Abstract
Ascorbate peroxidase, a haem protein (EC 1.11.1.11), efficiently scavenges hydrogen peroxide (H2O2) in cytosol and chloroplasts of plants. In this study, a full-length coding sequence of thylakoid-bound ascorbate peroxidase cDNA (TatAPX) was cloned from a drought tolerant wheat cultivar C306. Homology modeling of the TatAPX protein was performed by using the template crystal structure of chloroplastic ascorbate peroxidase from tobacco plant (PDB: 1IYN). The model structure was further refined by molecular mechanics and dynamic methods using various tools such as PROCHECK, ProSA and Verify3D. The predicted model was then tested for docking with H2O2, the substrate for TatAPX enzyme. The results revealed that Arg233 and Glu255 in the predicted active site of the enzyme are two important amino acid residues responsible for strong hydrogen bonding affinity with H2O2, which might play an important role in scavenging of H2O2 from the plant system.
Key words: ascorbate peroxidase, wheat, molecular modeling, docking
Introduction
Reactive oxygen species (ROS) are byproducts of aerobic metabolism and are produced in excess within plant cells under abiotic and biotic stresses 1., 2.. ROS are harmful to plant cellular metabolism and hence need to be scavenged from the plant system 3., 4.. ROS scavenging enzymes, particularly ascorbate peroxidase (APX, EC 1.11.1.11), play a major role in scavenging H2O2 in chloroplasts, cytosol and glyoxysomes 5., 6.. Chloroplastic APXs (chAPX) including thylakoid-bound (TatAPX) and stromal (sAPX) isoforms scavenge the H2O2 produced during photosynthesis. Scavenging of the ROS produced in chloroplast is essentially required to avoid damage of the photosynthetic machinery. The TatAPX enzyme located at the stromal thylakoid membranes locally removes the ROS produced at the donor side of photosystem I (PS-I). TatAPX is the primary target for inactivation of ROS under oxidative stress 4., 7., 8..
TatAPX has a binding affinity with H2O2 and is also involved in the scavenging of H2O2 outside the thylakoid membrane. This hypothesis has been validated by using computational tools and techniques. Computational characterization of the chloroplastic isoform has been performed by analysis of the hydrophobicity of wheat TatAPX protein (9), which showed that predicted wheat TatAPX protein has one major hydrophobic region at C-terminus. This might be the domain for binding of the protein to the thylakoid membrane. Similar hydrophobic domain at C-terminus has also been reported in pumpkin TatAPX (10) and spinach glyoxysome bound APX (11).
We have cloned a coding sequence of TatAPX gene (NCBI accession No. EF184291) from a drought tolerant wheat cultivar C306 12., 13.. To understand the functional correlation of TatAPX with H2O2, we have carried out homology-based comparative modeling and molecular docking studies. Since model structure of TatAPX of wheat is not yet available in the Protein Data Bank (PDB), homology searching was carried out and the best homologous structure (PDB: 1IYN) was used as a template structure for the construction of TatAPX 3D model structure using various bioinformatics tools. The predicted structure was optimized by replacement of missing atom loop refinement, energy minimization and structure validation using various modeling servers such as PROCHECK, Verify3D and ProSA (14). Interactions of the substrate (H2O2) with the model TatAPX protein were deciphered by docking analysis (15). The H2O2-TatAPX complex structure was used to predict the binding orientation and to determine the affinity and important conserved amino acid residues involved in the interaction of the H2O2 with TatAPX.
Results and Discussion
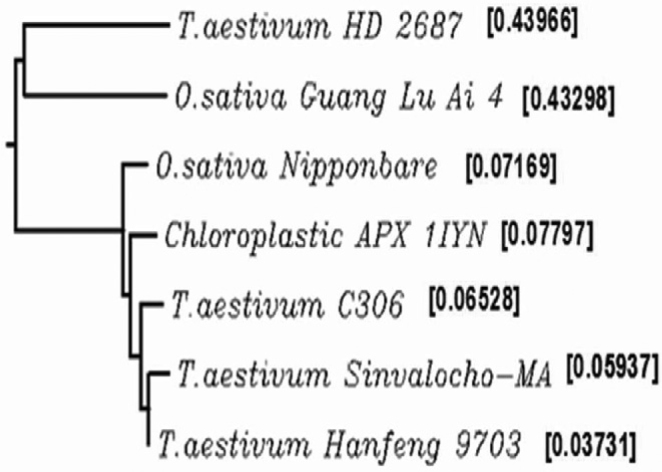
The TatAPX cDNA cloned from a drought tolerant wheat cv. C306 contains a single open reading frame (nucleotide number 20 to 1,314) that could potentially code for a single polypeptide. Hence, the cDNA cloned in this study contains a full-length coding sequence. Multiple sequence alignment of the TatAPX cDNA (EF184291) with that of EST from winter wheat (TC250985) showed 96.3% and 96.2% similarity at nucleotide and amino acid level, respectively. The fast minimum evolution of gene as produced by BLASTN for highly similar sequence (megablast) pairwise alignments indicated that TatAPX is closely related to wheat and rice APX. The sequence similarity among different wheat and rice APX indicated that the cloned TatAPX in this study is more closely related to drought tolerant wheat genotypes such as Triticum aestivum cv. Sinvalocho MA and T. aestivum w. Hanfeng 9703 (Figure 1).
