. J., Theis J. D., Bergen H.R. 3rd, and Dogan A. (2009) Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood 114, 4957–4959 [DOI] [PubMed] [Google Scholar]
- 29. Nomura F. (2015) Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): A revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta 1854, 528–537 [DOI] [PubMed] [Google Scholar]
- 30. Buchan B. W., and Ledeboer N. A. (2014) Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev. 27, 783–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leung N., Nasr S. H., and Sethi S. (2012) How I treat amyloidosis: the importance of accurate diagnosis and amyloid typing. Blood 120, 3206–3213 [DOI] [PubMed] [Google Scholar]
- 32. Schwamborn K. (2012) Imaging mass spectrometry in biomarker discovery and validation. J. Proteomics 75, 4990–4998 [DOI] [PubMed] [Google Scholar]
- 33. Wei Y., Tong J., Taylor P., Strumpf D., Ignatchenko V., Pham N. A., Yanagawa N., Liu G., Jurisica I., Shepherd F. A., Tsao M. S., Kislinger T., and Moran M. F. (2011) Primary tumor xenografts of human lung adeno and squamous cell carcinoma express distinct proteomic signatures. J. Proteome Res. 10, 161–174 [DOI] [PubMed] [Google Scholar]
- 34. Bose A., Teh M. T., Mackenzie I. C., and Waseem A. (2013) Keratin k15 as a biomarker of epidermal stem cells. Int. J. Mol. Sci. 14, 19385–19398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y., Lyle S., Yang Z., and Cotsarelis G. (2003) Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J. Invest. Dermatol. 121, 963–968 [DOI] [PubMed] [Google Scholar]
- 36. Abbas O., and Bhawan J. (2011) Expression of stem cell markers nestin and cytokeratin 15 and 19 in cutaneous malignancies. J. Eur. Acad. Dermatol. Venereol. 25, 311–316 [DOI] [PubMed] [Google Scholar]
- 37. Kim H. C., Sohng S. H., Shin D. H., Choi J. S., and Bae Y. K. (2016) Immunohistochemical expression of cytokeratin 15, cytokeratin 19, follistatin, and Bmi-1 in basal cell carcinoma. Int. J. Dermatol. 55, 36–44 [DOI] [PubMed] [Google Scholar]
- 38. Shi I., Hashemi Sadraei N., Duan Z. H., and Shi T. (2011) Aberrant signaling pathways in squamous cell lung carcinoma. Cancer Informatics 10, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boyero L., Sanchez-Palencia A., Miranda-Leon M. T., Hernandez-Escobar F., Gomez-Capilla J. A., and Farez-Vidal M. E. (2013) Survival, classifications, and desmosomal plaque genes in non-small cell lung cancer. Int. J. Med. Sci. 10, 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zoubeidi A., and Gleave M. (2012) Small heat shock proteins in cancer therapy and prognosis. Int. J. Biochem. Cell Biol. 44, 1646–1656 [DOI] [PubMed] [Google Scholar]
- 41. Garrido C., Paul C., Seigneuric R., and Kampinga H. H. (2012) The small heat shock proteins family: the long forgotten chaperones. Int. J. Biochem. Cell Biol. 44, 1588–1592 [DOI] [PubMed] [Google Scholar]
- 42. Rocchi P., So A., Kojima S., Signaevsky M., Beraldi E., Fazli L., Hurtado-Coll A., Yamanaka K., and Gleave M. (2004) Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 64, 6595–6602 [DOI] [PubMed] [Google Scholar]
- 43. Aloy M. T., Hadchity E., Bionda C., Diaz-Latoud C., Claude L., Rousson R., Arrigo A. P., and Rodriguez-Lafrasse C. (2008) Protective role of Hsp27 protein against gamma radiation-induced apoptosis and radiosensitization effects of Hsp27 gene silencing in different human tumor cells. Int. J. Radiation Oncol., Biol., Physics 70, 543–553 [DOI] [PubMed] [Google Scholar]
- 44. Marinova D. M., Slavova Y. G., Trifonova N., Kostadinov D., Maksimov V., and Petrov D. (2013) Stress protein Hsp27 expression predicts the outcome in operated small cell lung carcinoma and large cell neuroendocrine carcinoma patients. J. Balkan Union Oncol. 18, 915–920 [PubMed] [Google Scholar]
- 45. Guo H., Bai Y., Xu P., Hu Z., Liu L., Wang F., Jin G., Wang F., Deng Q., Tu Y., Feng M., Lu D., Shen H., and Wu T. (2010) Functional promoter -1271G>C variant of HSPB1 predicts lung cancer risk and survival. J. Clin. Oncol. 28, 1928–1935 [DOI] [PubMed] [Google Scholar]
- 46. Lopez Guerra J. L., Wei Q., Yuan X., Gomez D., Liu Z., Zhuang Y., Yin M., Li M., Wang L. E., Cox J. D., and Liao Z. (2011) Functional promoter rs2868371 variant of HSPB1 associates with radiation-induced esophageal toxicity in patients with non-small-cell lung cancer treated with radio(chemo)therapy. Radiotherapy Oncol. 101, 271–277 [DOI] [PubMed] [Google Scholar]
- 47. Pang Q., Wei Q., Xu T., Yuan X., Lopez Guerra J. L., Levy L. B., Liu Z., Gomez D. R., Zhuang Y., Wang L. E., Mohan R., Komaki R., and Liao Z. (2013) Functional promoter variant rs2868371 of HSPB1 is associated with risk of radiation pneumonitis after chemoradiation for non-small cell lung cancer. Int. J. Radiation Oncol., Biol., Physics 85, 1332–1339 [DOI] [PubMed] [Google Scholar]
- 48. Zimmermann M., Mueller T., Dieplinger B., Bekos C., Beer L., Hofbauer H., Dome B., and Ankersmit H. J. (2014) Circulating heat shock protein 27 as a biomarker for the differentiation of patients with lung cancer and healthy controls–a clinical comparison of different enzyme linked immunosorbent assays. Clin. Lab. 60, 999–1006 [DOI] [PubMed] [Google Scholar]
- 49. Zeng G. Q., Zhang P. F., Deng X., Yu F. L., Li C., Xu Y., Yi H., Li M. Y., Hu R., Zuo J. H., Li X. H., Wan X. X., Qu J. Q., He Q. Y., Li J. H., Ye X., Chen Y., Li J. Y., and Xiao Z. Q. (2012) Identification of candidate biomarkers for early detection of human lung squamous cell cancer by quantitative proteomics. Mol. Cell. Proteomics 11, M111 013946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang W., Xu X., Wang W., Shao W., Li L., Yin W., Xiu L., Mo M., Zhao J., He Q., and He J. (2011) The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumour Biol. 32, 1199–1208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.