Figure 1.
Phylogenetic tree showing sequence similarity of wheat ascorbate peroxidase cDNA (TatAPX) with other relatives.
Computational analysis of membrane bound nature of TatAPX protein
As the cloned gene showed closest similarity with thylakoid-bound ascorbate peroxidise, hydrophobicity of the deduced wheat TatAPX protein was analysed by ProtScale as described by Kyte and Doolittle (9). Hydrophobic analysis showed that the predicted wheat TatAPX protein has one major hydrophobic region at C-terminus. This might be the domain for binding of the protein to the thylakoid membrane. Similar hydrophobic domain at C-terminal end of pumpkin TatAPX was reported by Yamaguchi et al. (10). The predicted mature protein of spinach TatAPX has one major hydrophobic region (residue 380—415) at the C-terminal end (7). Ishikawa et al. (11) also observed a C-terminal membrane binding domain in spinach glyoxysome bound APX. A PROF prediction was carried out for TatAPX and the secondary structure composition of the protein was found to be 41.3% for assigning helix. Membrane bound nature of TatAPX was searched using various transmembrane protein prediction servers such as MINNOU (http://minnou.cchmc.org/), waveTM (http://athina.biol.uoa.gr/bioinformatics/waveTM/), HMMTOP (http://www.enzim.hu/hmmtop/) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) 16., 17., 18., 19.. Prediction of transmembrane region and helices in the TatAPX proteins showed that amino acid residues 410 to 431 belong to transmembrane helix category towards C-terminal end (Figure S1). Sequence comparisons among various thylakoid-bound ascorbate peroxidase proteins of Spinacia oleracea, Nicotina tabacum, Brassica oleracea, Arobidopsis thaliana and T. aestivum showed that transmembrane regions were conserved (data not shown). The above results clearly indicated the membrane bound nature of TatAPX in wheat.
Expression and synteny analysis
Wheat digital northern analysis showed that total 32 ESTs were found in 19 libraries for thylakoid-bound ascorbate peroxidase of T. aestivum (http://www.tigr.org/). Expression frequency was highest at vegetative growth stage (0.257%), whereas relatively lower expression was observed in spike, callus, flower, root and seed tissues. This analysis indicated that TatAPX might play a crucial role in scavenging H2O2 in photosynthetic tissues. Orthologous genomic location of this transcript can be found on Chromosome 2 of rice at location 20,862,392–20,866,211 (LOC_Os02g34810) on the wheat-rice synteny map. The rice orthologous location expressed in response to salinity stress (http://www.ncbi.nlm.nih.gov/geo/).
Homology modeling
A high level of sequence identity promises a more reliable alignment between the target sequence and the template structure. The query sequence from wheat TatAPX protein was selected for homology-based searching of the template structure by the BLAST program against the structural database of PDB (http://www.rcsb.org) 20., 21.. Sequences that showed maximum identity with high score and low Evalue were aligned (Figure S2) and the alignment was used to build a 3D model for TatAPX. According to the result of BLAST search against PDB, four reference proteins (PDB: 1IYN, 2VCF, 1OAF and 1APX) represented a high level of sequence identity, that is, 78%, 42%, 42% and 43%, respectively (22) (Table S1). Out of these four, the best homologous structure belonged to chloroplastic ascorbate peroxidase protein of N. tabacum (PDB: 1IYN) and was selected for comparative modeling. The initial model of TatAPX was built by homology modeling methods using Modeller 9v5 software on Linux x86_64 machine 23., 24., 25., 26.. The method was based on an input sequence alignment between the target amino acid sequence to be modeled and a template protein with already known structure. 1IYN was used as template structure for the construction of TatAPX model protein for wheat cv. C306. The Modeller software constructed five different structure models for TatAPX (wheat cv. C306), which was based on physiochemical properties such as van der Waals interaction, hydrophobicity, hydrophilicity, atomic charges and atomic energy. We found three different types of scores (molpdf, DOPE and GA341 score) related to above mentioned properties (Table 1); Model 4 (formally called TatAPX-4) was observed as the potential candidate for remaining experiments. This model was further refined by molecular dynamic (MD) method and its final stable structure was obtained as shown in Figure 2. MD simulates the movements of all the particles in a molecular system by iteratively solving Newton’s equations of motion (27). MD methods are governed by the system’s Hamiltonian and consequently Hamilton’s equations of motion are integrated to movable particles to new positions and to get new velocities at these new positions. Finally it leads to generate an account of information of a macromolecule with respect to its thermodynamic properties (28).
Table 1.
Summary of successfully produced models of TatAPX using Modeller 9v5
| Filename | molpdf score | DOPE score | GA341 score |
|---|---|---|---|
| apx.B99990001.pdb | 2495.93896 | —33340.20312 | 1.00000 |
| apx.B99990002.pdb | 2540.43481 | —33176.03125 | 1.00000 |
| apx.B99990003.pdb | 2602.05444 | —33321.75781 | 1.00000 |
| apx.B99990004.pdb | 2458.73145 | —33379.94922 | 1.00000 |
| apx.B99990005.pdb | 2326.41553 | —33342.69922 | 1.00000 |
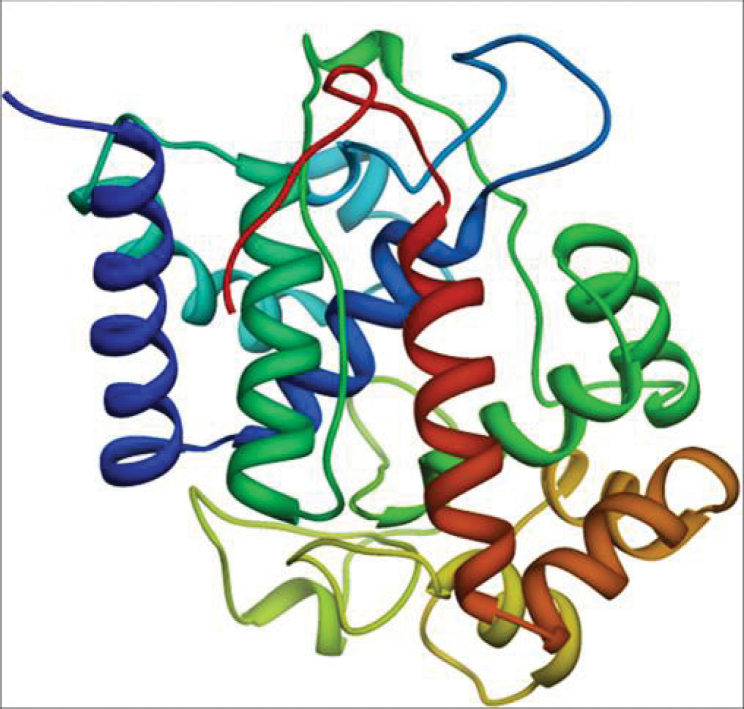
Figure 2.
The final 3D structure of TatAPX-4 model protein. The structure was obtained by Modeller 9v5 and verified using PROCHECK, Verify3D and ProSA servers (Ribbon shape model visualized in chimera).
Structure optimization and validation
TatAPX-4 structure was optimized with various strategies like reconstruction of protein missing side-chain atoms, protein missing backbone atoms, protein missing residues and energy minimization, and was followed by structure validation. The missing atoms’ fulfillment was done by Profix, a program that uses backbone rotamer library, side-chain rotamer library and distance geometry constraints to sample segment conformations to recover missing atoms either on side-chain or on backbone. Energy minimization was performed by Minst, a structure minimization program with constraints using the smoothing energy technique as well as CHARMM22 and AMBER94 force fields to minimize the structure.
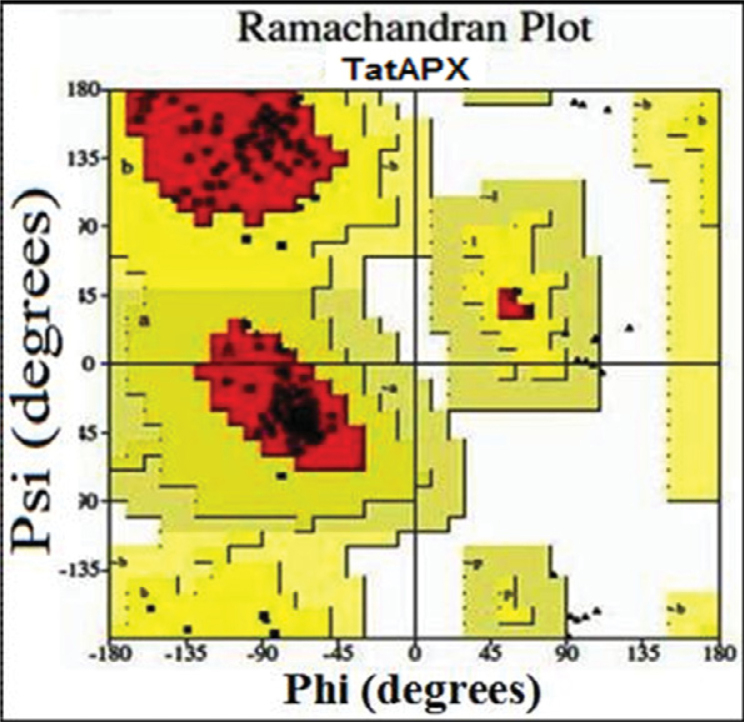
Finally, we got the refined model structure and designated it as TatAPX-4. The structure validation was then carried out by the reliable RCSB server (http://deposit.pdb.org/validate/). This considers primary checking for accuracy of the structure with respect to structure domain and structure units using PROCHECK V3.4.4, which is based on Ramachandran plot calculation 22., 29., 30.. This validation process puts 100% residues of TatAPX-4 in the most favored regions according to Ramachandran plot analysis (Figure 3). The reliability of the target proteins was examined by torsion angles Ø and Ψ and a percentage quality measurement of a protein structure was used, in which four sorts of occupancies were called “core”, “allowed”, “generously allowed” and “disallowed regions”, respectively. These values were calculated for TatAPX-4 and template protein 1IYN (Table S2). The overall RMS deviation for covalent bonds relative to the standard dictionary is 0.020Å and the overall RMS deviation for covalent angles relative to the standard dictionary is 2.5°. Altogether these findings revealed that all residues of TatAPX-4 were found in most favored and allowed regions according to Ramachandran plot analysis.
Figure 3.
Ramachandran plot calculation on 3D model of TatAPX-4. The plot calculations were done by PROCHECK program and the plot reveals that 100% of all residues of TatAPX-4 were found in the most favored regions according to Ramachandran plot analysis.
The final structure was further checked by Verify3D (http://nihserver.mbi.ucla.edu/Verify_3D/), a program that analyzes the compatibility of an atomic model (3D) with its own amino acid sequence (1D) (31). Each residue is assigned a structural class based on its location and environment (alpha, beta, loop, polar, nonpolar, etc.) using ProSA program (https://prosa.services.came.sbg.ac.at) (32). The compatibility score above zero in the Verify3D graph (Figure S3) corresponded to acceptable side chain environment. The energy profile obtained from the Verify3D program showed good score that was between 0.1 to 0.71 3D—1D profile score of the residues. 99.27% of the residues had an averaged 3D—1D score >0.2. A further comparison by the ProSA program between the unprocessed model (TatAPX) protein and refined model (TatAPX-4) protein showed that ProSA energy was more negative than that of the unprocessed structure (32) (Figure S4). This represented the residues’ interaction energy of the protein. ProSA also calculates an overall quality score (Z score) for a protein structure, which was found to be —8.75 for TatAPX-4 model protein (Figure S5). This score was better than the previously calculated Z score (—7.3) for the model structure (TatAPX) before attempting structure optimization and validation.
Structure alignment and superimposition
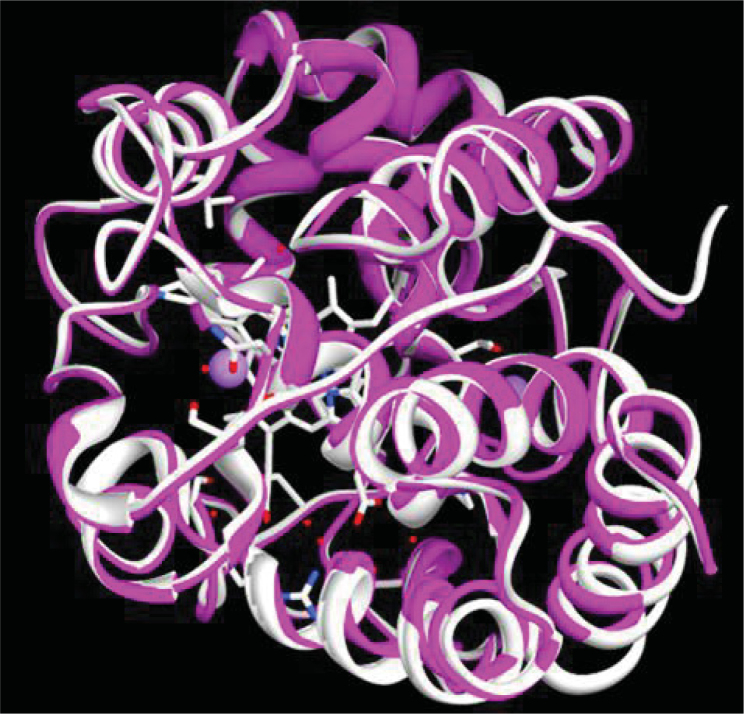
Structurally conserved regions (SCRs) between model TatAPX-4 and homologous proteins (PDB: 1IYN, 2VCF, 1OAF and 1APX) were determined by multiple sequence alignment using ClustalW (Figure S6). The root mean square deviation (RMSD) was measured by the average distance between the backbones of superimposed proteins (TatAPX-4 and best homologous protein, i.e., 1IYN) (33) (Figure 4). The RMSD of Cα trace between the template and final refined model was 0.433Å (between 273 atom pairs) with a significant score of 1276.6. This model was used for the identification of active sites and for docking of the substrate with the ascorbate peroxidase.
Figure 4.
Superimposition of template protein 1IYN (represented in white color) and refined model TatAPX-4 (represented in magenta color).
Pairwise 3D alignment
Amino acid sequences of template (1IYN) and the final TatAPX-4 protein were aligned using ClustalW (34). The secondary structures were also analyzed and compared by pairwise 3D alignment using MATRAS 1.2 (35) (http://biunit.aist-nara.ac.jp/matras/) (Figure S7). It was found that the structures of template and the final model protein TatAPX have 85.4% sequence and 98.5% secondary structure identity (Table S3), which showed that the final structure was highly reliable and could be further used for active site identification.
Active site identification of model protein TatAPX
After the final model was built, the possible binding sites of TatAPX were searched using various binding site prediction servers such as Q-site finder (http://bmbpcu36.leeds.ac.uk/qsitefinder/), CASTp (http://sts-fw.bioengr.uic.edu/castp/) and PINUP (http://sparks.informatics.iupui.edu/PINUP/) (36—38). The comparative study revealed a few number of active site residues such as Tyr89, Pro92, Ile93, His100, Ala132, Ala232, Arg233, Gly254 and Glu255. Binding site for template protein (1IYN) was also predicted using the same methodology. Predicted residues for 1IYN were Cys24, Pro26, Ile27, Val29, Arg30, His34, Ser97, Pro131, Phe146, Leu159, Ser160, Ala162, His163, Arg167 and His255. These studies showed that residues Pro, His, Ala, Ile, Arg and Glu were highly conserved in active site of both model and the template protein, and hence it could be predicted that their biological function would be identical. The above identified active site was chosen as the most favorable site for docking studies.
Molecular docking
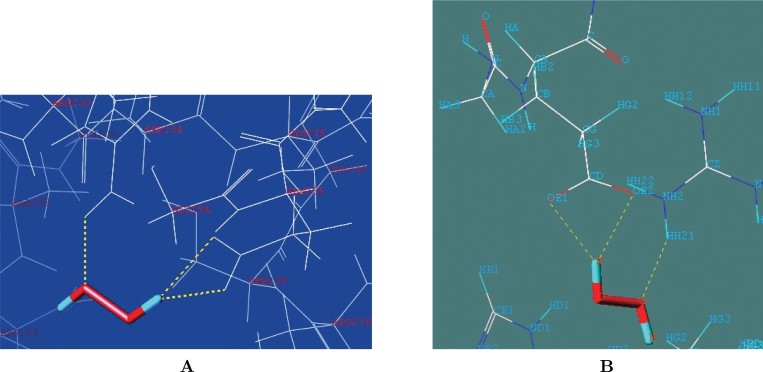
Molecular docking was performed by Sybyl 8.0 Surflex-Dock method (Tripos Inc., USA) on Linux x86_64 machine (39). This method is based on various scoring algorithms such as crash, polar, D-score, PMF-score, G-score, ChemSCO and CScore. The ligand H2O2 was obtained from pubchem (http://www.ncbi.nlm.nih.gov/sites/entrez?db=pccompound) and its energy was minimized with Sybyl 8.0 using Tripos fore field. We docked H2O2 small molecule inside the cavity of TatAPX-4 protein. Based on all the above mentioned scores, we found H2O2-001 is the highest scoring (Total score=3.2100) docked conformation (Table 2). To understand the molecular interaction between TatAPX-4 and H2O2, the substrate H2O2–TatAPX protein complex was analyzed using surflex-dock result browser. The data clearly demonstrated that H2O2 substrate was located in the active site area and was stabilized by hydrogen bonding interactions. It was also evident that H2O2 formed H bonds with two amino acids Arg233 and Glu255 (Figure 5A). One of the oxygen atoms of H2O2 formed a hydrogen bond with the hydrogen atom of Arg233, and the H atoms of H2O2 formed two H bonds with two oxygen atoms (Atom ID: 326, 327) of Glu255 of TatAPX (Figure 5B). The hydrogen bonds present in the enzyme-substrate complex along with their distance and angles are listed in Table 3.
Table 2.
Ten best docked conformations of the substrate based on crash, polar, D-score, PMF-score, G-score, ChemSCO and CScore
| Conformation | Total score | Crash | Polar | D-Score | PMF-Score | G-Score | ChemSCO | CScore |
|---|---|---|---|---|---|---|---|---|
| H2O2-001 | 3.2100 | —0.0700 | 2.8000 | —8.6871 | —7.8155 | —62.7806 | —8.9665 | 5 |
| H2O2-002 | 3.2100 | —0.0700 | 2.8000 | —8.5002 | —6.9858 | —61.7252 | —8.9498 | 5 |
| H2O2-003 | 3.0000 | —0.0500 | 2.2400 | —8.7117 | —7.7566 | —62.7705 | —8.9994 | 5 |
| H2O2-004 | 2.8300 | —0.1500 | 2.2300 | —8.3430 | —3.3868 | —29.4016 | —9.0286 | 2 |
| H2O2-005 | 2.7700 | —0.1000 | 2.2200 | —8.7098 | —9.4345 | —62.6957 | —7.9176 | 3 |
| H2O2-006 | 2.7700 | —0.1000 | 2.2200 | —7.0705 | —0.7750 | —32.9710 | —9.5580 | 1 |
| H2O2-007 | 2.5300 | —0.0000 | 2.2700 | —8.7518 | —9.3951 | —62.7909 | —7.9347 | 4 |
| H2O2-008 | 2.5300 | —0.0000 | 2.2700 | —9.7545 | —7.0284 | —24.0938 | —9.5922 | 3 |
| H2O2-009 | 2.5200 | —0.1300 | 1.6900 | —9.5994 | —6.9342 | —25.7850 | —9.5922 | 3 |
| H2O2-010 | 2.4800 | —0.0500 | 1.8400 | —6.7617 | —11.3541 | —57.1600 | —6.2557 | 2 |
Figure 5.
A. Molecular interaction between TatAPX-4 model protein and the substrate hydrogen peroxide (H2O2). B. Binding of the H2O2 substrate with the residues in the active site of TatAPX-4 using Sybyl 8.0 software suite.
Table 3.
Hydrogen bonds along with their distance and bond angles between the H2O2 substrate and active site residues of TatAPX using Sybyl 8.0 software suite
| Catalase enzyme |
H2O2 atom | Distance | Angle | |
|---|---|---|---|---|
| Residue | Atom | |||
| Arg233 | HH21 | Hydrogen peroxide O | 1.944Å | 131.15 Deg |
| Glu255 | OE1 | Hydrogen peroxide H | 2.169Å | 61.31 Deg |
| Glu255 | OE2 | Hydrogen peroxide H | 2.216Å | 59.14 Deg |
The side chain flexibility plays a crucial role at the ligand binding site of the receptor protein and it allows conformational changes of side chain residues after ligand binding. Yang et al. (40) suggested that for most proteins, which exhibit limited backbone motion, ligands tend to bind to low energy conformations of their binding sites. This opens the possibility of incorporating alternative binding site conformations to improve the efficacy of docking and structure based drug design algorithms. We have also adopted the similar approach to check the minimum free energy of the receptor protein structure before and after binding of the ligand in binding site residues by using PROCHECK. However, we have not found any significant variations at energy levels of side chain residues as the values of G-factor detected were same, i.e., 0.7 for receptor structure before and after binding with ligand.
Conclusion
In this study, we have cloned and computationally characterized thylakoid-bound ascorbate peroxidase cDNA (TatAPX) from a drought tolerant wheat cv. C306. The TatAPX cDNA is potentially useful for the development of transgenic crop plants tolerant to abiotic stresses. Computational studies confirmed the membrane bound nature of the TatAPX. We have constructed a 3D model of TatAPX protein by using the Modeller 9v5 software, and obtained a refined model after energy minimization. The final refined model was further assessed by Verify3D, ProSA and PROCHECK programs. The results showed that this modeled protein is highly reliable and stable. The modeled protein was further used for docking with the substrate (H2O2). We found that Arg233 and Glu255 are important residues for strong hydrogen bond interaction with the substrate H2O2. The results indicated that arginine and glutamic acid play an important role in scavenging H2O2 from the drought tolerant wheat cv. C306.
Materials and Methods
Cloning of TatAPX cDNA from drought tolerant wheat cultivar C306
Wheat seeds of cultivar C306 were sown in 4-inch-diameter pot and watered daily. After 15 days of germination, a single leaf was selected and treated with 100 μM ABA. Three hours later, good quality RNA was isolated from the ABA treated leaves using RNeasy Plant Mini Kit (Qiagen). TatAPX gene-specific primers (TatAPX-F: GACACTCCAAACCTCAGCCAT and TatAPX-R: TTAGTTCCCGGTCAGAGACGTC) were designed based on the EST (TC250985) sequence from winter wheat. By using total RNA as template and with TatAPX gene-specific primers, a one-step RT-PCR (Qiagen) reaction was carried out to amplify cDNA. The amplicon was cloned into pDRIVE vector (QIAGEN GmbH, Germany). E. coli XL1-Blue cells were transformed with the ligation products. The recombinant plasmids were confirmed by restriction analysis and sequenced. The nucleotide sequence data were submitted to the GenBank (Accession No. EF184291).
In silicocharacterization ofTatAPX
The cloned TatAPX coding sequence was searched for homology using BLASTN on NCBI server (http://www.ncbi.nlm.nih.gov) and the translated protein sequence for the complete ORF was searched for homology using BLASTP against the structural database of PDB (http://www.rcsb.org).
3D model building and validation
The model of TatAPX gene product was built by using homology-modeling methods of the Modeller 9v5, a program for homology or comparative modeling of protein 3D structures by using a template structure (1IYN) and homologous multiple protein alignment (ClustalW). Modeller implements comparative protein structure modeling by satisfaction of spatial restraints. The structure having the least modeler objective function obtained from the Modeller was improved by energy minimization. The final model obtained was further assessed by PROCHECK V3.4.4, a program for checking the stereochemical quality of a protein structure 22., 29., 30., Verify3D, a program that analyzes the compatibility of an atomic model (3D) with its own amino acid sequence (1D) (31) and ProSA (Protein Structure Analysis) server, a program used for refinement and validation of protein model (32). This model was further refined by MD method and the final stable structure of the TatAPX-4 was obtained. The TatAPX model thus generated was used for the identification of active site and for docking of the substrate (H2O2) with the enzyme.
Structure alignment and active site prediction
2D and 3D structure alignment was carried out using ClustalW and MATRAS 1.2, respectively 34., 35.. SCRs between model TatAPX-4 and homologous proteins (PDB: 1IYN, 2VCF, 1OAF and 1APX) were determined by multiple sequence alignment. Active site of model protein (TatAPX) from wheat cv. C306 was identified by using various binding site prediction servers, including Q-site finder, a program that works by binding hydrophobic (CH3) probes to the protein and finding clusters of probes with the most favorable binding energy, and CASTp, a program that uses weighted Delaunay triangulation and the alpha complex for shape measurements. It provides identification and measurements of surface accessible pockets as well as interior inaccessible cavities and PINUP server 36., 37., 38..
Docking studies
The refined protein model (TatAPX-4) was used to study its ligand binding mechanism. The ligand structure (H2O2) was minimized with Sybyl 8.0 using Tripos fore field (Tripos Inc., USA). Docking analysis was performed by Sybyl 8.0 molecular modeling tool to identify active sites on protein structure where favorable protein-ligand interactions can occur (39). The small substrate molecule (H2O2) was docked inside the cavity of TatAPX-4 protein. We checked the minimum free energy of the receptor protein structure by taking into account the flexibility of the side chains before and after binding of the ligand in binding site residues using PROCHECK. Sybyl generated 10 best docked conformations of the substrate based on various scoring algorithms such as crash, polar, D-score, PMF-score, G-score, ChemSCO and CScore. The best docked conformation was selected based on consensus score for molecular interaction analysis. The strength of individual scoring functions was combined to produce a consensus that is more robust and accurate than any single function for evaluating ligand-receptor interactions.
Authors’ contributions
AK carried out model refinement, energy minimization and docking studies and co-wrote the manuscript. SKL carried out in silico characterization, homology modeling, conceived the idea of modeling and docking studies in APX and co-wrote the manuscript. LK cloned the TatAPX. VC participated in the design of the study and helped to draft the manuscript. KCB participated in the design of the study, overall coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
SKL and KL gratefully acknowledge University Grants Commission (UGC) and Council of Scientific and Industrial Research (CSIR) for CSIR-UGC Junior Research Fellowship grant. This work was supported by the Indian Council of Agricultural Research (ICAR)-sponsored Network Project on Transgenics in Crops (NPTC).
Supporting Online Material
Figures S1–S7 and Tables S1–S3
References
- 1.Hong C.Y. Expression of ascorbate peroxidase 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 2007;58:3273–3283. doi: 10.1093/jxb/erm174. [DOI] [PubMed] [Google Scholar]
- 2.Miller G. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 4.Asada K. Ascorbate peroxidase—a hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 1992;85:235–241. [Google Scholar]
- 5.Danna C.H. Thylakoid-bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiol. 2003;132:2116–2125. doi: 10.1104/pp.103.021717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa T. Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol. 1998;39:23–34. doi: 10.1093/oxfordjournals.pcp.a029285. [DOI] [PubMed] [Google Scholar]
- 8.Seki M. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi K. cDNA cloning of thylakoid-bound ascorbate peroxidase in pumpkin and its characterization. Plant Cell Physiol. 1996;37:405–409. doi: 10.1093/oxfordjournals.pcp.a028961. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa T. cDNAs encoding spinach stromal and thylakoid-bound ascorbate peroxidase differing in the presence or absence of their 3’-coding regions. FEBS Lett. 1996;384:289–293. doi: 10.1016/0014-5793(96)00332-8. [DOI] [PubMed] [Google Scholar]
- 12.Bansal K.C., Sinha S.K. Assessment of drought resistance in 20 accessions of Triticum aestivum and related species. I. Total dry matter and grain yield stability. Euphytica. 1991;56:7–14. [Google Scholar]
- 13.Bansal K.C., Sinha S.K. Assessment of drought resistance in 20 accessions of Triticum aestivum and related species. II. Stability in yield components. Euphytica. 1991;56:15–26. [Google Scholar]
- 14.Luo Q. The 3D structure of the defense-related rice protein Pir7b predicted by homology modeling and ligand binding studies. J. Mol. Model. 2008;14:559–569. doi: 10.1007/s00894-008-0310-3. [DOI] [PubMed] [Google Scholar]
- 15.Sekhar P.N. In silico modeling and hydrogen peroxide binding study of rice catalase. In Silico Biol. 2006;6:435–447. [PubMed] [Google Scholar]
- 16.Cao B. Enhanced recognition of protein transmembrane domains with prediction-based structural profiles. Bioinformatics. 2006;22:303–309. doi: 10.1093/bioinformatics/bti784. [DOI] [PubMed] [Google Scholar]
- 17.Pashou E.E. WaveTM: wavelet-based transmembrane segment prediction. In Silico Biol. 2004;4:127–131. [PubMed] [Google Scholar]
- 18.Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 19.Krogh A. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Jenuth J.P. The NCBI. Publicly available tools and resources on the web. Methods Mol. Biol. 2000;132:301–312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 21.Johnson M. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski R.A. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 23.Eswar N. Comparative protein structure modeling with Modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5 Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marti-Renom M.A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 25.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 26.Fiser A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berendsen H.J.C. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 28.Joshi G. β chain of T-cell receptor protein structure reduces total energy in simulation process. Adv. Biotech. 2007;2007:17–19. [Google Scholar]
- 29.Morris A.L. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 30.Kleywegt G.J., Jones T.A. Phi/psichology: Ramachandran revisited. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg D. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 32.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen E.F. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J.D. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2 Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 35.Kawabata T. MATRAS: a program for protein 3D structure comparison. Nucleic Acids Res. 2003;31:3367–3369. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurie A.T., Jackson R.M. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 37.Liang J. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang S. Protein binding site prediction with an empirical scoring function. Nucleic Acids Res. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homer R.W. SYBYL line notation (SLN): a single notation to represent chemical structures, queries, reactions, and virtual libraries. J. Chem. Inf. Model. 2008;48:2294–2307. doi: 10.1021/ci7004687. [DOI] [PubMed] [Google Scholar]
- 40.Yang A.Y. Molecular modelling prediction of ligand binding site flexibility. J. Comput. Aided Mol. Des. 2004;18:235–250. doi: 10.1023/b:jcam.0000046820.08222.83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S7 and Tables S1–S3